Trans-differentiation of human mesenchymal stem cells generates functional hepatospheres on poly(L-lactic acid)-co-poly(ε-caprolactone)/collagen nanofibrous scaffolds†
Received 19th February 2013, Accepted 7th June 2013
First published on 10th June 2013
Abstract
Mesenchymal stem cell (MSC)-based liver tissue engineering on nanofibrous scaffold holds great promise for cell-based therapy in liver injuries and end-stage liver failure treatments. We investigated the hepatic trans-differentiation potential of human MSCs on a biocomposite poly(L-lactic acid)-co-poly (ε-caprolactone)/collagen (PLACL/collagen) nanofibrous scaffold. The nanofibrous scaffolds comprised of PLACL, collagen and a PLACL/collagen blend (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were fabricated by electrospinning and also evaluated for fiber morphology, surface wettability, functional groups, porosity and tensile properties. Hepatic trans-differentiation of human bone marrow-derived MSCs (hMSCs) was carried out on these scaffolds over a period of 28 days using sequential induction with hepatogenic growth factors. Hepatogenesis was confirmed by scanning electron microscopy (SEM), cell phenotype tracking dye expression, quantitative expression of hepatic genes, immunofluorescence staining of hepatocyte-specific markers and albumin release. The results proved that the porous PLACL/collagen nanofibrous scaffold supported enhanced hMSC proliferation and hepatic trans-differentiation compared to individual PLACL and collagen scaffolds as well as a monolayer culture on tissue culture plate (TCP). Interestingly, hMSC-derived hepatocyte-like cells on PLACL/collagen nanofibrous scaffolds could aggregate to form functional ‘hepatospheres’ similar to normal hepatic spheroids. The present study concludes that PLACL/collagen nanofibrous scaffolds are potentially biomimetic and upon sequential induction with hepatogenic growth factors/cytokines, it augments trans-differentiation of hMSCs towards functional hepatosphere formation. Such bioengineered nanofibrous scaffold hepatic construct provides a promising approach for cellular therapy of damaged livers in end-stage liver failure treatments.
1) were fabricated by electrospinning and also evaluated for fiber morphology, surface wettability, functional groups, porosity and tensile properties. Hepatic trans-differentiation of human bone marrow-derived MSCs (hMSCs) was carried out on these scaffolds over a period of 28 days using sequential induction with hepatogenic growth factors. Hepatogenesis was confirmed by scanning electron microscopy (SEM), cell phenotype tracking dye expression, quantitative expression of hepatic genes, immunofluorescence staining of hepatocyte-specific markers and albumin release. The results proved that the porous PLACL/collagen nanofibrous scaffold supported enhanced hMSC proliferation and hepatic trans-differentiation compared to individual PLACL and collagen scaffolds as well as a monolayer culture on tissue culture plate (TCP). Interestingly, hMSC-derived hepatocyte-like cells on PLACL/collagen nanofibrous scaffolds could aggregate to form functional ‘hepatospheres’ similar to normal hepatic spheroids. The present study concludes that PLACL/collagen nanofibrous scaffolds are potentially biomimetic and upon sequential induction with hepatogenic growth factors/cytokines, it augments trans-differentiation of hMSCs towards functional hepatosphere formation. Such bioengineered nanofibrous scaffold hepatic construct provides a promising approach for cellular therapy of damaged livers in end-stage liver failure treatments.
Introduction
The liver, the organ with the highest regenerative potential, cannot replenish its damaged hepatocytes in patients with end-stage liver failure, and liver transplantation is the only alternative. However, the growing demand for liver homografts for transplantation is on the rise due to the shortage of organ donors and graft incompatibility.1 Regeneration of the liver tissue by potential hepatogenic precursor cells is a therapeutic possibility, provided they mimic the functionality of native hepatocytes in the liver microenvironment. Stem cells of varying stemness and tissue origin such as embryonic stem cells,2 hematopoietic stem cells,3 induced pluripotent stem cells,4 adipose tissue-derived stem cells5 and cord blood stem cells6 have been proven as promising candidates for hepatocyte differentiation. Embryonic stem cells (ESCs), being pluripotent, possess the highest hepatogenic differentiation potential, but controversies related to teratoma formation, immune rejection and ethics involved in deriving them limit their therapeutic roles.7,8 Bone marrow-derived MSCs, though comparatively less plastic, can give rise to functional hepatocytes9 and if injected in an autologous manner can serve as the best therapeutically effective cell source for treating liver failure.As clinical transplantation of MSCs for liver tissue regeneration becomes a promising tool, an ideal hepatic tissue engineering strategy needs to be optimized before the in vitro regenerated tissue or organ can be taken from bench to bedside. In this regard, MSCs have been explored for fine-tuned trans-differentiation towards hepatic lineages, with a defined milieu of chemicals and growth factors in a sequential manner, of course, on extracellular matrix (ECM) scaffolds.10 Growth factors in combination with ECM enhance the attachment, proliferation and differentiation of stem cells, thus mimicking the in vivo microenvironment.11 Most of the studies investigating MSC differentiation to hepatocytes have been performed on monolayer cultures.9,12 However, such methodologies result in inefficient and heterogenous differentiation, as the two-dimensional (2D) cultures do not mimic the natural liver microenvironment. Three-dimensional (3D) culture systems support an effective and homogenous differentiation by providing structural integrity, promoting improved cell–cell and cell–ECM interactions due to high surface area and porosity thus ensuring growing hepatic tissue remodelling.13,14 The fate of MSCs during differentiation is known to be modulated by nanotopographical cues.15 In fact, hepatocytes, because of their anchorage dependence, proliferate better on 3D nanofibrous scaffolds and form functional aggregates.16,17
Electrospinning is a simple and potentially effective methodology that has successfully delivered fabrication of nanofibrous structures of micro- to nanoscale dimensions comparable to those of natural ECM.18 These electrospun nanofibrous scaffolds possess high volume-to-surface areas and porosity, thus facilitating effective cell–cell and cell–ECM communications and 3D tissue regeneration. A variety of biodegradable and biocompatible polymers of natural and synthetic origins have been employed to fabricate nanofibrous scaffolds for tissue engineering applications. Biosynthetic hybrid scaffolds that provide suitable biological and mechanical features and mimic the ECM architecture of liver tissue can be used as a suitable substrate for hepatic tissue engineering. PLACL is a synthetic, biodegradable and non-toxic copolymer of poly-L-lactic acid (PLLA) and polycaprolactone (PCL), which has been widely studied as a biomaterial for surgery and drug delivery applications.19 On blending with collagen, this copolymer becomes preferable for various tissue engineering applications, as collagen possess unique integrin binding sites on its surface for cell adhesion and proliferation.20,21 Stem cells have been differentiated towards hepatocyte-like cells effectively on various nanofibrillar surfaces in a proof-of-concept style.22–26 However, optimization is still needed to modulate stem cell differentiation towards generation of a functional 3D hepatic construct on nanofibrous scaffolds. In the present study, we have attempted to evaluate the in vitro hepatomimetic potential of a well-established biosynthetic nanofibrous scaffold fabricated from a PLACL and collagen blend (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) by inducing hepatic trans-differentiation of human bone marrow MSCs. This study focused on assessing the impact of 3D biosynthetic nanofibrous scaffolds from a PLACL/collagen blend upon hepatic trans-differentiation of hMSCs as compared to individual PLACL and collagen nanofibrous scaffolds as well as a conventional monolayer culture system on tissue culture plate (TCP).
1) by inducing hepatic trans-differentiation of human bone marrow MSCs. This study focused on assessing the impact of 3D biosynthetic nanofibrous scaffolds from a PLACL/collagen blend upon hepatic trans-differentiation of hMSCs as compared to individual PLACL and collagen nanofibrous scaffolds as well as a conventional monolayer culture system on tissue culture plate (TCP).
Experimental section
Materials
For differentiation experiments, hMSCs were acquired from Lonza (Allendale, NJ, USA). Poly(L-lactic acid)-co-poly-(ε-caprolactone) (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, Mw 150 kDa) was obtained from Boehringer Ingelheim Pharma, GmbH & Co., Ingelheim, Germany. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), antibiotics and trypsin–EDTA, CMFDA (5-chloromethylfluorescein diacetate), secondary antibody Alexa Fluor 488 and 594, Iscove's modified Dulbecco's medium (IMDM), TRIzol™ reagent, SuperScript™ first strand synthesis system and 4′-6-diamidino-2-phenylindole (DAPI) were procured from Invitrogen (Life Technologies, Carlsbad, CA, USA). Bovine serum albumin (BSA), 1,1,1,3,3,3-hexafluor-2-propanol (HFP), anti-human anti-albumin antibody raised in goat, anti-CY-18, anti-AFP, hexamethyldisilazane (HMDS), hepatocyte growth factor (HGF), oncostatin M (OSM), insulin-transferring selenium premix (ITS), dexamethasone, nicotinamide, epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) were from Sigma-Aldrich (St. Louis, MO, USA). Collagen type I was obtained from Koken Company Ltd. (Tokushima-ku, Tokyo, Japan). SYBR green real-time PCR mastermix was purchased from Applied Biosystems (Foster City, CA, USA).
30, Mw 150 kDa) was obtained from Boehringer Ingelheim Pharma, GmbH & Co., Ingelheim, Germany. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), antibiotics and trypsin–EDTA, CMFDA (5-chloromethylfluorescein diacetate), secondary antibody Alexa Fluor 488 and 594, Iscove's modified Dulbecco's medium (IMDM), TRIzol™ reagent, SuperScript™ first strand synthesis system and 4′-6-diamidino-2-phenylindole (DAPI) were procured from Invitrogen (Life Technologies, Carlsbad, CA, USA). Bovine serum albumin (BSA), 1,1,1,3,3,3-hexafluor-2-propanol (HFP), anti-human anti-albumin antibody raised in goat, anti-CY-18, anti-AFP, hexamethyldisilazane (HMDS), hepatocyte growth factor (HGF), oncostatin M (OSM), insulin-transferring selenium premix (ITS), dexamethasone, nicotinamide, epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) were from Sigma-Aldrich (St. Louis, MO, USA). Collagen type I was obtained from Koken Company Ltd. (Tokushima-ku, Tokyo, Japan). SYBR green real-time PCR mastermix was purchased from Applied Biosystems (Foster City, CA, USA).Fabrication of electrospun nanofibrous scaffolds
Polymer solutions of PLACL (10%, w/v), collagen (8% w/v) and a PLACL/collagen blend (10% w/v, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w ratio) were prepared by dissolving them in HFP. For electrospinning, the respective polymer solutions were fed into a 3 ml standard syringe attached to a blunted stainless steel needle (22½ G) using a syringe pump (KD-100, KD Scientific Inc., Holliston, MA, USA) at an optimized flow rate of 1 ml h−1. On application of a high voltage (Gamma High Voltage Research, Ormond Beach, FL, USA) of 18.5 kV, the polymer solution was spun as nanofibers of random orientation, which were collected on coverslips (diameter, 15 mm; thickness, 0.13–0.16 mm, Menzel-Glaser, Thermo Scientific, Germany). The ambient conditions of the spinning apparatus were controlled with a vacuum dehumidifier machine (KNF Neuberger Inc, Trenton, NJ, USA). The nanofiber mats spun on the cover slips were dried overnight in vacuum to remove the residual solvents and subsequently used for characterization and cell culture experiments.
