 Open Access Article
Open Access ArticleRecyclable cellulose-containing magnetic nanoparticles: immobilization of cellulose-binding module-tagged proteins and a synthetic metabolon featuring substrate channeling
Suwan
Myung
ab,
Chun
You
a and
Y.-H. Percival
Zhang
*abcd
aBiological Systems Engineering Department, Virginia Tech, 304 Seitz Hall, Blacksburg, Virginia 24061, USA. E-mail: ypzhang@vt.edu; Fax: +1-540-231-3199; Tel: +1-540-231-7414
bInstitute for Critical Technology and Applied Science (ICTAS), Virginia Tech, Blacksburg, Virginia 24061, USA
cCell Free Bioinnovations Inc., Blacksburg, Virginia 24060, USA
dGate Fuels Inc., Blacksburg, Virginia 24060, USA
First published on 1st July 2013
Abstract
Easily recyclable cellulose-containing magnetic nanoparticles were developed for immobilizing family 3 cellulose-binding module (CBM)-tagged enzymes/proteins and a self-assembled three-enzyme complex called the synthetic metabolon. Avicel (microcrystalline cellulose)-containing magnetic nanoparticles (A-MNPs) and two controls of dextran-containing magnetic nanoparticles (D-MNPs) and magnetic nanoparticles (MNPs) were prepared by a solvothermal method. Their adsorption ability was investigated by using CBM-tagged green fluorescence protein and phosphoglucose isomerase. A-MNPs had higher adsorption capacity and tighter binding on CBM-tagged proteins than the two control MNPs because of the high-affinity adsorption of CBM on cellulose. In addition, A-MNPs were used to purify and co-immobilize a three-enzyme metabolon through a CBM-tagged scaffoldin containing three different cohesins. The three-enzyme metabolon comprised of dockerin-containing triosephosphate isomerase, aldolase, and fructose 1,6-bisphosphatase was self-assembled because of the high-affinity interaction between cohesins and dockerins. Thanks to spatial organization of the three-enzyme metabolon on the surface of A-MNPs, the metabolon exhibited a 4.6 times higher initial reaction rate than the non-complexed three-enzyme mixture at the same enzyme loading. These results suggested that the cellulose-containing MNPs were new supports for immobilizing enzymes, which could be selectively recycled or removed from other biocatalysts by a magnetic force, and the use of enzymes immobilized on A-MNPs could be very useful to control the On/Off process in enzymatic cascade reactions.
Introduction
Enzyme immobilization has been widely used for prolonging the lifetime of immobilized enzymes and separating and recycling immobilized enzymes from soluble substrates and products.1,2 Numerous enzyme immobilization technologies have been developed, such as physical adsorption,3 covalent bonding,4,5 encapsulation/entrapment,6 cross-linking enzyme aggregate (CLEA),7 and so on. The physical adsorption of an enzyme onto an insoluble support is attractive because it is simple, quick, cheap, and causes no or little damage to enzyme activity. However, this technology may suffer from the leakage of the enzyme from the support, non-specific binding, and steric hindrance by the support. Cellulose-binding module (CBM) tags, in particular, family 3, have been applied as affinity tags for the purification and immobilization of CBM-tagged fusion proteins on cellulosic supports because of their high affinity to cellulose, which is less costly, inert, stable, and biodegradable.8,9 A one-step proteinpurification and immobilization method has been developed to selectively adsorb CBM-tagged proteins on cellulose.10,11Magnetic nanoparticles (MNPs) are widely used in biotechnological and biomedical applications.12–14 Numerous synthesis and surface functionalization methods for iron oxide MNPs have been developed, such as co-precipitation,15thermal decomposition,16hydrothermal synthesis,17 microemulsion,18 and sonochemical synthesis.19 Also, MNPs can be applied to an easy separation of target materials in a liquid phase reaction. By using a magnetic force, MNPs are considered as controllable carriers for target materials, such as enzymes,20drugs,21 antibodies,22 and so on. For enzyme immobilization on MNPs, it is important to functionalize the surface of MNPs for the selective attachment of target biomolecules. Huang et al. studied the properties of surface functional groups, biocompatibility, and bioapplication of three MNPs prepared using dextran, chitosan, or polyacrylic acid as surfactants while not including cellulose.23
Cell-free biosystems comprised of (non-natural) synthetic enzymatic pathways can implement complicated biochemical reactions that microbes and catalysts cannot do,24,25 for example, high-yield hydrogen generation from sugars,26 enzymatic conversion of cellulose to starch.27 For the purpose of biomanufacturing, these cell-free biosystems could be used to produce high-yield hydrogen,28–30alcohols,31organic acids,32 jet fuel,33proteins,34 electricity,35,36 fine chemicals,37 saccharide drugs,38 and even to fix CO2.39,40 Cell-free biomanufacturing could be economically competitive with microbial fermentation for the production of biocommodities only when all of the enzymes in systems have high total turn-over numbers and low-cost bulk enzyme production and purification are available.25 However, it is impossible that all enzymes in cell-free biosystems have the same lifetime because their turn-over numbers often vary by several orders of magnitude. It could be too costly to eliminate all enzymes when only a fraction of them with a short lifetime is deactivated. Therefore, it is essential to selectively remove deactivated enzymes or re-use active enzymes from other enzymes.
