 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembly of three bacterially-derived bioactive lipopeptides†
Ian W. Hamley*a,
Ashkan Dehsorkhia,
Paula Jauregia,
Jani Seitsonenb,
Janne Ruokolainenb,
François Couttec,
Gabrielle Chataignéc and
Philippe Jacquesc
aSchool of Chemistry, Pharmacy and Food Biosciences, University of Reading, Whiteknights, Reading, RG6 6AD, UK. E-mail: I.W.Hamley@reading.ac.uk
bDepartment of Applied Physics, Aalto University School of Science, P.O. Box 15100, FI-00076 Aalto, Finland
cProBioGEM, Laboratoire de Procédés Biologiques, Génie Enzymatique et Microbien, UPRES EA 1026, Polytech'Lille, IUTA, Université Lille 1, Sciences et Technologies, F-59655 Villeneuve d'Ascq Cedex, France
First published on 21st August 2013
Abstract
The self-assembly in aqueous solution of three lipopeptides obtained from Bacillus subtilis has been investigated. The lipopeptides surfactin, plipastatin and mycosubtilin contain distinct cyclic peptide headgroups as well as differences in alkyl chain length, branching and chain length distribution. Cryogenic transmission electron microscopy and X-ray scattering reveal that surfactin and plipastatin aggregate into 2 nm-radius spherical micelles, whereas in complete contrast mycosubtilin self-assembles into extended nanotapes based on bilayer ordering of the lipopeptides. Circular dichroism and FTIR spectroscopy indicate the presence of turn structures in the cyclic peptide headgroup. The unexpected distinct mode of self-assembly of mycosubtilin compared to the other two lipopeptides is ascribed to differences in the surfactant packing parameter. This in turn is due to specific features of the conformation of the peptide headgroup and alkyl chain branching.
Introduction
Bioactive lipopeptides are attracting attention due to their ready production using fermentation methods.1 Bacillus subtilis mainly produces three families of lipopeptidic biosurfactant: the surfactins, the fengycins (or plipastatins) and the iturins (including mycosubtilin).2 These lipopeptides have attracted interest as an alternative to chemically synthesized surfactants and also have potential applications due to their antimicrobial and antifungal properties.1a,2 They also have other applications in agriculture, for example the lipopeptide surfactin produced by B. subtilis has the ability to induce systemic resistance in plants, avoiding adverse environmental effects associated with conventional synthetic pesticides.2,3 These compounds also have potential uses in biomedicine,1a,4 for instance exhibiting anti-tumor properties as well as the afore-mentioned antimicrobial and antifungal activity.Lipopeptides may be classified as peptide amphiphiles (PAs) which combine bioactivity from the peptide headgroup with a strong amphiphilic character from the lipid tail. This molecular structure leads to the potential for self-assembly into a variety of nanostructures such as cylindrical fibrils, nanosheets, vesicles and micelles.5 The spontaneous self-assembly of peptide amphiphiles in water has so far focussed on PAs with linear peptide chains.5d These typically create extended nanostructures where the lipid tails are packed into the core and the peptide headgroups are displayed at exterior surface. This unique self-assembled structure may play a role in the demonstrated in vitro bioactivity of these materials, and may also be relevant to potential use in tissue engineering and regenerative medicine.5b,6 Relatively little research has examined the self-assembly of PAs incorporating cyclic peptide headgroups.
The self-assembly of surfactin at interfaces and in bulk solution has previously been investigated using neutron scattering techniques.7 In bulk, small-angle neutron scattering revealed the formation of spherical micelles with a 25 Å radius at 1 mM concentration (pH 7.5).7 On the other hand, Ishigami et al. reported the formation of large rod-shaped micelles by surfactin, with concomitant β-sheet formation.8 Micelle formation has been inferred from dynamic light scattering measurements on surfactin and mycosubtilin and their mixtures.3c The aggregation behaviour including micellization of iturin A have been studied using time-resolved fluorescence methods and by transmission electron microscopy. These results showed that the structure formed by this lipopeptide is clearly dependent on concentration: micelles are formed at the critical micellar concentration and larger vesicular structures at higher concentration.9 We are not aware of any prior studies on the self-assembly of plipastatin (fengycin). In the present manuscript, we use a wide range of physico-chemical methods to compare the self-assembled nanostructures of the three B. subtilis-derived lipopeptides surfactin, mycosubtilin and plipastatin.