1 w/w ratio) were prepared by dissolving them in HFP. For electrospinning, the respective polymer solutions were fed into a 3 ml standard syringe attached to a blunted stainless steel needle (22½ G) using a syringe pump (KD-100, KD Scientific Inc., Holliston, MA, USA) at an optimized flow rate of 1 ml h−1. On application of a high voltage (Gamma High Voltage Research, Ormond Beach, FL, USA) of 18.5 kV, the polymer solution was spun as nanofibers of random orientation, which were collected on coverslips (diameter, 15 mm; thickness, 0.13–0.16 mm, Menzel-Glaser, Thermo Scientific, Germany). The ambient conditions of the spinning apparatus were controlled with a vacuum dehumidifier machine (KNF Neuberger Inc, Trenton, NJ, USA). The nanofiber mats spun on the cover slips were dried overnight in vacuum to remove the residual solvents and subsequently used for characterization and cell culture experiments.Characterization of nanofibrous scaffolds
The morphology of electrospun nanofibers on coverslips was examined under a field emission scanning electron microscope (FESEM) (FEI-QUANTA 200F, The Netherlands) at an accelerating voltage of 10 kV, after sputter-coating with gold (JEOL JFC-1200 fine coater, Japan). The average diameter of the nanofibers was determined by measuring at 100 different points in 512 × 471 pixel scanning electron microscopic (SEM) images (with a scale of 9.6 pixels per μm) using image analysis software (ImageJ, National Institutes of Health, Bethesda, MD, USA). The wettability of the nanofibrous scaffolds was measured by sessile drop-water contact angle measurement using a VCA Optima Surface Analysis system (AST products, Billerica, MA). The hydrophobic PLACL nanofibrous scaffolds were made hydrophilic by air plasma treatment with electrodeless radio frequency glow discharge plasma cleaner (Model: PDC-001, Harrick Scientific Corporation, USA). Attenuated total reflectance Fourier transform infrared (ATR-FTIR) spectroscopic analysis of electrospun nanofibrous scaffolds was performed on an Avatar 380 instrument, (Thermo Nicolet, Waltham, MA, USA) over a range of 450–4500 cm−1 at a resolution of 2 cm−1. Porosity as well as pore size distribution of the nanofibrous scaffolds was measured by a capillary flow porometer (CFP-1200-A, Ithaca, NY, USA) by wet up/dry up method and the analysis was done using automated capillary flow porometer system software. The thickness of PLACL, collagen and PLACL/collagen nanofibrous scaffolds were measured by a micrometer (Mitutoyo, Japan) and their apparent density and porosity were calculated according to He et al.27 Tensile properties of electrospun nanofibrous scaffolds were determined with a tabletop tensile tester (Instron 3345, Canton, MA, USA). Initially, the electrospun membrane mat of 20–30 μm thickness was cut into rectangular specimens of 10 mm (breadth) × 20 mm (length) dimensions. The specimen ends were gripped with the clips of tensile testing machines and with an applied load (10 N) via automated software (Bluehill® 2 Materials Testing Software for Universal Testing Systems, Norwood, MA, USA) at a crosshead speed of 5 mm min−1, and the data were recorded every 50 μs. The generated tensile stress–strain curve data was calculated to find out the tensile stress–strain and elastic modulus of the nanofibrous scaffolds. A minimum of six specimens of individual scaffolds of PLACL, collagen and PLACL/collagen membranes having equal length, breadth and thickness were analysed for the tensile strength measurements.Culture and maintenance of hMSCs on nanofibrous scaffolds
Cryopreserved hMSCs of passage 2 were revived and cultured in DMEM–high glucose medium supplemented with 10% FBS, 1% antibiotic and antimycotic solution under humidified conditions of 37 °C and 5% CO2. Cells were maintained till they reached confluence and fresh culture medium was replenished twice per week. Meanwhile, the coverslips coated with nanofibrous scaffolds were subjected to UV sterilization for 3 h and were placed in 24-well tissue culture plates with a stainless steel ring placed over them to prevent the scaffold lifting off the glass coverslips. The scaffolds were washed thrice with phosphate-buffered saline (PBS) to remove any residual cytotoxic solvent and soaked in DMEM overnight before cell seeding. Confluent hMSCs were detached by 0.25% trypsin–EDTA and seeded on the scaffolds (PLACL, collagen and PLACL/collagen) as well as on TCP at a density of 1 × 104 cells per cm2. The cell–scaffold constructs were maintained under standard culture conditions before initiating the hepatic trans-differentiation experiments.Hepatic trans-differentiation protocol
Hepatic trans-differentiation of hMSCs was performed by sequential exposure to a cocktail of inducers9 after 70–80% confluence on the various scaffolds and TCP. The cells were serum-deprived for 2 days and placed in IMDM supplemented with 20 ng ml−1 EGF and 10 ng ml−1 bFGF, prior to induction by a two-step protocol. These preconditioned cells were treated with step 1 differentiation medium, consisting of IMDM supplemented with 20 ng ml−1 HGF and 10 ng ml−1 bFGF, 0.61 g L−1 nicotinamide for 7 days, followed by step 2 maturation medium, consisting of IMDM supplemented with 20 ng ml−1 oncostatin M, 1 μmol L−1 dexamethasone, and 50 mg ml−1 insulin–transferrin–selenium (ITS+) premix. The differentiating cells were maintained in a standard humidified atmosphere for up to 28 days and replenished with fresh differentiation/maturation media twice per week. The extent of hepatogenesis was assessed on the 14th and 28th days by observing the morphological changes, hepatic marker expression and functional assays.Cell proliferation
The hMSC proliferation rates on various nanofibrous scaffolds and TCP during hepatic differentiation were evaluated at the 7th, 14th and 28th days using colorimetric MTS assay (CellTiter 96 Aqueous One solution, Promega, Madison, WI). The assay is based on the principle that mitochondrial dehydrogenase enzymes of metabolically active cells reduce the yellow tetrazolium salt [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2(4-sulfophenyl)-2H-tetrazolium] in MTS reagent to form purple formazan crystals and the amount of crystals formed is directly proportional to the number of live cells. In this experiment, cells were rinsed with PBS and incubated for 3 h in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio mixture of MTS reagent and serum-free DMEM medium. After the incubation period, the samples were pipetted out into 96-well plates and the absorbance was measured at 490 nm using a microplate reader (Fluostar Optima, BMG Lab Technologies, Germany).
5 ratio mixture of MTS reagent and serum-free DMEM medium. After the incubation period, the samples were pipetted out into 96-well plates and the absorbance was measured at 490 nm using a microplate reader (Fluostar Optima, BMG Lab Technologies, Germany).Cell phenotype tracking by fluorescent dye expression
The phenotypic changes of the hMSCs during the process of hepatic differentiation were tracked using the fluorescent dye CMFDA, which on cleavage of its acetates by cytosolic esterases produces a brightly fluorescent CMFDA derivative. At various time points (7th day onwards until the 28th day), the differentiating cells were incubated with fresh DMEM medium containing 25 μM CMFDA dye. After incubation, the cells were replenished with fresh culture medium and incubated overnight. Next day, the cells were washed with PBS in the dark and observed under an inverted Leica DM IRB laser scanning microscope (Leica DC 300F) at 488 nm.Cell morphology analysis by scanning electron microscopy
Cell attachment, proliferation and morphological changes of the differentiating hMSCs on the nanofibrous scaffolds as well as on TCP were analysed on the 7th and 28th days by SEM. The cells were fixed in 3% glutaraldehyde for 3 h followed by washing in deionized water and dehydration with upgrading concentrations of ethanol (50%, 70% 90%, 100%) twice for 15 min each. The final washing with 100% ethanol was followed by air-drying with hexamethyldisilazane (HMDS) overnight in a fume hood. The cell–scaffold constructs on the coverslips were sputter-coated with gold and the morphologies of undifferentiated and differentiated cells were scanned under FESEM at an accelerating voltage of 10 kV.Immunofluorescence staining for hepatic marker expression
Hepatocyte specific marker expression in trans-differentiated hMSCs cultured on different substrates after the 28th day of induction was analysed by immunofluorescence. The cells were fixed with 100% ice-cold methanol for 10 min and permeabilized with 0.1% Triton-X 100. To avoid non-specific antibody binding, cells were blocked with 3% BSA followed by incubation with mouse monoclonal primary antibody for human albumin (ALB)/α-fetoprotein (AFP)/cytokeratin-18 (CY-18) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) overnight at 4 °C. The cells were washed thrice and incubated with Alexa Fluor 488 and 594 (1
100) overnight at 4 °C. The cells were washed thrice and incubated with Alexa Fluor 488 and 594 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) for 1 h and the nuclei were stained with DAPI (0.5 μg ml−1) for another 30 min. The cells were finally washed and mounted on a glass slide with Vectashield (Vector Laboratories, Burlingame, CA, USA) and fluorescent marker expression was observed under a laser confocal scanning microscope (Olympus FV1000).