Spatial organization of cascade enzymes could greatly accelerate reaction rates.41,42 This phenomenon is called substrate channeling, a process of transferring the product catalyzed by one enzyme as the substrate to the adjacent cascade enzyme without fully equilibrating the bulk phase.41 For example, the optimized distance between the two enzymes controlled by DNA scaffolds results in over 20-fold improvement of the reaction rate compared to the free enzyme mixture.43 Enzyme complexes on specific DNA origami tiles with the controlled inter-enzyme spacing and position enhanced the activity more than 15 times higher than the free enzyme mixture.44 In addition to facilitating reaction rates, synthetic enzyme complexes called metabolons may avoid the degradation of labile metabolites.41,45
As natural cellulase complexes called cellulosomes were formed through the high-affinity interaction between cohesins and dockerins, Bayer et al.46 proposed to construct designed enzyme complexes by utilizing species-specific dockerins and cohesins, where they can bind tightly at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Later, a few synthetic mini-cellulosomes containing various extracellular glycoside hydrolases were constructed.47–50 Few efforts were made for constructing synthetic enzyme complexes containing cascade enzymes from a metabolic pathway by using dockerins and cohesins.51,52
1. Later, a few synthetic mini-cellulosomes containing various extracellular glycoside hydrolases were constructed.47–50 Few efforts were made for constructing synthetic enzyme complexes containing cascade enzymes from a metabolic pathway by using dockerins and cohesins.51,52
In this paper, we prepared cellulose-containing MNPs for the immobilization of CBM-tagged proteins and a synthetic metabolon for the first time. Avicel-containing MNPs (A-MNPs), dextran-containing MNPs (D-MNPs), and MNPs were prepared by a solvothermal method. Also substrate channeling of the synthetic metabolon on the surface of A-MNPs was investigated.
Materials and methods
Chemicals and strains
All chemicals were of reagent grade, purchased from Sigma-Aldrich (St. Louis, MO) and Fisher Scientific (Pittsburgh, PA), unless otherwise noted. Avicel PH105, microcrystalline cellulose, was purchased from FMC (Philadelphia, PA). Thermotoga maritima (ATCC 43589) genomic DNA was purchased from the American Type Culture Collection (Manassas, VA). Oligonucleotides were synthesized by Integrated DNA Technologies (Coraville, IA). Liquid glucose reagent based on hexokinase/glucose-6-phosphate dehydrogenase was purchased from Pointe Scientific Inc. (Canton, MI). Escherichia coli BL21 Star (DE3) (Invitrogen, Carlsbad, CA) containing a protein expression plasmid was used for producing the desired recombinant protein. Luria–Bertani (LB) medium supplemented with 100 μg mL−1ampicillin or 50 μg mL−1kanamycin was used for E. coli cell growth and recombinant protein expression.Plasmid construction
The plasmids used are summarized in Table 1. Plasmid pNT02 encoding the thioredoxin-GFP-CBM (TGC) fusion protein was described elsewhere.53 Plasmid pET20b-tim has an expression cassette containing the T. maritima tim gene.33 Plasmid pET28a-ald whose expression cassette contains T. maritima ald gene was described elsewhere.54 The pCIF plasmid encoding the CBM-intein-FBP fusion protein45 and pCIP plasmid encoding the CBM-intein-PGI fusion protein10 were described elsewhere. pET20b-mini-scaf, pET20b-tim-ctdoc, pET20b-ald-ccdoc, and pET20b-fbp-rfdoc made by simple cloning55 were described elsewhere.51| Plasmid | Characteristics | Purified protein and purification methods | Ref. |
|---|---|---|---|
| pNT02 | AmpR, gfp-cbm expression cassette cloned, green fluorescence protein (gfp) gene, cellulose-binding module (cbm) gene from C. thermocellum | GFP-CBM (TGC), bio-specific adsorption of CBM-tagged GFP on RAC followed by ethylene glycol elusion | 53 |
| pCP | AmpR, cbm-pgi expression cassette cloned, cbm gene from C. thermocellum, pgi gene from C. thermocellum | CBM-PGI, bio-specific adsorption of CBM on RAC followed by ethylene glycol elusion | 10 |
| pET33b-tim | KanR, tim expression cassette cloned, tim gene from T. thermophilus | TIM, heat treatment and ammonium sulfate precipitation | 51 |
| pET20a-ald | KanR, ald expression cassette cloned, ald gene from T. maritima | ALD, heat treatment and ammonium sulfate precipitation | 33 |
| pCIF | AmpR, cbm-intein-fbp expression cassette cloned, fbp gene from T. maritima | FBP, bio-specific adsorption of CBM tagged intein-FBP on RAC followed by intein self-cleavage | 51 |