Experimental
Materials
Surfactin and plipastatin were produced and purified as described recently.10 Strain B. subtilis BBG131 was used for surfactin production and strain B. subtilis Bs2504 was used for plipastatin production.10, 11 Mycosubtilin was produced by the strain B. subtilis BBG125 as described recently.12 Molecular weights and purities of the lipopeptides are listed in Tables 1 and 2.| Mycosubtilin | Plipastatin | ||||||
|---|---|---|---|---|---|---|---|
| m/z | % | Form | m/z | % | Form | ||
| 1057.56 | 1.8 | C15 | 1434.79 | 1.3 | Plipastatin B C12 | Plipastatin A C14 | |
| 1071.58 | 27.9 | n C16 | 1448.79 | 5.6 | Plipastatin B C13 | Plipastatin A C15 | |
| iso C16 | 1462.80 | 6.6 | Plipastatin B C14 | ||||
| 1085.59 | 58.1 | n or iso C17 | 1462.80 | 12.2 | Plipastatin A C16 | ||
| anteiso C17 | 1476.82 | 17.4 | Plipastatin B C15 | ||||
| 1099.61 | 12.2 | n or iso C18 | 1476.82 | 19.9 | Plipastatin A C17 | ||
| Surfactin A | 1490.84 | 10.3 | Plipastatin B C16 | ||||
| m/z | % | Form | 1490.84 | 25.1 | Plipastatin A C18 | ||
| 1046.65 | 11.5 | C13 | 1504.85 | 0.6 | Plipastatin B C17 | ||
| 1060.66 | 40.9 | C14 | 1504.85 | 1.0 | Plipastatin B C19 | ||
| 1074.68 | 43.5 | C15 | |||||
| 1088.68 | 1.3 | C16 | |||||
| 1102.68 | 1.1 | C17 | |||||
| Sample | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
Purity (%) |
|---|---|---|
| Surfactin | 3.4 | 94% |
| Plipastatin | −3.65 | 91% |
| Mycosubtilin | −0.42 | 99% |
Solution preparation
PA solutions were dissolved in Milli-Q water to the desired concentration and mixed using sonication in an ultrasonic bath at ∼50 °C for 5–10 minutes.Liquid chromatography coupled to mass spectrometry (HPLC-MS) analysis
The analyses were performed on an Accela UHPLC system acquired from Fisher Scientific (Thermo Fisher Scientific, Bremen, Germany). The chromatographic separation was performed with a Vydac C18 250 × 3 mm 5 μm column stored in an oven at 40 °C (Grace Discovery Sciences, Deerfield, IL). The column was operated at a flow rate 600 μL min−1. Solvent A contained water with 0.1% formic acid and Solvent B was a mixture of acetonitrile and 0.1% formic acid. 10 μL of sample was injected in a no waste injection mode and eluted with a linear gradient of solvent B at 55 to 65 % B over 20 min for plipastatin analysis and with an isocratic system for surfactin and mycosubtilin analyses with 80 % B and 40 % B respectively. The Accela UHPLC system was hyphenated with an Orbitrap Exactive mass spectrometry system (Thermo Fisher Scientific, Bremen, Germany). A flow divider called “'splitter” is used as the output rate of HPLC is higher than that tolerated by the mass spectrometer. The flow was split to give approx 100 μL min−1 directed into mass spectrometer via the electro-spray interface. The system was equipped with an ESI interface that was used in positive ionization mode with the following conditions: capillary temperature and voltage at 275 °C and 9 V respectively, ion spray voltage at 3.3 kV and tube lens voltage at 95 V. Nitrogen (N2) was used as the sheath gas and helium (He) as auxiliary gas with a flow rate of 25 and 5 arbitrary units. For the MS experiments, full MS scans were applied, for which the spectra were recorded in the range of m/z 200–2000 with a resolution of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. An external calibration of the equipment for mass accuracy was carried out the day before the analysis according to the manufacturer's guidelines.