250) for 1 h and the nuclei were stained with DAPI (0.5 μg ml−1) for another 30 min. The cells were finally washed and mounted on a glass slide with Vectashield (Vector Laboratories, Burlingame, CA, USA) and fluorescent marker expression was observed under a laser confocal scanning microscope (Olympus FV1000).RNA isolation, semi-quantitative reverse transcription polymerase reaction (RT-PCR) and quantitative real-time RT-PCR for hepatic gene expression
Total RNA was isolated from hMSCs differentiated in various substrates as well as the control group on days 14 and 28 using TRIzol™ reagent and the quality and quantity of RNA were measured with a Nanodrop™ spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The total RNA isolated from an equal number of untreated hMSCs was used as a negative control and that of HepG2 cells as the positive control in this experiment. All RNA samples (10 μg) were treated with DNase-I and first-strand cDNA synthesis was carried out with random hexamers using the SuperScript™ first-strand synthesis system according to the manufacturer's protocol. Semi-quantitative RT-PCR for early hepatic genes such as AFP, hepatocyte nuclear factor-4α (HNF-4α) and late hepatic gene ALB was carried out by amplifying the cDNA with specific primers (Table 2) under optimised thermal cycling conditions. The amplified products were separated by electrophoresis on a 1.2% agarose gel and stained with ethidium bromide for visualization. Quantitative real-time PCR for hepatic genes (AFP, ALB and HNF-4α) was carried out with ABI PRISM 7500 Fast Real-Time PCR System using SYBR green master mix. The experiment was optimized with the addition of 10 μl of 2× SYBR Green PCR Master Mix, 30 ng template cDNA and 5 pmol forward and reverse primers, in a total volume of 25 μl for a total of 40 cycles at an annealing temperature of 60 °C. Melting curve analysis was performed at the end of the reaction to determine the melting temperature (Tm) of a single nucleic acid target sequence in an unknown sample. Relative quantification of gene expression in unknown samples was performed using the comparative CT (2−ΔΔCT) method by normalizing with a reference gene (glyceraldehyde-3-phosphate dehydrogenase) and untreated MSCs of third passage were used as the calibrator. All reactions were performed in triplicate for statistical validation.
Table 1 Characterization of the electrospun nanofibers; water contact angles, tensile properties and porosity analysis
| Parameters | Contact angle (°) | Tensile stress (MPa) | Tensile strain (%) | Modulus of elasticity (MPa) | Mean flow pore diameter (μm) | Maximum pore size distribution | Porosity (%) |
|---|
| After plasma treatment. |
|---|
| PLACL | 129.3 ± 2.8 | 1.98 ± 0.64 | 88.92 ± 12.16 | 1.02 ± 0.44 | 1.24 | 654.82 | 83 ± 12 |
| 52.6 ± 9.6a |
| PLACL/Collagen | 46.7 ± 5.5 | 0.52 ± 0.12 | 50.29 ± 19.28 | 0.88 ± 0.28 | 0.58 | 354.47 | 89 ± 16 |
Table 2 The genes and primer sequences used in RT-PCR and real-time PCR
| Gene namea | Primer sequence (5′–3′) | Product size (bp) | NCBI accession number |
|---|
| F: forward primer; R: reverse primer; ALB: albumin; AFP: α-fetoprotein; HNF 4α: hepatocyte nuclear factor 4α; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; β-ACT: β-actin. * denotes primers used in real time PCR. |
|---|
| ALB | F: TGAGAAAACGCCAGTAAGTGAC | 265 | NM_000477.5 |
| R: TGCGAAATCATCCATAACAGC |
| ALB* | F: CGCTATTAGTTCGTTACACCA | 101 | NM_000477.5 |
| R: TTTACAACATTTGCTGCCCA |
| AFP | F: GCTTGGTGGTGGTGGATGAAACA | 157 | NM_001134.1 |
| R: TCCTCTGTTATTTGTGGCTTTTG |
| AFP* | F: AAATGCGTTTCTCGTTGC | 136 | NM_001134.1 |
| R: GCCACACGGCCAATAGTTTGT |
| HNF 4α | F: GAGCAGGAATGGGAAGAATG | 205 | NM_000457.3 |
| R: GGCTGTCCTTTGGGATGAAG |
| HNF 4α* | F: ATGACAATGAGTATGCCTACCT | 131 | NM_178850.1 |
| R: GGTCGTTGATGTAGTCCTCC |
| GAPDH* | F: GCACCGTCAAGGCTGAGAAC | 73 | NM_002046.3 |
| R: GGATCTCGCTCCTGGAAGATG |
| β-ACT | F: AGAGCTACGAGCTGCCTGAC | 184 | NM_001101.2 |
| R: AGCACTGTGTTGGCGTACAG |
Albumin ELISA
To evaluate the functionality of the hMSCs-derived hepatocytes on various substrates, we assessed the amount of albumin secreted into the culture media at various time points (days 7, 14, 21 and 28). HepG2 conditioned media prepared by culturing an equal number HepG2 cells as the number of hMSCs used for the differentiation experiment was used as a positive control. Albumin secretion was determined using a human albumin ELISA quantitation set (Bethyl Laboratory, Montgomery, TX, USA) according to the manufacturer's instructions. Briefly, the wells of an ELISA plate were coated with affinity-purified human albumin coating antibody for 1 h followed by blocking for 30 min. Each standard (serially diluted), control and sample were loaded into the wells and incubated for 1 h followed by the addition of a HRP-conjugated human albumin detection antibody. Tetramethylbenzidine (TMB) substrate solution was added for 15 min and color reaction was stopped by the stop solution (0.16 M sulfuric acid). Absorbance was measured at 450 nm using a microplate reader (Fluostar Optima, BMG Lab Technologies, Germany). The amount of albumin released was normalised to the total number of cells determined from each well.Statistical analysis
All the experiments were done in triplicate for statistical validation and reproducibility was assessed by repeating each experiment at least thrice. The differences in hMSC proliferation rate and hepatic gene expression as well as albumin release during the process of differentiation on various scaffolds were compared and the average value represented as mean ± standard deviation (SD). Statistical differences within and between the groups were analysed using one- or two-way analysis of variance (ANOVA) followed by Tukey's post hoc test with statistical package for social sciences (IBM SPSS statistics, version 19, NY, USA). Differences were considered statistically significant at p ≤ 0.05.Results
Characterization of electrospun nanofibrous scaffolds
The optimised process of electrospinning generated nanofibrous scaffolds (PLACL, collagen and PLACL/collagen blend) with uniform, bead-free, randomly oriented fibers of unpatterned topography and interconnected pores. The SEM images of the nanofibers revealed an average fiber diameter of 563.58 ± 21.89, 283 ± 12.68 and 352 ± 16.28 nm for PLACL, collagen and PLACL/collagen, respectively (Fig. 1). The fibers (>60%) are of nanoscale dimensions, mostly in the range 200–600 nm. Hydrophilicity studies of the scaffolds by water contact angle measurement (Table 1) shows that PLACL was initially highly hydrophobic, and was made hydrophilic by plasma treatment. The collagen and PLACL/collagen nanofibrous scaffolds were adsorbent, revealing the presence of hydrophilic groups on their surfaces.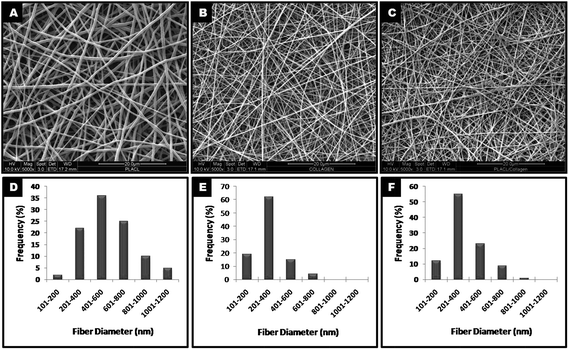 |
| | Fig. 1 FESEM images of electrospun nanofibers of PLACL (A), collagen (B) and a PLACL/collagen blend (C) at high magnification (5000×) showing unpatterned topography with their respective size distributions (D–F). | |
The ATR-FTIR spectra of nanofibrous scaffolds reveals chemical characteristics and functional groups present on their surface that are responsible for their hydrophilicity. The PLACL/collagen nanofibrous scaffold shows characteristic absorption peaks common to PLACL alone along with unique peaks at 3287 cm−1 and 3018 cm−1, representing an N–H stretch of amide A (AA) and C–H stretch of amide B (AB), respectively, suggesting the presence of amino groups on their surfaces (Fig. 2). Peaks at 1649 and 1550 cm−1 for amide I (AI) and amide II (AII) bands represent collagen's secondary structure and the peak at 1287 cm−1 for amide III (AIII) represents the triple helical structure of collagen. The pore size distributions as well as the average pore size of the PLACL/collagen nanofibrous scaffold shows varying porosity as detailed in Table 1. Mechanical strength measurements of PLACL and PLACL/collagen nanofibrous scaffolds showed a typical stress–strain curve (Fig. S1, ESI†) and three zones could be distinguished. In the first zone, stress increased non-linearly with strain (up to 5% strain). In the second zone, stress increased linearly with strain (between 5% and 45% strain) and finally in the third zone (after 45% strain) deformation was observed. The modulus of elasticity was determined from the linear part of the curve. Other tensile parameters are given in Table 1. The tensile properties of PLACL/collagen (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) blended nanofibrous scaffolds were lower than that of PLACL. The elastic modulus of the PLACL/collagen nanofibrous scaffolds was comparable to that of PLACL nanofibrous scaffolds.
1) blended nanofibrous scaffolds were lower than that of PLACL. The elastic modulus of the PLACL/collagen nanofibrous scaffolds was comparable to that of PLACL nanofibrous scaffolds.