| pCIP | AmpR, cbm-intein-pgi expression cassette cloned, pgi gene from T. maritima | PGI, bio-specific adsorption of CBM tagged intein-PGI on RAC followed by intein self-cleavage | 45 |
| pET20b-mini-scaf | AmpR, mini-scaffoldin expression cassette cloned, containing a CBM module from C. thermocellum and three different cohesins from C. thermocellum, C. cellulovorans and R. flavefaciens | CBM-CtCoh-CcCoh-RfCoh (mini-scaf), bio-specific adsorption of CBM on RAC followed by ethylene glycol elusion | 10 |
| pET20b-tim-ctdoc | AmpR, tim-ctdoc expression cassette cloned, tim gene from T. thermophiles, the C. thermocellum dockerin module | TIM-CtDoc, bio-affinity interaction between CtDoc and mini-scaf followed by CBM adsorption on RAC and ethylene glycol elusion | 51 |
| pET20b-ald-ccdoc | AmpR, ald-ccdoc expression cassette cloned, ald gene from T. maritime, the C. cellulovorans dockerin module | ALD-CcDoc, bio-affinity interaction between CcDoc and mini-scaf followed by CBM adsorption on RAC and ethylene glycol elusion | 51 |
| pET20b-fbp-rfdoc | AmpR, fbp-rfdoc expression cassette cloned, fbp gene from T. maritime FBP, the R. flavefaciens dockerin module | FBP-RfDoc, bio-affinity interaction between RfDoc and mini-scaf followed by CBM adsorption on RAC and ethylene glycol elusion | 51 |
Recombinant protein expression and purification
For the preparation of recombinant proteins: TIM and ALD, two hundred milliliters of the LB culture containing 50 μg mL−1 of kanamycin was incubated in 1 L Erlenmeyer flasks with a rotary shaking rate of 250 rpm at 37 °C. When the absorbance (A600) reached ca. 1.2, the recombinant protein expression was induced by adding isopropyl beta-D-1-thiogalactopyranoside (IPTG, 0.1 mM, final concentration). The culture was induced at 37 °C for 4 h. The cells were harvested by centrifugation at 4 °C, washed twice with 50 mM Tris–HCl buffer (pH 7.5), and re-suspended in 15 mL of 30 mM Tris–HCl buffer (pH 7.5) containing 0.5 M NaCl and 1 mM EDTA. The cell pellets were lysed using a Fisher Scientific Sonic Dismembrator Model 500 (5 s pulse on and off, total 360 s, at 20% amplitude) in an ice bath. After centrifugation, the target proteins (TIM and ALD) were purified through heat treatment at 60 °C for 20 min followed by gradient ammonium sulfate precipitation.56 The expression and purification of tag-free FBP,45 PGI and CBM-PGI10 were described elsewhere.56 The expression and purification conditions of mini-scaffoldin, TIM-CtDoc, ALD-CcDoc, and FBP-RfDoc were described elsewhere.51 The TGC protein containing thioredoxin, green fluorescent protein, and cellulose-biding module was purified as described elsewhere.53Preparation of MNPs
A-MNPs and D-MNPs were synthesized by the solvothermal method.19,23,57 Briefly, 0.338 g of iron chloride (FeCl3·6H2O, 1.25 mmol) was completely dissolved in 10 mL of ethylene glycol to yield a clear yellow solution, followed by the addition of 1.36 g of sodium acetate (NaAc·3H20, 10 mmol) and 0.125 of Avicel or dextran. The mixture was stirred vigorously for 30 min. The mixture was sealed in a pressure container and then heated at 200 °C for 12 h. The container was cooled down slowly at room temperature. The products collected by a magnet were washed with ethanol and then dried at 60 °C for 6 h. The MNPs without cellulose were synthesized as described above.TEM and SEM of MNPs
Morphology and structure images of MNPs were examined with a FEI TITAN 300 field emission analytical electron microscope (FEI, Hillsboro, OR) and the LEO 1550 as a high-performance Schottky field-emission scanning electron microscope (Carl Zeiss Microscopy, Jena, Germany).Adsorption of TGC or CBM-PGI on three MNPs
For determining the maximum adsorption capacity of MNPs, the crude cell lysate containing TGC was mixed with MNPs at room temperature for 10 min. The protein mass concentration of the unbound protein and total protein was measured by the Bradford assay. The maximum adsorption capacity of MNPs was calculated following the Langmuir isotherm.53The leakage of adsorbed CBM-PGI on MNPs was investigated as follows. The adsorbed CBM-PGI on MNPs was mixed by vortexing for 3 seconds in a 100 mM HEPES buffer (pH 7.5) at room temperature. After MNPs were removed by using a magnet, the supernatant was decanted. The MNPs were re-suspended with a fresh HEPES buffer at a v/v ratio of 50. These washing steps were repeated several times. A small amount of the re-suspended MNPs containing adsorbed enzyme was withdrawn for the PGI activity assay.