000. An external calibration of the equipment for mass accuracy was carried out the day before the analysis according to the manufacturer's guidelines.
MALDI-TOF and MS–MS analysis
Mass profile experiments were analyzed with an Ultraflex MALDI-ToF/ToF mass spectrometer (Bruker, Bremen, Germany) equipped with a smartbeam laser. Spectra were acquired in reflector positive mode in the range of 800 at 3000 Da. Each spectrum was the result of 1000 laser shots per m/z segment per sample delivered in 10 sets of 50 shots distributed in three different locations on the surface of the matrix spot. The instrument was externally calibrated in positive reflector mode using Bradykinin (1–7) [M + H]+ 757.3991, angiotensin II [M + H]+ 1046.5418, angiotensin I [M + H]+ 1296.6848, substance P [M + H]+ 1347.7354, bombesin [M + H]+ 1619.8223, ACTH1–17 [M + H]+ 2093.0862.Circular dichroism (CD)
Spectra were recorded using a Chirascan spectropolarimeter (Applied Photophysics, UK). CD was performed on solutions of the three lipopeptides in water and placed in cover slip cuvettes (0.1 mm thick). Spectra are presented with absorbance A < 2 at any measured point with a 0.5 nm step, 1 nm bandwidth and 1 second collection time per step at 20 °C.Fourier transform infra-red (FTIR) spectroscopy
Spectra were measured on a Nicolet Nexus spectrometer with DTGS detector. FTIR data was measured for PA solutions in D2O at a concentration of 1 wt%. Samples were sandwiched between two CaF2 plate windows (spacer 0.025 mm). Spectra were scanned 128 times over the range of 4000–900 cm−1. Data was corrected by baseline subtraction.X-ray diffraction (XRD)
X-ray diffraction was performed on stalks prepared by suspending drops of PA solutions (1 wt% in water) between the ends of wax-coated capillaries, and allowing them to dry. The stalk was mounted vertically onto the four axis goniometer of a RAXIS IV++ X-ray diffractometer (Rigaku) equipped with a rotating anode generator. The XRD data was collected using a Saturn 992 CCD camera.Small-angle X-ray scattering (SAXS)
SAXS experiments were performed on the EMBL BioSAXS beamline P12 on storage ring PETRA III at the Deutsches Elektronen-Synchtrotron (DESY), Hamburg, Germany. Samples were loaded in PCR tubes in a multi-well plate in a robotic sample changer and delivered automatically into a flow-through capillary tube. SAXS patterns were recorded using a Pilatus 2M detector with a sample-detector distance of 3.1 m. The energy was 10 keV with a wavelength of 1.24 Å. Data were reduced to one-dimensional form and background subtraction was performed using the software PRIMUS.13Cryo-transmission electron microscopy (cryo-TEM)
Imaging was carried out using a field emission cryo-electron microscope (JEOL JEM-3200FSC), operating at 200 kV voltage. Images were taken in bright field mode and using zero loss energy filtering (omega type) with the slit with of 20 eV. Micrographs were recorded using a Gatan Ultrascan 4000 CCD camera. The specimen temperature was maintained at −187 °C during the imaging. Vitrified specimens were prepared using an automated FEI Vitrobot device using Quantifoil 3.5/1 holey carbon copper grids with the hole size of 3.5 μm. Just prior to use, grids were plasma cleaned using a Gatan Solarus 9500 plasma cleaner and then transferred into the environmental chamber of a FEI Vitrobot at room temperature and 100% humidity. Thereafter 3 μl of sample solution (2 wt% concentration) was applied on the grid and it was blotted twice for 5 seconds and then vitrified in a 1/1 mixture of liquid ethane and propane at temperature of −180 °C. The most viscous gel (5 wt% concentration) was blotted 4 times for 5 s. The grids with vitrified sample solution were maintained at liquid nitrogen temperature and then cryo-transferred to the microscope.Dynamic light scattering (DLS)