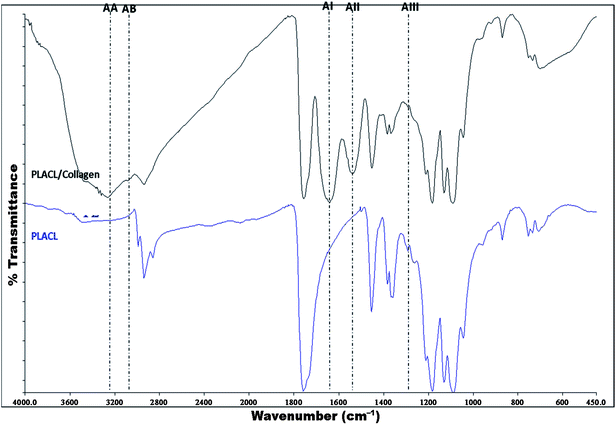 |
| | Fig. 2 FT-IR spectra of PLACL and PLACL/collagen nanofibers with peaks representing specific functional groups (AA: amide A; AB: amide B; AI: amide I, AII: amide II and AIII: amide III). | |
Proliferation of hMSCs
MTS assay results showed that there was a significant increase in hMSC proliferation (p ≤ 0.05) on collagen and PLACL/collagen nanofibrous scaffolds compared to PLACL and TCP after 7 days of culture (Fig. 3). This indicates that the initial cell attachment and cell–scaffold interactions were better on collagen and PLACL/collagen. The cell number increased significantly on all the scaffolds at day 14 and also on day 28 compared to day 7. Additionally, increases within the group appear to be statistically significant between TCP and collagen, and TCP and PLACL/collagen at D7 and D14. SEM analysis of hMSCs seeded on various nanofibrous scaffolds on day 7 reveals effective cell–scaffold interactions and spreading with higher efficiency on collagen and PLACL/collagen compared to PLACL (Fig. 4). Although, higher cell proliferation was observed on PLACL/collagen and collagen scaffolds at day 14 compared to other substrates, no further increase in proliferation was observed at day 28. Interestingly, there was no significant difference in cell growth between PLACL/collagen and collagen scaffolds at different time points. On the other hand, a significant increase in cell number (p ≤ 0.05) was observed on TCP at day 28 compared to day 14. PLACL scaffolds were comparably less supportive for cell attachment and proliferation at various time points.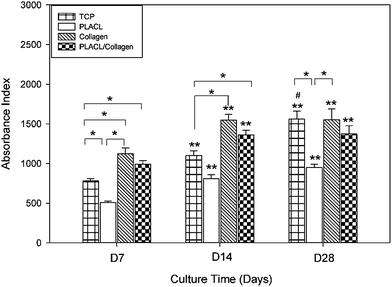 |
| | Fig. 3 MTS assay for hMSCs proliferation on nanofibrous scaffolds of PLACL, collagen, PLACL/collagen and TCP during hepatic differentiation. The differences in cell number within the groups are significant between TCP and collagen, TCP and PLACL/collagen, PLACL and collagen and PLACL and TCP at D7 and D14 (*p ≤ 0.05). The cell numbers in TCP, PLACL, collagen and PLACL/collagen nanofibrous scaffolds show an increase when comparing between D7 vs. D14 and D7 vs. D28 (**p ≤ 0.05). No significant increase in cell number was observed during hepatic maturation on any of nanofibrous scaffolds except TCP at day 28 compared to day 14 (#p ≤ 0.05). | |
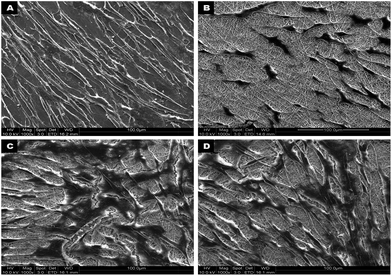 |
| | Fig. 4 FESEM micrographs showing hMSCs adhesion and growth after 7 days of culture on TCP (A), PLACL (B), collagen (C) and PLACL/collagen nanofibrous scaffolds (D) at 1000× magnification. Interaction of hMSCs with nanofibers allowed better adhesion and proliferation on collagen and PLACL/collagen nanofibrous scaffolds. | |
Hepatic trans-differentiation of hMSCs
During hepatic differentiation, it was observed that the spindle-shaped fibroblastic morphology of hMSCs was maintained during initial cell spreading as well as the preconditioning step in the first week. However, the morphology had changed after 7 days of hepatic induction and they possessed broad, flattened morphologies with a high nucleo-cytoplasmic ratio. Upon further maturation with oncostatin M, dexamethasone and ITS premix for two more weeks up to day 28, the differentiated cells acquired cuboidal to polygonal shape (Fig. 5A–C) with accumulations of abundant cytoplasmic granules. On collagen scaffolds, the hMSC-derived hepatocyte-like cells acquired polygonal shapes (Fig. 5C) as of normal hepatocytes, whereas the extent of differentiation was less on PLACL, which showed a flat irregular morphology (Fig. 5B) after 28 days of differentiation. TCP supported minimal differentiation potential with few proliferating hMSCs still retaining the spindle shaped morphology (Fig. 5A). Interestingly, the differentiating hMSCs on PLACL/collagen nanofibrous scaffolds accumulated to form spheroid-like aggregates as happens in hepatocyte primary cultures on 3D culture system.17 The hepatospheres thus formed (5 ± 2 per cm2 area) are evenly distributed (Fig. 5D) and are of different sizes with a range of 10–100 μm diameter (Fig. 5D–F) as assessed by ImageJ analysis software. Such spheroids are associated with higher cellular metabolism, which correlates with the 28 day MTS assay data of the PLACL/collagen nanofibrous scaffolds. The hepatospheres formed on this scaffold very effectively with proper integration with the scaffold matrix, and abundant secretion of ECM components was quite evident (Fig. 5E). Phenotypical changes during hepatic differentiation tracked by CMFDA fluorescent dye expression reveal a noticeable difference in the cellular morphology on the PLACL/collagen nanofibrous scaffold. Until day 7, the hMSCs maintained a spindle-shaped fibroblastic morphology and then acquired a flat, polygonal morphology on day 18, with retracting ends. On day 21 of induction, the differentiating cells started aggregating, only on the PLACL/collagen nanofibrous scaffolds, with a spheroid-like morphology being prominent on day 24. Fully formed hepatospheres were observed on day 28 with more cells accumulating to form spheroids (Fig. 6).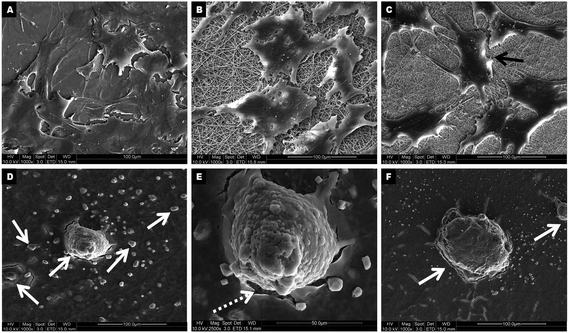 |
| | Fig. 5 FESEM micrographs showing hMSCs-derived hepatocyte-like cells at day 28 on TCP (A), PLACL (B) and collagen (C) as well as hepatospheres of different dimensions on PLACL/collagen nanofibrous scaffolds (D) at 1000× magnification. Representative micrographs showing hepatospheres formed on PLACL/collagen nanofibrous scaffolds are of different size, approximately 50 μm (E) to ∼95 μm diameter (F) at 2500× and 1000× magnification respectively. hMSCs adapted better on PLACL/collagen nanofibrous scaffolds for hepatic trans-differentiation to form hepatospheres. The white arrows in (D and F) represents hepatospheres of different diameters, whereas the white dotted arrow in (E) indicates the excessive ECM proteins secreted by hepatospheres spread over nanofibers. The black arrow in (C) indicates hepatocyte-like cells formed on the collagen nanofibrous scaffolds. | |
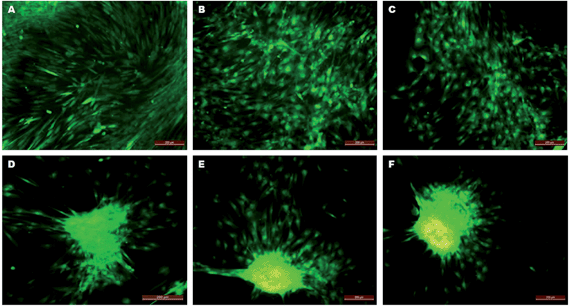 |
| | Fig. 6 CMFDA fluorescent dye expression in trans-differentiating hMSCs on PLACL/collagen nanofibrous scaffolds depicting phenotypic progress in hepatosphere formation. Fluorescent micrographs show an undifferentiated fibroblastic morphology on day 7 (A), a cuboidal morphology on day 14 (B) and day 18 (C), cell migration on day 21 (D), aggregate formation on day 24 (E) and mature hepatosphere formation on day 28 (F) (scale bar = 200 μm). | |
Semi-quantitative and quantitative expression of hepatic genes
Positive expression of AFP, HNF-4α and ALB at days 14 and 28 on various substrates at the mRNA level validates the trans-differentiation of hMSCs towards a hepatic lineage (Fig. 7). Semi-quantitative RT-PCR results reveal that the expression of ALB and HNF-4α was greater on day 28 compared to day 14, whereas AFP expression remained either same or declined after day 14 during the process of differentiation (Fig. 7A). Differentiation on PLACL scaffolds was characterized by the low level of expression of all three hepatic genes on both day 14 and 28 compared to other scaffolds. The untreated MSCs did not express any of hepatic genes. Quantitative real-time PCR analysis of hMSCs differentiated on various scaffolds at day 14 showed a higher level of AFP, HNF-4α and ALB expression on PLACL/collagen nanofibrous scaffolds compared to other substrates (Fig. 7B–D). The expression level of HNF-4α and ALB gradually increased from day 14 to 28, whereas AFP expression declined significantly (p ≤ 0.05) at day 28 compared to day 14 on PLACL/collagen nanofibrous scaffolds (Fig. 7B). However, there was no significant decline in AFP expression (p ≤ 0.05) after day 14 in trans-differentiated hepatocyte-like cells on TCP, PLACL and collagen scaffolds. Also, there was a significant difference (p ≤ 0.05) in hepatic gene expression between hepatocyte-like cells on collagen and hepatospheres on PLACL/collagen at day 28. The quantitative expression of all three genes by hepatospheres formed on PLACL/collagen at day 28 was comparable to that of HepG2 cells.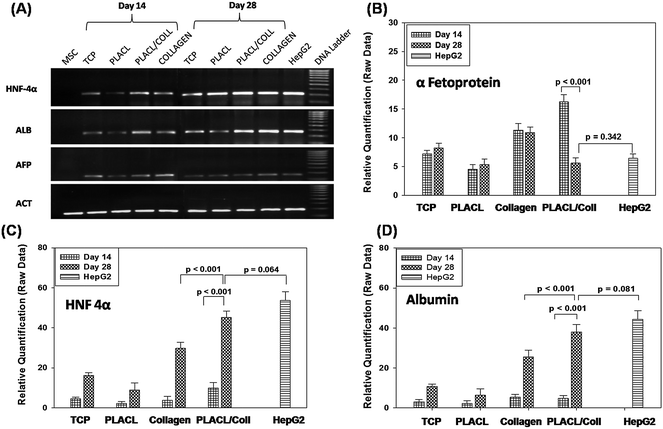 |
| | Fig. 7 Hepatocyte-specific gene expression profile of hMSCs-derived hepatocyte-like cells/hepatospheres by semi-quantitative RT-PCR analysis (A) and relative quantification of α-fetoprotein, HNF-4α and albumin mRNA expression by RT-PCR (B, C and D respectively) on various scaffolds and TCP on days 14 and 28. Untreated hMSCs were used as a negative control, whereas HepG2 cells were used as a positive control for hepatic gene expression. Differences in gene expression between the groups were statistically significant at p ≤ 0.05. | |
Immunofluorescence analysis of hepatic markers
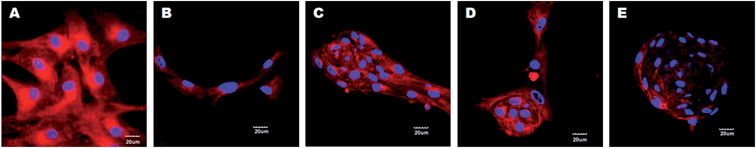
The trans-differentiation efficiency of hMSCs to hepatocytes was further confirmed by immunofluorescence staining for the expression of hepatocyte-specific markers AFP, ALB and CY-18 protein levels at day 28 (Fig. 8). Cells induced to differentiate into hepatocytes on PLACL/collagen and collagen scaffolds showed enhanced fluorescent expression of all three markers compared to cells on PLACL scaffolds and TCP. The three dimensional hepatospheres formed on the PLACL/collagen nanofibrous scaffolds showed a higher expression of albumin and cytokeratin-18, whereas the expression of AFP was down-regulated at day 28. We assessed the expression pattern of AFP on the PLACL/collagen nanofibrous scaffolds to trace its down-regulation with the progress of maturation during hepatosphere formation. On day 14, the expression of AFP was strongest and as maturation progressed, AFP expression declined with hepatosphere formation on day 28 (Fig. 9) which correlates with the quantitative real time gene expression data.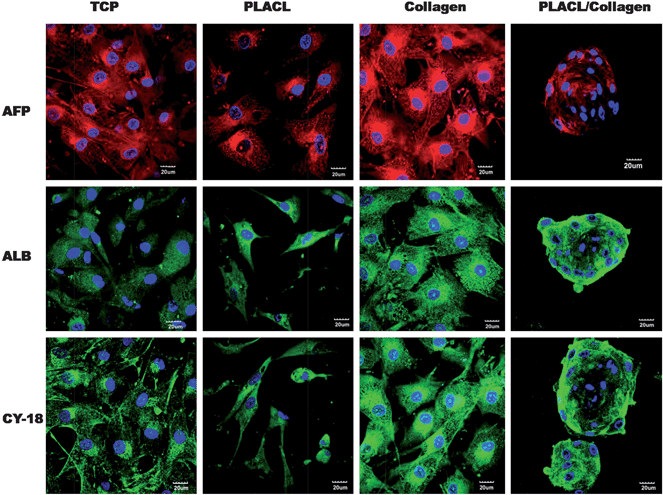 |