Enzyme activity assays
The activity assay of enzymes was conducted based on the initial reaction velocity. For the TIM assay, glyceraldehyde-3-phosphate (G3P) was the substrate and dihydroxyacetone phosphate (DHAP) was the product. DHAP was measured by using glycerol 3-phophate dehydrogenase (GPDH) in the presence of NADH and the consumption of NADH was measured at 340 nm. Because thermophilic glycerol-3-phophate dehydrogenase was not available and NADH was not stable at high temperatures, thermophilic TIM activity was measured by using a discontinuous means. Specifically, the generation of DHAP by TIM was measured on 2 mM G3P in 100 mM HEPES buffer (pH 7.5) containing 10 mM MgCl2 and 0.5 mM MnCl2 at 60 °C. The reaction was stopped by adding 5.8 M HClO4 (final, 0.65 M) in an ice-water bath for 5 min followed by the addition of 5 M KOH until pH ∼ 7 (ref. 51). After centrifugation, the supernatant was mixed with 0.2 mM NADH and GPDH. The consumption of NADH was measured at 340 nm.The ALD activity was measured by a continuous cascade reaction along with sufficient TIM, FBP, and PGI.51G3P and DHAP were substrates and F16P was the product. After the cascade reaction, the reaction was stopped by the addition of HClO4.51 The final product of G6P was measured by the Pointe Scientific liquid enzymatic glucose kit at 37 °C. The absorbance was read at 340 mM with a reference of the blank ALD solution.
FBP and PGI activities were measured as described elsewhere.10,45
For the 3-enzyme cascade reaction assay, G3P was the substrate and F6P was the product. The product F6P can be measured by the liquid glucose reagent kit supplemented with PGI at 37 °C for 3 min. Specifically, the generation of F6P can be done by using cascade enzymes with 2.5 mM of G3P in 200 mM HEPES buffer (pH 7.5) containing 10 mM MgCl2, 0.5 mM MnCl2, 0.5 mM thiamine pyrophosphate and 1 mM CaCl2 at 60 °C. The reaction was stopped by adding HClO4 in an ice-water bath followed by the neutralization of 5 M KOH. After centrifugation, the product F6P in the supernatant was assayed by the liquid glucose reagent supplemented with PGI at 37 °C for 3 min.
Other assays
The mass concentration of protein was measured using the Bio-Rad modified Bradford protein kit with bovine serum albumin as a standard protein.58 12% SDS-PAGE was performed in the Tris–glycine buffer as described elsewhere.11 The fitting curves of adsorption data in the Langmuir isotherm was obtained with software named Curve Expert 1.4 (http://www.curveexpert.net/). The error bars in the figures represent standard deviations.Results and discussion
Preparation and characterization of MNPs
Three types of magnetic particles (A-MNPs, D-MNPs, and MNPs) were synthesized by using a solvothermal method. FeCl3, ethylene glycol, and sodium acetate were the source of magnetite, a solvent and high boiling point reducing agent, and an electrostatic stabilizer to prevent particle agglomeration, respectively.23,57 The addition of dextran as a surfactant can prevent particle agglomeration and facilitate the formation of nanocrystal particles.23,57 In this study, insoluble Avicel (microcrystalline nano-cellulose) powders with an average aggregated particle size of 20 μm were used instead of water-soluble dextran. Under optimized preparation conditions, the weights of A-MNPs, MNPs and D-MNPs were 97.5 ± 0.6, 94.4 ± 0.6 and 95.1 ± 0.8 mg, respectively, from 335 mg of FeCl3 (1.25 mmol). A-MNPs, D-MNPs, and MNPs were characterized by using SEM (Fig. 1a, c and e) and TEM (Fig. 1b, d and f). D-MNPs and MNPs had typical spherical shapes, as described elsewhere.23,57 The particle sizes of A-MNPs (Fig. 1a & b) were a little larger and they exhibited broader size distribution compared to MNPs (Fig. 1c & d) and D-MNPs (Fig. 1e & f). Their average sizes were ∼600 nm for A-MNPs, ∼300 nm for MNPs, and ∼300 nm for D-MNPs (Fig. 1). Small cracks/holes were observed on the surface of A-MNPs and D-MNPs while MNPs have a smooth surface. These results implied that polymers (Avicel or dextran) were present at both the surface and the interior of the particles, which was reported in the cases of dextran, chitosan, or polyacrylic acid as surfactants.23 | ||
| Fig. 1 Images of different types of MNPs under SEM (a, c, and e) and TEM (b, d, and f). A-MNPs (a & b); MNPs (c & d); and D-MNPs (e & f). The bars indicate 100 nm in SEM and 500 nm in TEM images, respectively. | ||
Protein adsorption and immobilization on MNPs
The proteinadsorption profiles on MNPs were studied based on CBM-tagged proteins: thioredoxin-GFP-CBM protein (TGC)53 and CBM-tagged phosphoglucose isomerase (PGI).10 It is well-known that family 3 CBM tags can be bound on the surface of cellulose through biospecific adsorption (Fig. 2a & c).11 The green aqueous solution containing TGC turned colorless after A-MNPs were added and a magnet was applied, suggesting that A-MNPs can bind with TGC (Fig. 2b). TGC adsorption by A-NMPs was very fast, reaching equilibrium within 5 min (data not shown). | ||
| Fig. 2 Scheme of CBM-tagged proteins adsorbed on A-MNPs. (a) TGC adsorbed on A-MNPs, (b) the process of collecting TGC adsorbed on A-MNPs by using a magnetic field and (c) PGI adsorbed on A-MNPs. | ||
The adsorption profiles of TGC were examined on A-MNPs, MNPs, and D-MNPs. The TGC adsorption profiles on MNPs and D-MNPs obeyed typical Langmuir isotherms due to simple physical adsorption (Fig. 3). The maximum binding capacity of MNPs and D-MNPs was 9.6 and 5.4 mg g−1, respectively. In contrast, the adsorption of TGC onto the surface of A-MNPs could be attributed to a combination of two adsorption forces: (1) simple physical adsorption between a protein and the large surface of MNPs and (2) high-affinity binding between the CBM tag and cellulosic materials.10,59 The lumped adsorption profile of TGC on A-MNPs may be fitted into a simple Langmuir isotherm with a relatively large deviation. The maximum binding capacity of A-MNPs was 13.1 mg g−1, nearly 1.4 and 2.4 times those of MNPs and D-MNPs.