The size of plipastatin micelles was measured using a Zetasizer Nano ZS system courtesy (Malvern Instruments Ltd, Worcestershire, U.K.) at 90° angle and 298 K. This instrument determines the intensity of the light scattered at a certain angle, which is in turn related to the Brownian motion of the particles, their diffusion coefficient and finally their size. It is able to detect particles ranging from 0.6 nm to 6 μm. Plipastatin buffered solutions were prepared at 0.05% in Tris buffer 50 mM, pH = 8.5. Each sample was analyzed 5 times and the size distribution by intensity, by number and by volume was recorded.Results
Lipopeptides of B. subtilis are usually produced in a mixture containing different homologous forms regarding the sequence of amino acid residues and the nature, length and the branching of the fatty acid chain. The length and isomeric nature of the fatty acid chain has an impact on the activity of the surfactant molecule and on the intermolecular hydrophobic interaction, which leads to micelle formation.2,14 We characterize the composition of the mixture of each lipopeptide families by combined HPLC-MS and MALDI-TOF analysis and molecular mass data is presented in Table 1. The chemical structures of the molecules are shown in Scheme 1. The mycosubtilin mixture mainly contains C16 (m/z = 1071.58) and C17 (m/z = 1085.59) fatty acid (85.98%) chains. The surfactin mixture mainly contains fatty acid chains of C14 (m/z = 1060.66) and C15 (m/z = 1074.68) (84.4%). The plipastatin mixture is quite complex and characterized by the presence of a mix of A and B forms, which show predominantly fatty acid chains to C18 (91.1%). | ||
| Scheme 1 Representative molecular structures of the three peptide amphiphiles. (a) Surfactin, (b) plipastatin, (c) mycosubtilin. As presented in Table 1, the samples comprise a mixture of lipid chain lengths. | ||
We set out to probe whether the headgroups in the three lipopeptides surfactin, plipastatin and mycosubtilin obtained from B. subtilis exhibit defined secondary structure. Specifically we wished to examine whether any of them exhibit β-sheet structures, since this is commonly observed for lipopeptides based on linear peptide headgroups.5d,15 FTIR and CD spectroscopy were used to investigate this, along with X-ray diffraction (discussed later). Fig. 1 shows FTIR spectra measured in the amide I′ and amide II′ regions from 1 wt% solutions of the three lipopeptides in D2O. All three spectra comprise broad peaks in the amide I′ region centred at 1640 cm−1 for surfactin and mycosubtilin and 1644 cm−1 for plipastatin. Peak positions in this range are typically ascribed to random coil structure.16 However, the cyclic nature of the peptide headgroups suggests that turn structures are likely to be present, as confirmed by CD spectroscopy to be discussed shortly.