| | Fig. 8 Confocal microscopy images (merged) showing expression of hepatocyte specific markers, AFP (Alexa fluor 594: red colour), ALB (Alexa fluor 488: green colour) and CY-18 (FITC: green colour) for hepatocyte-like cells formed on TCP, PLACL and collagen and hepatospheres formed on PLACL/collagen nanofibrous scaffolds at day 28. Nuclei were stained with DAPI (blue colour). Three dimensional hepatospheres on PLACL/collagen nanofibrous scaffolds showed strong expression of albumin and cytokeratin-18 and weak expression of α-fetoprotein (scale bar = 20 μm). | |
 |
| | Fig. 9 Confocal microscopy images (merged) for AFP expression (Alexa Fluor 594: red colour) during hepatosphere formation on PLACL/collagen nanofibrous scaffolds on day 14 (A), day 18 (B), day 21 (C), day 24 (D) and day 28 (E). Nuclei were stained with DAPI (blue colour). A decline in fluorescence intensity for AFP expression was observed on hepatospheres at day 28 (scale bar = 20 μm). | |
Functional assay of the trans-differentiated hepatocytes on nanofibrous scaffolds
The amount of albumin released into the culture media was negligible on day 7 and increased as differentiation progressed. The amount of albumin secretion was significantly higher on day 28 (p ≤ 0.05) on all substrates compared to day 14 and day 21 (Fig. 10). Differences in albumin concentration were observed between TCP vs. collagen and TCP vs. PLACL/collagen at days 14, 21 and 28. Although there was no difference in albumin release from hepatocyte-like cells on collagen and PLACL/collagen at day 21 (p = 0.605), the hepatospheres formed on PLACL/collagen at day 28 released more albumin (p ≤ 0.05) compared to the collagen scaffolds as well as TCP and PLACL. No significant increase in albumin release was observed between day 21 and 28 on TCP, PLACL and collagen scaffolds. The progressive increase in albumin concentration is in accordance with real-time PCR data and is higher on the PLACL/collagen nanofibrous scaffolds on day 28 compared to other time points. Untreated MSCs showed a negligible amount of albumin release. Albumin released from day 28 hepatospheres on PLACL/collagen nanofibrous scaffolds was comparable to that of HepG2 cells (p = 0.081).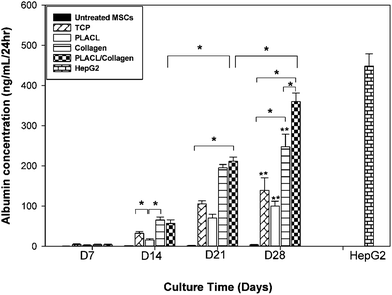 |
| | Fig. 10 Albumin assay for culture media collected during hepatic trans-differentiation of hMSCs on various substrates and TCP at different time points. HepG2 conditioned medium was used as a positive control which was independent of substrate parameters as well as time points. The differences in albumin concentration within the groups were significant between TCP and collagen, TCP and PLACL/collagen, collagen and PLACL/collagen at day 28 (*p ≤ 0.05). No significant increase in albumin release was observed between days 21 and 28 on TCP, PLACL and collagen nanofibrous scaffolds (**p ≤ 0.05). However, hepatospheres on PLACL/collagen nanofibrous scaffolds showed a higher amount of albumin release at day 28 compared to days 14 and 21. | |
Discussion
An ideal strategy for stem cells and nanotechnology based liver tissue engineering would involve suitable biocompatible scaffolds, an effective stem cell source and a hepatomimetic microenvironment. Electrospun nanofibrous scaffolds have proven to be effective in providing nanotopographical signals, mechanical stability, structural guidance and anchorage for cell integration with the surrounding tissues.18 MSCs, due to their easy accessibility and possible trans-differentiation potential towards hepatocytes,9,12 serve as a prospective cell source for liver tissue engineering. Piryaei et al.24 have successfully differentiated mouse bone marrow-derived MSCs into hepatocyte-like cells on ultra-web nanofibers (Ultra-Web™ Synthetic ECM; Donaldson Co., Minneapolis, MN, USA). Kazemnejad et al.22 could derive hepatocyte-like cells from hMSCs on a biocompatible nanofibrous scaffolds. Most of these studies report the generation of hepatocyte-like cells on electrospun nanofibrous scaffolds, but failed to achieve functional 3D hepatic aggregates, similar to those observed with normal hepatocytes on nanofibrous scaffolds. We have reported for the first time the generation of hMSCs-derived functional 3D hepatospheres on electrospun nanofibrous scaffolds.The present study evaluated the hepatic trans-differentiation potential of hMSCs seeded on electrospun nanofibrous scaffolds of a PLACL/collagen blend (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) by sequential induction with hepatogenic growth factors. Bioengineered scaffolds should be sufficiently hydrophilic to allow superior cell adhesion, spreading and infiltration similar to that of the native hepatic microenvironment, which is highly hydrated. Initially, PLACL was strongly hydrophobic, but blending it with collagen impregnated functional groups such as amine, hydroxyl and carboxyl groups to its surface to make it more hydrophilic for liver tissue engineering. FTIR analysis of PLACL/collagen reveals the presence of unique peaks of collagen representing amides A and B as well as amides I, II and III apart from common PLACL peaks (Fig. 2). This implies that blending a collagen component with PLACL enhances the bioactivity of the composite scaffolds, as it greatly favours cell adhesion and proliferation. Porosity of the scaffold is an important parameter for hepatic tissue engineering and PLACL/collagen nanofibrous scaffolds with porosities as high as 89% with maximum pore distribution allows proper cell attachment and infiltration, easy flow of nutrients, metabolites, hepatogenic growth factors and waste products in and out of cells, which is desirable for a liver tissue construct. The tensile properties of the nanofibrous scaffolds are an important attribute, while considering its in vivo implantation as a hepatic substitute. The mechanical stability of the substrate plays an important role, as the scaffold guides the growth, proliferation and differentiation of stem cells. Low stiffness may be a desirable property of the scaffolds with regard to integration with native tissue, since an extremely stiff matrix might hinder the proliferation and differentiation potential of stem cells. Ideally, less stiff and biodegradable scaffolds such as PLACL and collagen are better choices for generating tissue engineered hepatic constructs in vitro. In the present study, PLACL/collagen (2
1) by sequential induction with hepatogenic growth factors. Bioengineered scaffolds should be sufficiently hydrophilic to allow superior cell adhesion, spreading and infiltration similar to that of the native hepatic microenvironment, which is highly hydrated. Initially, PLACL was strongly hydrophobic, but blending it with collagen impregnated functional groups such as amine, hydroxyl and carboxyl groups to its surface to make it more hydrophilic for liver tissue engineering. FTIR analysis of PLACL/collagen reveals the presence of unique peaks of collagen representing amides A and B as well as amides I, II and III apart from common PLACL peaks (Fig. 2). This implies that blending a collagen component with PLACL enhances the bioactivity of the composite scaffolds, as it greatly favours cell adhesion and proliferation. Porosity of the scaffold is an important parameter for hepatic tissue engineering and PLACL/collagen nanofibrous scaffolds with porosities as high as 89% with maximum pore distribution allows proper cell attachment and infiltration, easy flow of nutrients, metabolites, hepatogenic growth factors and waste products in and out of cells, which is desirable for a liver tissue construct. The tensile properties of the nanofibrous scaffolds are an important attribute, while considering its in vivo implantation as a hepatic substitute. The mechanical stability of the substrate plays an important role, as the scaffold guides the growth, proliferation and differentiation of stem cells. Low stiffness may be a desirable property of the scaffolds with regard to integration with native tissue, since an extremely stiff matrix might hinder the proliferation and differentiation potential of stem cells. Ideally, less stiff and biodegradable scaffolds such as PLACL and collagen are better choices for generating tissue engineered hepatic constructs in vitro. In the present study, PLACL/collagen (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) scaffolds possess adequate tensile properties to withstand a stress of 0.52 ± 0.12 MPa and a strain of 50.29 ± 19.28% at maximum load (Table 1). The elastic modulus, which is a measure of resistance to deformation (stiffness), was around 0.88 MPa for the PLACL/collagen nanofibrous scaffolds, whereas that of normal liver tissue is below 10 kPa.28 Therefore, for engineering soft tissues like liver, the stiffness needs to be controlled to guide effective differentiation of hMSCs on scaffolds. PLACL/collagen nanofibrous scaffolds, being highly porous and biodegradable, might be effective in maintaining low stiffness upon implantation, which needs further assessment.