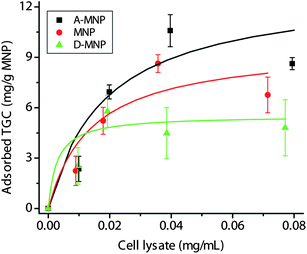 | ||
| Fig. 3 The adsorption profiles of TGC on A-MNPs, MNPs, and D-MNPs. The curves were fitted by the Langmuir equations. | ||
Because physically adsorbed proteins on MNPs may be washed out, the leakage of CBM-PGI immobilized on MNPs was investigated (Fig. 4). By using the same amounts of MNPs, excess cell lysate of CBM-PGI was mixed with different MNPs (i.e., MNPs were saturated by proteins). The absolute specific activities of A-MNPs, MNPs and D-MNPs before the first washing were 0.73 ± 0.01, 0.38 ± 0.02, and 0.30 ± 0.01 U mg−1 of MNPs, respectively. It was found that the remaining PGI activity on A-MNPs decreased by only 20% after 8 washes, while the remaining PGI activities on D-MNPs and MNPs were 34 and 46% of initial activities, respectively. This result suggested that CBM-tagged enzymes can be bound more tightly with A-MNPs through the CBM tag than physical adsorption on D-MNPs and MNPs. Similarly, immobilized CBM-tagged PGI on regenerated amorphous cellulose was strong enough to retain enzyme activity even after a number of washes.10
 | ||
| Fig. 4 Leakage testing of CBM-PGI immobilized on A-MNPs, MNPs, and D-MNPs. Remaining activities of immobilized CBM-PGI were measured after a number of washing steps. | ||
We further investigated the adsorption of CBM-tagged PGI by A-MNPs, MNPs and D-MNPs. At the same amounts of MNPs and CBM-PGI, the immobilized PGI on A-MNPs in terms of enzyme activity was 2.46 times that on D-MNPs and 1.94 times that on MNPs (data not shown). The above results suggested that A-MNPs not only had higher enzyme-immobilization capacity but also retained more enzyme activity compared to D-MNPs and MNPs. It was mainly due to the bio-specific affinity interaction between cellulose in MNPs and the CBM tag, which could decrease the possibility of random non-active adsorption. Although A-MNPs have larger particle sizes with more binding capacity than MNPs and D-MNPs, this result suggested that a significant fraction of the binding capacity of A-MNPs is internal, consistent with their porous structure observed in Fig. 1. Similarly, most of the binding capacity of Avicel is internal rather than external.53
Synthetic metabolon immobilized on A-MNPs
Triosephosphate isomerase (TIM, EC 5.3.1.1), aldolase (ALD, EC 4.1.2.13), and fructose 1,6-bisphosphatase (FBP, EC3.1.3.11) are cascade enzymes in the glycolytic and gluconeogenic pathways, which could be used for high-yield hydrogen production from sugars.28,29 TIM catalyzes the reversible conversion of glyceraldehyde-3-phosphate (G3P) to dihydroxyacetone phosphate (DHAP) (eqn (1)). ALD catalyzes the reversible aldol condensation of G3P and DHAP to fructose 1,6-bisphosphate (F16P) (eqn (2)). FBP catalyzes the irreversible conversion of F16P to fructose 6-phosphate (F6P) (eqn (3)). | (1) |
 | (2) |
 | (3) |
The immobilization of a self-assembled three-enzyme complex called metabolon containing TIM, ALD and FBP through a synthetic trifunctional scaffoldin was investigated on A-MNPs (Fig. 5a). This synthetic metabolon was comprised of a dockerin-containing Thermus thermophilus triose phosphate isomerase (TIM), a dockerin-containing Thermotoga maritima fructose bisphosphate aldolase (ALD), a dockerin-containing T. maritimafructose bisphosphatase (FBP) and a mini-scaffold containing a family 3 cellulose-binding module at the N-terminus followed by three different types of cohesins from the Clostridium thermocellum CipA, Clostridium cellulovorans CbpA and Ruminococcus flavefaciens ScaB.51 This synthetic three-enzyme complex was assembled in vitro through the high-affinity interaction between cohesins and dockerins at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 when the cell extracts containing four proteins were mixed.