 | ||
| Fig. 1 FTIR spectra in the amide I′/II′ regions obtained from 1 wt% solutions of the three lipopeptides, as indicated. | ||
Also notable in the FTIR spectra are differences in the spectra in the amide II′ region since both surfactin and plipastatin exhibit a broad peak centered near 1570 cm−1, this peak being absent for mycosubtilin. Also in the 1720–1760 cm−1 range, a peak is observed for surfactin and plipastatin but not for mycosubtilin. We ascribe these differences to the absence of acidic residues in mycosubtilin in constrast to surfactin and plipastatin. Aspartic acid and glutamic acid side chains produces a peak near 1720 cm−1 due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretches and these residues give peaks in the range 1550–1570 cm−1 due to CO2− asymmetric stretching.16,17 As will be shown shortly, this difference in the amino acid sequence, and charge, leads to pronounced differences in self-assembly. At pH 7, surfactin is predicted to have charge −2, mycosubtilin charge 0 and plipastatin charge −1 (peptide properties, http://www.%20Innovagen.com). In previous work we measured the surface charge of surfactin in terms of zeta potential and found that surfactin is strongly negatively charged in a wide range of pH.18 Of the three molecules, surfactin is the most hydrophobic (see calculated log
O stretches and these residues give peaks in the range 1550–1570 cm−1 due to CO2− asymmetric stretching.16,17 As will be shown shortly, this difference in the amino acid sequence, and charge, leads to pronounced differences in self-assembly. At pH 7, surfactin is predicted to have charge −2, mycosubtilin charge 0 and plipastatin charge −1 (peptide properties, http://www.%20Innovagen.com). In previous work we measured the surface charge of surfactin in terms of zeta potential and found that surfactin is strongly negatively charged in a wide range of pH.18 Of the three molecules, surfactin is the most hydrophobic (see calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values in Table 1). However, this alone does not account for the aggregation tendencies of the surfactants which depends on the chemical structure and in particular on the ratio of the areas of the hydrophobic to hydrophilic portions of the molecule, represented by the surfactant packing parameter.14
P values in Table 1). However, this alone does not account for the aggregation tendencies of the surfactants which depends on the chemical structure and in particular on the ratio of the areas of the hydrophobic to hydrophilic portions of the molecule, represented by the surfactant packing parameter.14
CD spectra are presented in Fig. 2. Both surfactin and plipastatin show peaks at 220 nm, although the sign of the CD signal is inverted. Mycosubtilin shows an additional positive maximum at 210 nm, and a negative minimum at 197 nm. The spectra are ascribed to a combination of random coil, polyproline II and turn structures, the latter arising from the constrained cyclic nature of the peptide headgroups.19 As discussed below, the distinct CD spectrum for mycosubtilin is due to its distinct mode of self-assembly into extended fibrillar nanostructures.
 | ||
| Fig. 2 CD spectra for 1 wt% solutions of the three lipopeptides, as indicated. | ||
Cryo-TEM comprises a very distinct nanostructure for mycosubtilin, compared to surfactin and plipastatin. As shown in Fig. 3a, the mycosubtilin sample comprises tape-like extended self-assemblies, with a variable width 15–80 nm. Some of the tapes containing an internal banded structure with a spacing of 5.1 nm, which on the basis of X-ray diffraction experiments discussed below, is assigned to the spacing of bilayer structures. In contrast, both plipastatin and surfactin form micellar structures (Fig. 3b and c) with a diameter of approximately 5 nm. The aggregation properties of the lipopeptides lead to differences in transparency of the samples as shown in ESI Fig. 3.† Solutions (1 wt%) of surfactin and plipastatin are transparent, whereas that of mycosubtilin is cloudy, consistent with the formation of extended fibrillar objects.