1) scaffolds possess adequate tensile properties to withstand a stress of 0.52 ± 0.12 MPa and a strain of 50.29 ± 19.28% at maximum load (Table 1). The elastic modulus, which is a measure of resistance to deformation (stiffness), was around 0.88 MPa for the PLACL/collagen nanofibrous scaffolds, whereas that of normal liver tissue is below 10 kPa.28 Therefore, for engineering soft tissues like liver, the stiffness needs to be controlled to guide effective differentiation of hMSCs on scaffolds. PLACL/collagen nanofibrous scaffolds, being highly porous and biodegradable, might be effective in maintaining low stiffness upon implantation, which needs further assessment.
With regard to initial cell adhesion and proliferation, the PLACL/collagen and collagen scaffolds provided a supportive microenvironment and there was an increase in cell number at day 7 as well as at increasing time points. Since the hMSCs adapted better to the scaffolds, cell number increased at day 14 and also on day 28 compared to day 7. The increase in cell number within the group between TCP and collagen, and TCP and PLACL/collagen at D7 and D14 may be due to the initial cell adhesion to the extracellular matrix (ECM) that is mediated by an integrin transmembrane receptor present in the collagen component.20 However, since the cells started differentiating, no significant increase in cell number was observed between day 14 and 28 cultures (p ≤ 0.05) on collagen and PLACL/collagen nanofibrous scaffolds. The increase in cell number from day 14 to 28 on TCP shows that the hMSCs still retained stemness and continued proliferation up to day 28 (Fig. 3). This implies that TCP was not supportive enough for differentiation. No significant difference in cell growth between the PLACL/collagen and collagen scaffolds at different time points implies that the PLACL/collagen bio-synthetic hybrid scaffolds were supportive enough for cell attachment, proliferation and differentiation to mimic natural polymers.
Hepatogenesis of hMSCs was achieved successfully on various nanofibrous scaffolds. On day 28, the cells attained complete differentiation on various scaffolds as evaluated by the morphological changes, hepatocyte-specific gene expression, immunofluorescence studies of hepatic markers and albumin release profiles. The extent of hepatogenesis is maximal on PLACL/collagen nanofibrous scaffolds compared to other nanofibrous scaffolds and TCP on day 28 and most importantly they could form functional hepatospheres, as would normal hepatocytes on nanofibrous scaffolds.17 The type of scaffold being employed for cell-based tissue engineering has a substantial effect upon the morphology of the cells.29 Hepatocytes are anchorage-dependent and their interactions with the ECM nanofibrous scaffolds induced them to attach and proliferate to form spheroids.30 Physical signals exerted by the nanofibers in terms of cell–substrate adhesion strength (Fig. 4) are known to induce directional migration as well as aggregate formation (Fig. 6) of hepatocytes.17,31 Formation of hepatospheres only on the PLACL/collagen-blended scaffolds and not on other nanofibrous scaffolds of PLACL or collagen (Fig. 5) may be due to the suitable fiber diameter, porosity, long-term stability of the cell–matrix interactions and guided trans-differentiation. PLACL scaffolds are not supportive enough for initial cell adhesion and hence differentiation. Although collagen nanofibrous scaffolds possess the best features to allow cell proliferation and hepatic differentiation, its weak mechanical profile does not support hepatosphere formation, rather forming flat hepatocyte-like cells (Fig. 5C). Hepatocyte spheroids are 3D aggregates that exhibit a high degree of cell–cell contact, biomatrix deposition and hence long-term maintenance with enhanced liver function.32,33 Cells in spheroids also have a morphology and ultra-structure similar to those found in a native liver lobule.34 The hMSCs-derived hepatospheres evidently showed superior hepatic features such as increased expression of mature hepatic genes and proteins (HNF-4α and ALB) and albumin secretion in culture media, compared to the hepatocyte-like cells formed on other nanofibrous scaffolds and TCP. The quantitative expression of all hepatic genes in hepatospheres was comparable to that of HepG2 cells, which implies that hepatospheres are functional entities. Although the number of positive cells was not countable due to technical difficulties with the nanofibers, apparently most of the cells contributed towards hepatosphere formation, as demonstrated by SEM (Fig. 5D) and immunofluorescence studies (Fig. 8). However, it is yet to be ascertained whether the majority of the cells within hepatospheres are indeed mature functional hepatocytes. Expression of the AFP gene as well as proteins declined on day 28 in the hepatosphere as compared to hepatocyte-like cells on day 14 (Fig. 7B and 9), which is a feature of differentiating hepatocytes which happens during normal liver development.35 This implies that the PLACL/collagen nanofibrous scaffolds support the hepatocyte differentiation process. Another important feature for appropriate hepatic functionality is the size of the hepatosphere; if it is below 100 μm, the cells in the interior of the spheroids do not become hypoxic.13,36 The hMSC-derived hepatospheres formed on the PLACL/collagen nanofibrous scaffolds are in the range 10–100 μm and hence can be appropriate hepatic constructs for further studies on in vivo liver tissue engineering. Also, hepatocytes grown on 2D monolayers tend to de-differentiate rapidly losing their liver specific functions37 and hence a 3D spheroid culture is suggested to be effective in preventing de-differentiation.38 Hepatospheres derived from hMSC trans-differentiation in this study show appropriate functional attributes such as albumin secretion and 3D spheroid formation. Due to the unique biodegradation profiles of the PLACL and collagen components of the biocomposite PLACL/collagen nanofibrous scaffolds, such bioengineered hepatic construct is expected to integrate easily with the host tissue upon implantation, which can be confirmed only after in vivo animal studies. However, the central mechanism elucidating the possible role of nanotopographical cues in guiding cellular remodelling and migration to form hepatospheres needs to be investigated further at a molecular level, particularly the formation of focal adhesions of hMSCs by nanofibers. It is known that the nanofibrous scaffolds due to its topographical stimuli promotes appropriate adhesion and structural remodelling to the growing hMSCs by forming focal adhesions39 and provides a tissue mimicking microenvironment. However, the role of soluble growth factors is more critical in augmenting hepatic trans-differentiation of hMSCs. Therefore, the synergistic effects of the biomaterial-based nanofibers (physical cues) and the optimized chemical soluble factors (humoral cues) improved the efficiency of trans-differentiation of hMSCs in this study. Recent studies suggest that the orientations or diameters of the nanofibers40 and surface charges41 are also crucial factors for regulating the function of cells resulting in controlled cell shapes. The observed results from the PLACL/collagen nanofibers with random orientation and average dimensions at a nano- to microscale (mostly of 200–600 nm diameter), promotes adhesion, migration and spheroid formation during hepatic trans-differentiation of hMSCs.
Conclusion
A tissue-engineered strategy for hepatic regeneration was the goal of the study, wherein hMSCs could be modulated to form functional hepatospheres on PLACL/collagen (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 blend) nanofibrous scaffolds with greater efficacy. The present study confirms the PLACL/collagen nanofibrous scaffolds augments trans-differentiation of hMSCs towards the generation of 3D hepatic constructs. The superior hepatic trans-differentiation potential of hMSCs on this blended biosynthetic composite may be attributed to the biophysical signals provided by the nanostructures and the mechanism involved needs further evaluation. Such an in vitro bioengineered hepatic construct might be a potential tool for future applications in hepatotoxicity testing, cellular therapy for end-stage liver failure and the development of tissue-engineered bioartificial liver, after addressing the long term maintenance of functionality in animal studies.
1 blend) nanofibrous scaffolds with greater efficacy. The present study confirms the PLACL/collagen nanofibrous scaffolds augments trans-differentiation of hMSCs towards the generation of 3D hepatic constructs. The superior hepatic trans-differentiation potential of hMSCs on this blended biosynthetic composite may be attributed to the biophysical signals provided by the nanostructures and the mechanism involved needs further evaluation. Such an in vitro bioengineered hepatic construct might be a potential tool for future applications in hepatotoxicity testing, cellular therapy for end-stage liver failure and the development of tissue-engineered bioartificial liver, after addressing the long term maintenance of functionality in animal studies.Acknowledgements
This study was supported by Frontier Lifeline Hospital, Chennai, India, NRF-Technion project, Nanoscience and Nanotechnology Initiative, Faculty of Engineering, National University of Singapore.Notes and references
- J. P. Iredale, Br. Med. J., 2003, 327, 143–147 CrossRef.
- J. Cai, Y. Zhao, Y. Liu, F. Ye, Z. Song, H. Qin, S. Meng, Y. Chen, R. Zhou, X. Song, Y. Guo, M. Ding and H. Deng, Hepatology, 2007, 45, 1229–1239 CrossRef CAS.
- S. Khurana, A. K. Jaiswal and A. Mukhopadhyay, J. Biol. Chem., 2009, 285, 4725–4731 CrossRef.
- G. J. Sullivan, D. C. Hay, I. H. Park, J. Fletcher, Z. Hannoun, C. M. Payne, D. Dalgetty, J. R. Black, J. A. Ross, K. Samuel, G. Wang, G. Q. Daley, J. H. Lee, G. M. Church, S. J. Forbes, J. P. Iredale and I. Wilmut, Hepatology, 2010, 51, 329–335 CrossRef CAS.
- H. Aurich, M. Sgodda, P. Kaltwasser, M. Vetter, A. Weise, T. Liehr, M. Brulport, J. G. Hengstler, M. M. Dollinger, W. E. Fleig and B. Christ, Gut, 2008, 58, 570–581 CrossRef.
- K. Nonome, X. K. Li, T. Takahara, Y. Kitazawa, N. Funeshima, Y. Yata, F. Xue, M. Kanayama, E. Shinno, C. Kuwae, S. Saito, A. Watanabe and T. Sugiyama, Am. J. Physiol.: Gastrointest. Liver Physiol., 2005, 289, G1091–G1099 CrossRef CAS.
- A. McLaren, Nature, 2001, 414, 129–131 CrossRef CAS.
- B. Blum and N. Benvenisty, in Advances in Cancer Research, ed. F. V. W. George and K. George, Academic Press, 2008, pp. 133–158 Search PubMed.
- K. D. Lee, T. K. C. Kuo, J. Whang-Peng, Y. F. Chung, C. T. Lin, S. H. Chou, J. R. Chen, Y. P. Chen and O. K. S. Lee, Hepatology, 2004, 40, 1275–1284 CrossRef CAS.