1 when the cell extracts containing four proteins were mixed.
 | ||
| Fig. 5 (a) Scheme of the cascade reactions catalysed by the multi-enzyme complex including TIM, ALD, and FBP on A-MNPs for the generation of f6p from g3p. (b) SDS-PAGE analysis of a purified multi-enzyme complex, three single enzymes, and additional enzyme for the product assay. Lane M, marker; Lane 1; enzyme complex including mini-scaffoldin, TIM-CtDoc, ALD-CcDoc, and FBP-RfDoc, Lane 2; purified TIM, Lane 3; purified ALD, Lane 4; purified FBP; Lane 5 purified PGI. | ||
When A-MNPs were applied, the synthetic three-enzyme complex was adsorbed onto the surface of A-MNPs for their fast purification and co-immobilization (Fig. 5a). The mini-scaffoldin, TIM, ALD, and FBP ratio was approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, as shown in SDS-PAGE gel (Fig. 5b, Lane 1). The activities of dockerin-containing TIM, ALD, and FBP in the presence of mini-scaffoldin were similar to those of dockerin-free enzymes (data not shown), suggesting that the dockerin addition did not influence the activity of each enzyme.51
1, as shown in SDS-PAGE gel (Fig. 5b, Lane 1). The activities of dockerin-containing TIM, ALD, and FBP in the presence of mini-scaffoldin were similar to those of dockerin-free enzymes (data not shown), suggesting that the dockerin addition did not influence the activity of each enzyme.51
Furthermore, the reaction rates of the synthetic metabolon immobilized on A-MNPs, the non-immobilized synthetic metabolon and the free enzyme mixture were investigated from 2.5 mM G3P at 60 °C (Fig. 6). The purified three free enzymes (Lanes 2, 3, and 4) are shown in Fig. 5. Another purified PGI (Lane 5) was used to measure fructose-6-phosphate formation. The metabolon immobilized on A-MNPs had an initial reaction rate of 0.285 μmol L−1 s, 4.6 times that of the free enzyme mixture (i.e., 0.062 μmol L−1 s). Such a rate enhancement was attributed to the fact that DHAP generated by TIM can be rapidly transferred to an adjacent ALD to yield F16P, as reported previously.51 Also, the immobilized synthetic metabolon exhibited 1.75 times higher initial reaction rate than that of the free synthetic metabolon (i.e., 0.163 μmol L−1 s), which was possibly due to shorter enzyme–enzyme distances when the metabolon was immobilized on the surface of the solid adsorbent than those in the aqueous solution. A similar observation was reported for metabolons immobilized on regenerated amorphous cellulose.11
 | ||
| Fig. 6 Comparison of the reaction activities of the enzyme complex immobilized on A-MNPs, the unbound enzyme complex, and the non-complexed enzyme mixture at the same enzyme loading. | ||
Whether substrate channelling among synthetic metabolons comprised of an enzyme cascade was observed or not was complicated, depending on a number of factors: enzymes used and reaction conditions (e.g., substrate concentration, enzyme loading, temperature, pH, etc.).41,51 In our another study, a two-enzyme complex containing cellobiose phosphorylase and photo glucan phosphorylase through the same synthetic scaffoldin was constructed and immobilized on A-MNPs. We did not observe any substrate channelling among adjacent cellobiose phosphorylase and potato glucan phosphorylase.27
Cell-free biosystems comprised of synthetic enzymatic pathways could become an innovative biomanufacturing platform. It was essential to ensure that all enzymes reached their maximum total turn-over numbers before their replacement. Immobilized CBM-tagged enzymes or their complexes on A-MNPs can be recycled easily from other free enzymes or immobilized enzymes by using a magnetic force. Therefore, it was highly operative to selectively separate deactivated enzymes from active enzymes. As a result, it could greatly decrease enzyme costs in cell-free biosystems. On the other hand, selective removal of enzymes immobilized on MNPs could be very effective in stopping cascade enzymatic reactions within a short time. For example, it will be important to stop and resume enzymatic hydrogen generation of biotransformers in hypothetical sugar-fuel cell vehicles.40 We envisioned that the selective removal and addition of some key enzymes immobilized on A-MNPs by a switchable magnetic force could stop and resume reactions rapidly.
Conclusions
The cellulose-containing magnetic nanoparticles were prepared for the immobilization of CBM-tagged proteins or the metabolons. Although A-MNPs exhibited higher adsorption capacity, they did not impair the activity of immobilized enzymes, and retained more enzyme activity after washing, compared to MNPs and D-MNPs because of the high-affinity binding of CBM tags on A-MNPs. Substrate channeling among the synthetic metabolons immobilized on the surface of A-MNPs could not only increase cascade reaction rates greatly but also decrease enzyme costs in cell-free biosystems. The use of enzymes immobilized on A-MNPs could be very useful in controlling the On/Off process in cascade enzymatic reactions.Acknowledgements
This work was supported by the Biological Systems Engineering Department of Virginia Tech, and partially supported by the Shell GameChanger Program and the CALS Biodesign and Bioprocessing Research Center. SM was partially supported by the ICTAS Scholar Program.References
- Z. Liu, H. Wang, B. Li, C. Liu, Y. Jiang, G. Yu and X. Mu, J. Mater. Chem., 2012, 22, 15085–15091 RSC.