SAXS confirms the findings from cryo-TEM concerning the self-assembled nanostructure, although it also provides more detailed information on the size and internal structure of the micelles. As shown in Fig. 4, the SAXS intensity profiles for surfactin and plipastatin could both be fitted using the form factor of a spherical shell, which represents a core–shell micellar structure. Further details of the models are provided in the ESI.† The low q data is dominated by structure factor effects, and was not included in the form factor fits. For surfactin, the form factor is indeed well described by a sphere model, supporting the findings of Shen et al.7 and contrary to the report by Ishigami et al. which indicated rod-like micelles.8 The fitted total micelle radius is Rtot = 2.0 nm with an inner with core radius Rin = 0.97 nm, with highly negative electron density. A similar model described the micelles of plipastatin which have Rout = 2.04 nm although the inner core was larger Rin = 1.41 nm and with a less negative scattering contrast. The radii obtained from SAXS are consistent with the dimensions of the micelles visible in the cryo-TEM images (Fig. 3b and c). In complete contrast, the SAXS intensity profile for mycosubtilin shows peaks from a bilayer structure. The data could be fitted using a model form factor of a Gaussian bilayer and a multi-layer structure factor based on the Caillé structure factor. This model, described fully elsewhere, has been used by us successfully to represent the scattering from other bilayer-based PA nanotape structures.15a The bilayer spacing is 5.4 nm, which can be obtained from the position of the three structure factor peaks as shown in Fig. 4b (the first is present as a shoulder on a form factor peak). This value is close to the spacing of the banded structures observed in some cryo-TEM images (Fig. 3a), and these striped structures are thus assigned to the spacing of bilayers within the tapes.
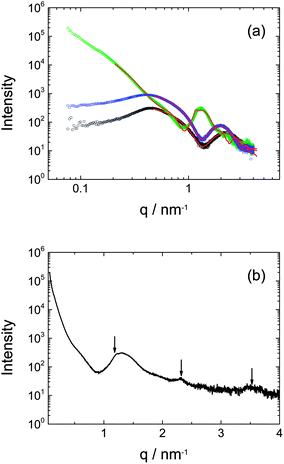 | ||
| Fig. 4 SAXS data for 1 wt% solutions of the three lipopeptides (a) comparison of data with model fits (red lines) as described in the text. Green triangles: mycosubtilin, blue squares: plipastatin, black circles: surfactin, (b) data for mycosubtilin (linear q scale) showing three orders of Bragg reflection from a bilayer structure. | ||
The difference in the ordering of mycosubtilin compared to the other two lipopeptides is also evident in X-ray diffraction data. Measurements were performed on stalks dried from 1 wt% solution and the X-ray diffraction patterns are shown in Fig. 5. The equatorial and meridional intensity profiles obtained by sector integration of the 2D patterns are shown in ESI Fig. 2.† Fig. 5 and ESI Fig. 2† show that, in contrast to surfactin and plipastatin, the XRD pattern for mycosubtilin contains three orders of Bragg reflection from a lamellar structure with period 5.5 nm, in good agreement with the value from SAXS. The alignment of these peaks indicates orientation of the bilayers with their normal along the fibre axis. Strong equatorial reflections at high angle are due to the lateral packing of the molecules within the bilayers, with a spacing of 4.2 Å (the 3.8 Å peak is assigned to the Cα–Cα spacing). These peaks due to the lateral packing of molecules are also observed for surfactin and plipastatin (the latter sample exhibited a higher degree of alignment) although in the case of these two lipopeptides, bilayer ordering is not observed. A further peak with d = 5.2 Å is observed for mycosubtilin, and is due to the packing of the lipid headgroups. Relative to the 4.2 Å peak this feature is weak. A peak with d = 1.4 nm for surfactin may be due to a periodicity arising from the length of the predominant C14/C15 lipid chains, which is estimated to have this length in a lipid ordered phase. The XRD data are consistent with the findings from FTIR and CD spectroscopy and point to a lack of defined secondary structure for surfactin and plipastatin, although mycosubtilin exhibits lamellar ordering with some α-helical content.
 | ||
| Fig. 5 XRD patterns obtained from stalks dried from 1 wt% solutions. (a) Surfactin, (b) plipastatin, (c) mycosubtilin. | ||
Dynamic light scattering results (ESI Fig. 4†) show that plipastatin forms micelles with an equivalent mean diameter of about 5 nm, consistent with the results from cryo-TEM and SAXS. This is similar to the micelle size reported for surfactin and mycosubtilin at the same concentrations and in buffered solutions.3c It is very interesting to note that the size of micelles measured by DLS technique is in agreement with those measured here by higher resolution techniques such as SAXS and TEM.