- S. Snykers, J. Kock, V. Tamara and V. Rogiers, Methods Mol. Biol., 2011, 698, 305–314 CAS.
- D. E. Discher, D. J. Mooney and P. W. Zandstra, Science, 2009, 324, 1673–1677 CrossRef CAS.
- R. E. Schwartz, M. Reyes, L. Koodie, Y. Jiang, M. Blackstad, T. Lund, T. Lenvik, S. Johnson, W. S. Hu and C. M. Verfaillie, J. Clin. Invest., 2002, 109, 1291–1302 CAS.
- R. Glicklis, L. Shapiro, R. Agbaria, J. C. Merchuk and S. Cohen, Biotechnol. Bioeng., 2000, 67, 344–353 CrossRef CAS.
- S. Y. Ong, H. Dai and K. W. Leong, Biomaterials, 2006, 27, 4087–4097 CrossRef CAS.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS.
- Z. Q. Feng, X. Chu, N. P. Huang, T. Wang, Y. Wang, X. Shi, Y. Ding and Z. Z. Gu, Biomaterials, 2009, 30, 2753–2763 CrossRef CAS.
- Z. Q. Feng, X. H. Chu, N. P. Huang, M. K. Leach, G. Wang, Y. C. Wang, Y. T. Ding and Z. Z. Gu, Biomaterials, 2010, 31, 3604–3612 CrossRef CAS.
- Q. P. Pham, U. Sharma and A. G. Mikos, Tissue Eng., 2006, 12, 1197–1211 CrossRef CAS.
- Y. Lemmouchi and E. Schacht, J. Controlled Release, 1997, 45, 227–233 CrossRef CAS.
- M. P. Prabhakaran, J. R. Venugopal and S. Ramakrishna, Biomaterials, 2009, 30, 4996–5003 CrossRef CAS.
- S. Mukherjee, J. Reddy Venugopal, R. Ravichandran, S. Ramakrishna and M. Raghunath, Adv. Funct. Mater., 2011, 21, 2291–2300 CrossRef CAS.
- S. Kazemnejad, A. Allameh, M. Soleimani, A. Gharehbaghian, Y. Mohammadi, N. Amirizadeh and M. Jazayery, J. Gastroenterol. Hepatol., 2009, 24, 278–287 CrossRef CAS.
- S. M. Hashemi, M. Soleimani, S. S. Zargarian, V. Haddadi-Asl, N. Ahmadbeigi, S. Soudi, Y. Gheisari, A. Hajarizadeh and Y. Mohammadi, Cells Tissues Organs, 2009, 190, 135–149 CrossRef CAS.
- A. Piryaei, M. Valojerdi, M. Shahsavani and H. Baharvand, Stem Cell Rev. Rep., 2010, 7, 103–118 CrossRef.
- Z. Farzaneh, B. Pournasr, M. Ebrahimi, N. Aghdami and H. Baharvand, Stem Cell Rev. Rep., 2010, 6, 601–610 CrossRef CAS.
- M. Ghaedi, M. Soleimani, I. Shabani, Y. Duan and A. Lotfi, Cell. Mol. Biol. Lett., 2011, 17, 89–106 CrossRef.
- W. He, Z. Ma, T. Yong, W. E. Teo and S. Ramakrishna, Biomaterials, 2005, 26, 7606–7615 CrossRef CAS.
- W. C. Yeh, P. C. Li, Y. M. Jeng, H. C. Hsu, P. L. Kuo, M. L. Li, P. M. Yang and P. H. Lee, Ultrasound Med. Biol., 2002, 28, 467–474 CrossRef.
- G. Catapano, L. De Bartolo, V. Vico and L. Ambrosio, Biomaterials, 2001, 22, 659–665 CrossRef CAS.
- E. L. LeCluyse, P. L. Bullock and A. Parkinson, Adv. Drug Delivery Rev., 1996, 22, 133–186 CrossRef CAS.
- M. J. Powers, R. E. Rodriguez and L. G. Griffith, Biotechnol. Bioeng., 1997, 53, 415–426 CrossRef CAS.
- J. Landry, D. Bernier, C. Ouellet, R. Goyette and N. Marceau, J. Cell Biol., 1985, 101, 914–923 CrossRef CAS.
- S. F. Abu-Absi, J. R. Friend, L. K. Hansen and W.-S. Hu, Exp. Cell Res., 2002, 274, 56–67 CrossRef CAS.
- A. Lazar, M. V. Peshwa, F. J. Wu, C. M. Chi, F. B. Cerra and W. S. Hu, Cell Transplant., 1995, 4, 259–268 CrossRef CAS.
- N. Shiojiri, J. M. Lemire and N. Fausto, Cancer Res., 1991, 51, 2611–2620 CAS.
- J. E. Babensee, J. M. Anderson, L. V. McIntire and A. G. Mikos, Adv. Drug Delivery Rev., 1998, 33, 111–139 CrossRef CAS.
- Y. A. Wen, D. Liu, Y. Y. Xiao, D. Luo, Y. F. Dong and L. P. Zhang, Toxicol. in Vitro, 2009, 23, 744–747 CrossRef CAS.
- K. S. Vasanthan, A. Subramanian, U. M. Krishnan and S. Sethuraman, Biotechnol. Adv., 2012, 30, 742–752 CrossRef CAS.
- Y. R. Shih, C. N. Chen, S. W. Tsai, Y. J. Wang and O. K. Lee, Stem Cells, 2006, 24, 2391–2397 CrossRef CAS.
- M. Chen, P. K. Patra, S. B. Warner and S. Bhowmick, Role of fiber diameter in adhesion and proliferation of NIH 3T3 fibroblast on electrospun polycaprolactone scaffolds, Tissue Eng., 2007, 13, 579–587 CrossRef CAS.
- J. Kim, D. H. Kim, K. T. Lim, H. Seonwoo, S. H. Park, Y. R. Kim, Y. Kim, Y. H. Choung, P. H. Choung and J. H. Chung, Charged nanomatrices as efficient platforms for modulating cell adhesion and shape, Tissue Eng., Part C, 2012, 18, 913–923 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Graphical image showing stress–strain curve of PLACL/collagen nanofibrous scaffolds. See DOI: 10.1039/c3tb20241k |
|
| This journal is © The Royal Society of Chemistry 2013 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were fabricated by electrospinning and also evaluated for fiber morphology, surface wettability, functional groups, porosity and tensile properties. Hepatic trans-differentiation of human bone marrow-derived MSCs (hMSCs) was carried out on these scaffolds over a period of 28 days using sequential induction with hepatogenic growth factors. Hepatogenesis was confirmed by scanning electron microscopy (SEM), cell phenotype tracking dye expression, quantitative expression of hepatic genes, immunofluorescence staining of hepatocyte-specific markers and albumin release. The results proved that the porous PLACL/collagen nanofibrous scaffold supported enhanced hMSC proliferation and hepatic trans-differentiation compared to individual PLACL and collagen scaffolds as well as a monolayer culture on tissue culture plate (TCP). Interestingly, hMSC-derived hepatocyte-like cells on PLACL/collagen nanofibrous scaffolds could aggregate to form functional ‘hepatospheres’ similar to normal hepatic spheroids. The present study concludes that PLACL/collagen nanofibrous scaffolds are potentially biomimetic and upon sequential induction with hepatogenic growth factors/cytokines, it augments trans-differentiation of hMSCs towards functional hepatosphere formation. Such bioengineered nanofibrous scaffold hepatic construct provides a promising approach for cellular therapy of damaged livers in end-stage liver failure treatments.
1) were fabricated by electrospinning and also evaluated for fiber morphology, surface wettability, functional groups, porosity and tensile properties. Hepatic trans-differentiation of human bone marrow-derived MSCs (hMSCs) was carried out on these scaffolds over a period of 28 days using sequential induction with hepatogenic growth factors. Hepatogenesis was confirmed by scanning electron microscopy (SEM), cell phenotype tracking dye expression, quantitative expression of hepatic genes, immunofluorescence staining of hepatocyte-specific markers and albumin release. The results proved that the porous PLACL/collagen nanofibrous scaffold supported enhanced hMSC proliferation and hepatic trans-differentiation compared to individual PLACL and collagen scaffolds as well as a monolayer culture on tissue culture plate (TCP). Interestingly, hMSC-derived hepatocyte-like cells on PLACL/collagen nanofibrous scaffolds could aggregate to form functional ‘hepatospheres’ similar to normal hepatic spheroids. The present study concludes that PLACL/collagen nanofibrous scaffolds are potentially biomimetic and upon sequential induction with hepatogenic growth factors/cytokines, it augments trans-differentiation of hMSCs towards functional hepatosphere formation. Such bioengineered nanofibrous scaffold hepatic construct provides a promising approach for cellular therapy of damaged livers in end-stage liver failure treatments.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) by inducing hepatic trans-differentiation of human bone marrow MSCs. This study focused on assessing the impact of 3D biosynthetic nanofibrous scaffolds from a PLACL/collagen blend upon hepatic trans-differentiation of hMSCs as compared to individual PLACL and collagen nanofibrous scaffolds as well as a conventional monolayer culture system on tissue culture plate (TCP).
1) by inducing hepatic trans-differentiation of human bone marrow MSCs. This study focused on assessing the impact of 3D biosynthetic nanofibrous scaffolds from a PLACL/collagen blend upon hepatic trans-differentiation of hMSCs as compared to individual PLACL and collagen nanofibrous scaffolds as well as a conventional monolayer culture system on tissue culture plate (TCP).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, Mw 150 kDa) was obtained from Boehringer Ingelheim Pharma, GmbH & Co., Ingelheim, Germany. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), antibiotics and trypsin–EDTA, CMFDA (5-chloromethylfluorescein diacetate), secondary antibody Alexa Fluor 488 and 594, Iscove's modified Dulbecco's medium (IMDM), TRIzol™ reagent, SuperScript™ first strand synthesis system and 4′-6-diamidino-2-phenylindole (DAPI) were procured from Invitrogen (Life Technologies, Carlsbad, CA, USA). Bovine serum albumin (BSA), 1,1,1,3,3,3-hexafluor-2-propanol (HFP), anti-human anti-albumin antibody raised in goat, anti-CY-18, anti-AFP, hexamethyldisilazane (HMDS), hepatocyte growth factor (HGF), oncostatin M (OSM), insulin-transferring selenium premix (ITS), dexamethasone, nicotinamide, epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) were from Sigma-Aldrich (St. Louis, MO, USA). Collagen type I was obtained from Koken Company Ltd. (Tokushima-ku, Tokyo, Japan). SYBR green real-time PCR mastermix was purchased from Applied Biosystems (Foster City, CA, USA).