- A. Liese and L. Hilterhaus, Chem. Soc. Rev., 2013, 42, 6236–6249 RSC.
- Y. Hu, W. M. Liu, B. Zou, S. S. Tang and H. Huang, Prog. Chem., 2010, 22, 1656–1664 CAS.
- D. S. Rodrigues, A. A. Mendes, W. S. Adriano, L. R. B. Goncalves and R. L. C. Giordano, J. Mol. Catal. B: Enzym., 2008, 51, 100–109 CrossRef CAS.
- Z. G. Zhu, Y. R. Wang, S. Minteer and Y.-H. P. Zhang, J. Power Sources, 2011, 196, 7505–7509 CrossRef CAS.
- R. Wang, B. Xia, B. J. Li, S. L. Peng, L. S. Ding and S. Zhang, Int. J. Pharm., 2008, 364, 102–107 CrossRef CAS.
- M. N. Gupta, S. Dalal and M. Kapoor, J. Mol. Catal. B: Enzym., 2007, 44, 128–132 CrossRef.
- O. Shoseyov, Z. Shani and I. Levy, Microbiol. Mol. Biol. Rev., 2006, 70, 283–295 CrossRef CAS.
- G. A. Velikodvorskaya, T. V. Tikhonova, I. D. Gurvits, A. S. Karyagina, N. V. Lavrova, O. V. Sergienko, V. N. Tashlitskii, N. A. Lunina and V. G. Lunin, Appl. Environ. Microbiol., 2010, 76, 8071–8075 CrossRef CAS.
- S. Myung, X. Z. Zhang and Y. H. P. Zhang, Biotechnol. Prog., 2011, 27, 969–975 CrossRef CAS.
- C. You and Y.-H. P. Zhang, ACS Synth. Biol., 2013, 2, 102–110 CrossRef CAS.
- A. M. G. C. Dias, A. Hussain, A. S. Marcos and A. C. A. Roque, Biotechnol. Adv., 2011, 29, 142–155 CrossRef CAS.
- A. H. Lu, E. L. Salabas and F. Schuth, Angew. Chem., Int. Ed., 2007, 46, 1222–1244 CrossRef CAS.
- S. Wang, P. Su, J. Huang, J. Wu and Y. Yang, J. Mater. Chem. B, 2013, 1, 1749–1754 RSC.
- C. Tassa, S. Y. Shaw and R. Weissleder, Acc. Chem. Res., 2011, 44, 842–852 CrossRef CAS.
- X. H. Sun, C. M. Zheng, F. X. Zhang, Y. L. Yang, G. J. Wu, A. M. Yu and N. J. Guan, J. Phys. Chem. C, 2009, 113, 16002–16008 CAS.
- S. Giri, S. Samanta, S. Maji, S. Ganguli and A. Bhaumik, J. Magn. Magn. Mater., 2005, 285, 296–302 CrossRef CAS.
- J. Vidal-Vidal, J. Rivas and M. A. Lopez-Quintela, Colloids Surf., A, 2006, 288, 44–51 CrossRef CAS.
- H. Deng, X. L. Li, Q. Peng, X. Wang, J. P. Chen and Y. D. Li, Angew. Chem., Int. Ed., 2005, 44, 2782–2785 CrossRef CAS.
- H. W. Gu, K. M. Xu, C. J. Xu and B. Xu, Chem. Commun., 2006, 941–949 RSC.
- H. Kempe and M. Kempe, Biomaterials, 2010, 31, 9499–9510 CrossRef CAS.
- X. H. Fu, Biochem. Eng. J., 2008, 39, 267–275 CrossRef CAS.
- X. L. Huang, J. Zhuang, D. Chen, H. Y. Liu, F. Q. Tang, X. Y. Yan, X. W. Meng, L. Zhang and J. Ren, Langmuir, 2009, 25, 11657–11663 CrossRef CAS.
- Y.-H. P. Zhang, S. Myung, C. You, Z. G. Zhu and J. Rollin, J. Mater. Chem., 2011, 21, 18877–18886 RSC.
- Y.-H. P. Zhang, Biotechnol. Bioeng., 2010, 105, 663–677 CAS.
- J. A. Rollin, W. Tam and Y. H. P. Zhang, Green Chem., 2013, 15, 1708–1719 RSC.
- C. You, H. Chen, S. Myung, N. Sathitsuksanoh, H. Ma, X.-Z. Zhang, J. Li and Y.-H. P. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7182–7187 CrossRef CAS.
- Y.-H. P. Zhang, B. R. Evans, J. R. Mielenz, R. C. Hopkins and M. W. W. Adams, PLoS One, 2007, 2, e456 Search PubMed.
- X. Ye, Y. Wang, R. C. Hopkins, M. W. W. Adams, B. R. Evans, J. R. Mielenz and Y.-H. P. Zhang, ChemSusChem, 2009, 2, 149–152 CrossRef CAS.