Summary and discussion
The self-assembly of lipopeptides containing cyclic peptide headgroups is to date relatively unexplored. We have compared for the first time, the self-assembly of three lipopeptides obtained from Bacillus subtilis. Surfactin and plipastatin both form spherical micelles whereas mycosubtilin self-assembles into extended nanotapes based on the stacking of lipopeptide bilayers. The bilayers can be imaged directly by cryo-TEM in some projections of the tapes as stripe structures. The spacing corresponds to 5.5 nm, in good agreement with the value obtained from SAXS.The formation of a bilayer structure by mycosubtilin indicates a surfactant packing parameter p = v/al ≈1 where v is the volume of the hydrophobic chain, a is the effective area per headgroup and l is the lipid chain length. In contrast, p is smaller for surfactin and plipastatin, such that spherical micelles are formed. This may be due to larger l values for the unbranched lipid chains, although the effective headgroup area will also differ due to the distinct conformations and electrostatics of the cyclic peptide units. Indeed contrary to the two other lipopeptides, mycosubtilin is a neutral compound. On the other hand, the molecular structure of mycosubtilin contains aromatic residues (including in the backbone) which may constrain conformation. Although plipastatin also contains aromatic residues (in the backbone also), the lipid chain is separated from the cyclic headgroup by more flexible linking units, this may increase conformational freedom and enable packing into spherical micelles. This may also be favoured by the considerable distribution in chain length for this sample compared to the other two.
Our findings highlight the subtle influence of peptide and alkyl chain conformation on the self-assembled nanostructure of bioderived lipopeptides. These findings may have relevance to the understanding of the bioactivity of these important classes of lipopeptides with antimicrobial and antifungal properties among others. The distinct self-assembled motifs present the functional peptide groups at a different density and morphology which in turn can influence delivery and activity. This will be the subject of future research.
Acknowledgements
This work was supported by EPSRC grant EP/G067538/1 to IWH. Beamtime at DESY was awarded under references SAXS-03, -04 (2013) and we are grateful for local support from Alexey Kikhney. This work received also the financial support of the European Funds of INTERREG IV PhytoBio Project and EGIDE with the ALLIANCE Project no. 19406WE.References
- (a) J. M. Bonmatin, O. Laprevote and F. Peypoux, Comb. Chem. High Throughput Screening, 2003, 6, 541 CrossRef CAS; (b) I. M. Banat, A. Franzetti, I. Gandolfi, G. Bestetti, M. G. Martinotti, L. Fracchia, T. J. Smyth and R. Marchant, Appl. Microbiol. Biotechnol., 2010, 87, 427 CrossRef CAS PubMed.
- P. Jacques, in Biosurfactants Microbiology Monographs, ed. G. Soberon-Chavez, Springer-Verlag, Berlin, 2011, vol. 20 Search PubMed.
- (a) M. Ongena and P. Jacques, Trends Microbiol., 2008, 16, 115 CrossRef CAS PubMed; (b) J. M. Raaijmakers, I. de Bruijn, O. Nybroe and M. Ongena, FEMS Microbiol. Rev., 2010, 34, 1037 CAS; (c) P. Jauregi, F. Coutte, L. Catiau, D. Lecouturier and P. Jacques, Sep. Purif. Technol., 2013, 104, 175 CrossRef CAS PubMed.
- P. Singh and S. S. Cameotra, Trends Biotechnol., 2004, 22, 142 CrossRef CAS PubMed.
- (a) H. G. Cui, M. J. Webber and S. I. Stupp, Pept. Sci., 2010, 94, 1 CrossRef CAS PubMed; (b) X. B. Zhao, F. Pan, H. Xu, M. Yaseen, H. H. Shan, C. A. E. Hauser, S. G. Zhang and J. R. Lu, Chem. Soc. Rev., 2010, 39, 3480 RSC; (c) F. Versluis, H. R. Marsden and A. Kros, Chem. Soc. Rev., 2010, 39, 3434 RSC; (d) I. W. Hamley, Soft Matter, 2011, 7, 4122 RSC.