30, Mw 150 kDa) was obtained from Boehringer Ingelheim Pharma, GmbH & Co., Ingelheim, Germany. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), antibiotics and trypsin–EDTA, CMFDA (5-chloromethylfluorescein diacetate), secondary antibody Alexa Fluor 488 and 594, Iscove's modified Dulbecco's medium (IMDM), TRIzol™ reagent, SuperScript™ first strand synthesis system and 4′-6-diamidino-2-phenylindole (DAPI) were procured from Invitrogen (Life Technologies, Carlsbad, CA, USA). Bovine serum albumin (BSA), 1,1,1,3,3,3-hexafluor-2-propanol (HFP), anti-human anti-albumin antibody raised in goat, anti-CY-18, anti-AFP, hexamethyldisilazane (HMDS), hepatocyte growth factor (HGF), oncostatin M (OSM), insulin-transferring selenium premix (ITS), dexamethasone, nicotinamide, epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) were from Sigma-Aldrich (St. Louis, MO, USA). Collagen type I was obtained from Koken Company Ltd. (Tokushima-ku, Tokyo, Japan). SYBR green real-time PCR mastermix was purchased from Applied Biosystems (Foster City, CA, USA).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w ratio) were prepared by dissolving them in HFP. For electrospinning, the respective polymer solutions were fed into a 3 ml standard syringe attached to a blunted stainless steel needle (22½ G) using a syringe pump (KD-100, KD Scientific Inc., Holliston, MA, USA) at an optimized flow rate of 1 ml h−1. On application of a high voltage (Gamma High Voltage Research, Ormond Beach, FL, USA) of 18.5 kV, the polymer solution was spun as nanofibers of random orientation, which were collected on coverslips (diameter, 15 mm; thickness, 0.13–0.16 mm, Menzel-Glaser, Thermo Scientific, Germany). The ambient conditions of the spinning apparatus were controlled with a vacuum dehumidifier machine (KNF Neuberger Inc, Trenton, NJ, USA). The nanofiber mats spun on the cover slips were dried overnight in vacuum to remove the residual solvents and subsequently used for characterization and cell culture experiments.
1 w/w ratio) were prepared by dissolving them in HFP. For electrospinning, the respective polymer solutions were fed into a 3 ml standard syringe attached to a blunted stainless steel needle (22½ G) using a syringe pump (KD-100, KD Scientific Inc., Holliston, MA, USA) at an optimized flow rate of 1 ml h−1. On application of a high voltage (Gamma High Voltage Research, Ormond Beach, FL, USA) of 18.5 kV, the polymer solution was spun as nanofibers of random orientation, which were collected on coverslips (diameter, 15 mm; thickness, 0.13–0.16 mm, Menzel-Glaser, Thermo Scientific, Germany). The ambient conditions of the spinning apparatus were controlled with a vacuum dehumidifier machine (KNF Neuberger Inc, Trenton, NJ, USA). The nanofiber mats spun on the cover slips were dried overnight in vacuum to remove the residual solvents and subsequently used for characterization and cell culture experiments.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio mixture of MTS reagent and serum-free DMEM medium. After the incubation period, the samples were pipetted out into 96-well plates and the absorbance was measured at 490 nm using a microplate reader (Fluostar Optima, BMG Lab Technologies, Germany).
5 ratio mixture of MTS reagent and serum-free DMEM medium. After the incubation period, the samples were pipetted out into 96-well plates and the absorbance was measured at 490 nm using a microplate reader (Fluostar Optima, BMG Lab Technologies, Germany).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) overnight at 4 °C. The cells were washed thrice and incubated with Alexa Fluor 488 and 594 (1
100) overnight at 4 °C. The cells were washed thrice and incubated with Alexa Fluor 488 and 594 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) for 1 h and the nuclei were stained with DAPI (0.5 μg ml−1) for another 30 min. The cells were finally washed and mounted on a glass slide with Vectashield (Vector Laboratories, Burlingame, CA, USA) and fluorescent marker expression was observed under a laser confocal scanning microscope (Olympus FV1000).
250) for 1 h and the nuclei were stained with DAPI (0.5 μg ml−1) for another 30 min. The cells were finally washed and mounted on a glass slide with Vectashield (Vector Laboratories, Burlingame, CA, USA) and fluorescent marker expression was observed under a laser confocal scanning microscope (Olympus FV1000).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) blended nanofibrous scaffolds were lower than that of PLACL. The elastic modulus of the PLACL/collagen nanofibrous scaffolds was comparable to that of PLACL nanofibrous scaffolds.
1) blended nanofibrous scaffolds were lower than that of PLACL. The elastic modulus of the PLACL/collagen nanofibrous scaffolds was comparable to that of PLACL nanofibrous scaffolds.








![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) by sequential induction with hepatogenic growth factors. Bioengineered scaffolds should be sufficiently hydrophilic to allow superior cell adhesion, spreading and infiltration similar to that of the native hepatic microenvironment, which is highly hydrated. Initially, PLACL was strongly hydrophobic, but blending it with collagen impregnated functional groups such as amine, hydroxyl and carboxyl groups to its surface to make it more hydrophilic for liver tissue engineering. FTIR analysis of PLACL/collagen reveals the presence of unique peaks of collagen representing amides A and B as well as amides I, II and III apart from common PLACL peaks (Fig. 2). This implies that blending a collagen component with PLACL enhances the bioactivity of the composite scaffolds, as it greatly favours cell adhesion and proliferation. Porosity of the scaffold is an important parameter for hepatic tissue engineering and PLACL/collagen nanofibrous scaffolds with porosities as high as 89% with maximum pore distribution allows proper cell attachment and infiltration, easy flow of nutrients, metabolites, hepatogenic growth factors and waste products in and out of cells, which is desirable for a liver tissue construct. The tensile properties of the nanofibrous scaffolds are an important attribute, while considering its in vivo implantation as a hepatic substitute. The mechanical stability of the substrate plays an important role, as the scaffold guides the growth, proliferation and differentiation of stem cells. Low stiffness may be a desirable property of the scaffolds with regard to integration with native tissue, since an extremely stiff matrix might hinder the proliferation and differentiation potential of stem cells. Ideally, less stiff and biodegradable scaffolds such as PLACL and collagen are better choices for generating tissue engineered hepatic constructs in vitro. In the present study, PLACL/collagen (2
1) by sequential induction with hepatogenic growth factors. Bioengineered scaffolds should be sufficiently hydrophilic to allow superior cell adhesion, spreading and infiltration similar to that of the native hepatic microenvironment, which is highly hydrated. Initially, PLACL was strongly hydrophobic, but blending it with collagen impregnated functional groups such as amine, hydroxyl and carboxyl groups to its surface to make it more hydrophilic for liver tissue engineering. FTIR analysis of PLACL/collagen reveals the presence of unique peaks of collagen representing amides A and B as well as amides I, II and III apart from common PLACL peaks (Fig. 2). This implies that blending a collagen component with PLACL enhances the bioactivity of the composite scaffolds, as it greatly favours cell adhesion and proliferation. Porosity of the scaffold is an important parameter for hepatic tissue engineering and PLACL/collagen nanofibrous scaffolds with porosities as high as 89% with maximum pore distribution allows proper cell attachment and infiltration, easy flow of nutrients, metabolites, hepatogenic growth factors and waste products in and out of cells, which is desirable for a liver tissue construct. The tensile properties of the nanofibrous scaffolds are an important attribute, while considering its in vivo implantation as a hepatic substitute. The mechanical stability of the substrate plays an important role, as the scaffold guides the growth, proliferation and differentiation of stem cells. Low stiffness may be a desirable property of the scaffolds with regard to integration with native tissue, since an extremely stiff matrix might hinder the proliferation and differentiation potential of stem cells. Ideally, less stiff and biodegradable scaffolds such as PLACL and collagen are better choices for generating tissue engineered hepatic constructs in vitro. In the present study, PLACL/collagen (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) scaffolds possess adequate tensile properties to withstand a stress of 0.52 ± 0.12 MPa and a strain of 50.29 ± 19.28% at maximum load (Table 1). The elastic modulus, which is a measure of resistance to deformation (stiffness), was around 0.88 MPa for the PLACL/collagen nanofibrous scaffolds, whereas that of normal liver tissue is below 10 kPa.28 Therefore, for engineering soft tissues like liver, the stiffness needs to be controlled to guide effective differentiation of hMSCs on scaffolds. PLACL/collagen nanofibrous scaffolds, being highly porous and biodegradable, might be effective in maintaining low stiffness upon implantation, which needs further assessment.
1) scaffolds possess adequate tensile properties to withstand a stress of 0.52 ± 0.12 MPa and a strain of 50.29 ± 19.28% at maximum load (Table 1). The elastic modulus, which is a measure of resistance to deformation (stiffness), was around 0.88 MPa for the PLACL/collagen nanofibrous scaffolds, whereas that of normal liver tissue is below 10 kPa.28 Therefore, for engineering soft tissues like liver, the stiffness needs to be controlled to guide effective differentiation of hMSCs on scaffolds. PLACL/collagen nanofibrous scaffolds, being highly porous and biodegradable, might be effective in maintaining low stiffness upon implantation, which needs further assessment.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 blend) nanofibrous scaffolds with greater efficacy. The present study confirms the PLACL/collagen nanofibrous scaffolds augments trans-differentiation of hMSCs towards the generation of 3D hepatic constructs. The superior hepatic trans-differentiation potential of hMSCs on this blended biosynthetic composite may be attributed to the biophysical signals provided by the nanostructures and the mechanism involved needs further evaluation. Such an in vitro bioengineered hepatic construct might be a potential tool for future applications in hepatotoxicity testing, cellular therapy for end-stage liver failure and the development of tissue-engineered bioartificial liver, after addressing the long term maintenance of functionality in animal studies.
1 blend) nanofibrous scaffolds with greater efficacy. The present study confirms the PLACL/collagen nanofibrous scaffolds augments trans-differentiation of hMSCs towards the generation of 3D hepatic constructs. The superior hepatic trans-differentiation potential of hMSCs on this blended biosynthetic composite may be attributed to the biophysical signals provided by the nanostructures and the mechanism involved needs further evaluation. Such an in vitro bioengineered hepatic construct might be a potential tool for future applications in hepatotoxicity testing, cellular therapy for end-stage liver failure and the development of tissue-engineered bioartificial liver, after addressing the long term maintenance of functionality in animal studies.