- J. S. Martín del Campo, J. Rollin, S. Myung, Y. Chun, S. Chandrayan, R. Patiño, M. W. W. Adams and Y. H. P. Zhang, Angew. Chem., Int. Ed., 2013, 52, 4587–4590 CrossRef.
- J.-K. Guterl, D. Garbe, J. Carsten, F. Steffler, B. Sommer, S. Reiße, A. Philipp, M. Haack, B. Rühmann, U. Kettling, T. Brück and V. Sieber, ChemSusChem, 2012, 5, 2165–2172 CrossRef CAS.
- X. Ye, K. Honda, T. Sakai, K. Okano, T. Omasa, R. Hirota, A. Kuroda and H. Ohtake, Microb. Cell Fact., 2012, 11, 120 CrossRef CAS.
- Y. R. Wang, W. D. Huang, N. Sathitsuksanoh, Z. G. Zhu and Y. H. P. Zhang, Chem. Biol., 2011, 18, 372–380 CrossRef CAS.
- J. R. Swartz, AIChE J., 2011, 58, 5–13 CrossRef.
- Z. G. Zhu, F. Sun, X. Zhang and Y.-H. P. Zhang, Biosens. Bioelectron., 2012, 36, 110–115 CrossRef CAS.
- S. Xu and S. D. Minteer, ACS Catal., 2011, 1, 91–94 Search PubMed.
- M. Bujara, M. Schümperli, R. Pellaux, M. Heinemann and S. Panke, Nat. Chem. Biol., 2011, 7, 271–277 CrossRef CAS.
- Y. Xu, S. Masuko, M. Takieddin, H. Xu, R. Liu, J. Jing, S. A. Mousa, R. J. Linhardt and J. Liu, Science, 2011, 334, 498–501 CrossRef CAS.
- X. D. Tong, B. El-Zahab, X. Y. Zhao, Y. Y. Liu and P. Wang, Biotechnol. Bioeng., 2011, 108, 465–469 CrossRef CAS.
- Y.-H. P. Zhang and W.-D. Huang, Trends Biotechnol., 2012, 30, 301–306 CrossRef CAS.
- Y.-H. P. Zhang, Biotechnol. Adv., 2011, 29, 715–725 CrossRef CAS.
- A. H. Chen and P. A. Silver, Trends Cell Biol., 2012, 22, 662–670 CrossRef CAS.
- O. I. Wilner, Y. Weizmann, R. Gill, O. Lioubashevski, R. Freeman and I. Willner, Nat. Nanotechnol., 2009, 4, 249–254 CrossRef CAS.
- J. L. Fu, M. H. Liu, Y. Liu, N. W. Woodbury and H. Yan, J. Am. Chem. Soc., 2012, 134, 5516–5519 CrossRef CAS.
- S. Myung, Y. Wang and Y. H. P. Zhang, Process Biochem., 2010, 45, 1882–1887 CrossRef CAS.
- E. A. Bayer, E. Morag and R. Lamed, Trends Biotechnol., 1994, 12, 379–386 CrossRef CAS.
- F. Mingardon, A. Chanal, C. Tardif, E. A. Bayer and H.-P. Fierobe, Appl. Environ. Microbiol., 2007, 73, 7138–7149 CrossRef CAS.
- C. You, X.-Z. Zhang, N. Sathitsuksanoh, L. R. Lynd and Y.-H. P. Zhang, Appl. Environ. Microbiol., 2012, 78, 1437–1444 CrossRef CAS.
- S.-L. Tsai, N. A. DaSilva and W. Chen, ACS Synth. Biol., 2013, 2, 14–21 CrossRef CAS.
- J. Sun, F. Wen, T. Si, J.-H. Xu and H. Zhao, Appl. Environ. Microbiol., 2012, 78, 3837–3845 CrossRef CAS.
- C. You, S. Myung and Y.-H. P. Zhang, Angew. Chem., Int. Ed., 2012, 51, 8787–8790 CrossRef CAS.
- F. Liu, S. Banta and W. Chen, Chem. Commun., 2013, 49, 3766–3768 RSC.
- J. Hong, X. H. Ye and Y. H. P. Zhang, Langmuir, 2007, 23, 12535–12540 CrossRef CAS.
- S. Y. Huang, Y.-H. P. Zhang and J. J. Zhong, Appl. Microbiol. Biotechnol., 2012, 93, 2403–2410 CrossRef CAS.
- C. You, X. Z. Zhang and Y. H. P. Zhang, Appl. Environ. Microbiol., 2012, 78, 1593–1595 CrossRef CAS.
- S. Myung and Y.-H. P. Zhang, PLoS One, 2013, 8, e61500 CAS.
- Y. Li, J. S. Wu, D. W. Qi, X. Q. Xu, C. H. Deng, P. Y. Yang and X. M. Zhang, Chem. Commun., 2008, 564–566 RSC.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- J. Hong, Y. Wang, X. Ye and Y.-H. P. Zhang, J. Chromatogr., A, 2008, 1194, 150–154 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