- (a) M. J. Webber, J. Tongers, M.-A. Renault, J. G. Roncalli, D. W. Losordo and S. I. Stupp, Acta Biomater., 2010, 6, 3 CrossRef CAS PubMed; (b) J. B. Matson, R. H. Zha and S. I. Stupp, Curr. Opin. Solid State Mater. Sci., 2011, 15, 225 CrossRef CAS PubMed; (c) J. B. Matson and S. I. Stupp, Chem. Commun., 2012, 48, 26 RSC.
- H. H. Shen, R. K. Thomas, C. Y. Chen, R. C. Darton, S. C. Baker and J. Penfold, Langmuir, 2009, 25, 4211 CrossRef CAS.
- Y. Ishigami, M. Osman, H. Nakahara, Y. Sano, R. Ishiguro and M. Matsumoto, Colloids Surf., B, 1995, 4, 341 CrossRef CAS.
- (a) A. Grau, J. C. Gomez-Fernandez, F. Peypoux and A. Ortiz, Peptides, 2001, 22, 1 CrossRef CAS; (b) I. Harnois, D. Genest, J. C. Brochon and M. Ptak, Biopolymers, 1988, 27, 1403 CrossRef CAS PubMed.
- F. Coutte, D. Lecouturier, V. Leclère, M. Béchet, P. Jacques and P. Dhulster, Process Biochem., 2013, 48, 25 CrossRef CAS PubMed.
- M. Ongena, E. Jourdan, A. Adam, M. Paquot, A. Brans, B. Joris, J. L. Arpigny and P. Thonart, Environ. Microbiol., 2007, 9, 1084 CrossRef CAS PubMed.
- M. Béchet, J. Castéra-Guy, J. S. Guez, N. E. Chibib, F. Coucheney, F. Coutte, P. Fickers, V. Leclère, B. Wathelet and P. Jacques, Bioresour. Technol., 2013 DOI:10.1016/j.biortech.2013.03.123.
- P. V. Konarev, V. V. Volkov, A. V. Sokolova, M. H. J. Koch and D. I. Svergun, J. Appl. Crystallogr., 2003, 36, 1277 CrossRef CAS.
- I. W. Hamley, Introduction to Soft Matter. Revised Edition, Wiley, Chichester, 2007 Search PubMed.
- (a) V. Castelletto, G. Cheng, C. Stain, C. J. Connon and I. W. Hamley, Langmuir, 2012, 28, 11599 CrossRef CAS PubMed; (b) V. Castelletto, I. W. Hamley, J. Perez, L. Abezgauz and D. Danino, Chem. Commun., 2010, 46, 9185 RSC; (c) S. E. Paramonov, H.-W. Jun and J. D. Hartgerink, J. Am. Chem. Soc., 2006, 128, 7291 CrossRef CAS PubMed.
- B. Stuart, Biological Applications of Infrared Spectroscopy, Wiley, Chichester, 1997 Search PubMed.
- (a) S. Krimm and J. Bandekar, Adv. Protein Chem., 1986, 38, 181 CrossRef CAS; (b) A. Barth and C. Q. Zscherp, Q. Rev. Biophys., 2002, 35, 369 CrossRef CAS.
- M. H. M. Isa, D. E. Coraglia, R. A. Frazier and P. Jauregi, J. Membr. Sci., 2007, 296, 51 CrossRef CAS PubMed.
- (a) L. G. Pease and C. Watson, J. Am. Chem. Soc., 1978, 100, 1279 CrossRef CAS; (b) C. A. Bush, S. K. Sarkar and K. D. Kopple, Biochemistry, 1978, 17, 4951 CrossRef CAS; (c) R. W. Woody, Methods Enzymol., 1995, 246, 34 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3sm51514a |
| This journal is © The Royal Society of Chemistry 2013 |

