 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The bacterial type IV pilus system – a tunable molecular motor
Berenike
Maier
*
Department of Physics and Biocenter, University of Cologne, Zülpicher Str. 77, 50937 Köln, Germany. E-mail: berenike.maier@uni-koeln.de
First published on 22nd May 2013
Abstract
Bacteria have evolved surface-exposed polymers called pili with remarkable physical properties. This highlight describes recent advances in the biophysics of type IV pilus systems. They are strong molecular motors whose velocity and force are tunable by external inputs. Coordination of multiple pili for mediating pilus-driven surface motility depends on cell shape, surface interaction, and cooperation with other motors. Rational design of surfaces can control pilus-mediated surface movement and potentially biofilm architecture.
Many bacterial species live at surfaces, and there they encounter a conflict. On one hand they have to strongly adhere to the surface to avoid being flushed away in the fluid flow, but on the other hand they have to be motile to colonize the surface. To solve this problem, bacteria have evolved surprisingly different molecular mechanisms. They generate pili, polymers that extend from the bacterial surface by ≈1 μm. Pili use different mechanical tricks enabling bacteria to colonize surfaces while remaining associated with the surface. Whereas type I pili have evolved a catch-bond mechanism1 in conjunction with force-induced unfolding,2 type IV pili are grappling hooks that enable bacteria move over surfaces. In this highlight, we will concentrate on the biophysics of type IV pili.
Molecular mechanism of force generation – grab and pull
Type IV pili (T4P) are helical polymers with a diameter of 6 nm (ref. 3) and a length on the order of 1 μm (Fig. 1a).4 Their main constituent is the major pilin (with the protein name PilE or PilA). Within the pilus, the effective length of a pilin subunit is 0.8 nm;5 thus a single pilus polymer consists of thousands of pilins. Pilin subunits are stored in the inner membrane of the bacterium.6 T4P polymerization is supported by an ATPase (PilF or PilB) located in the cytoplasm (Fig. 1b).7 The surface properties are determined by insertion of minor pilins and by post-translational modification.8–10 For the details of the molecular mechanism of pilus assembly the reader is referred to a recent review by Giltner et al.11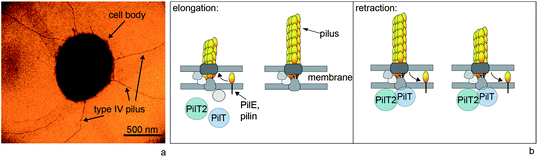 | ||
| Fig. 1 Type IV pili are dynamic polymers. (a) Transmission electron micrograph of Neisseria gonorrhoeae. (b) Simplified molecular mechanism of T4P elongation and retraction. The major pilin subunits are stored in the inner membrane. During polymerization, they are integrated into the elongating pilus. Supported by the retraction ATPase PilT, the T4P depolymerizes and most likely the pilins are reinserted into the inner membrane. | ||
Type IV pili can retract by depolymerization (Fig. 1b). During retraction, the pilin subunits are most likely recycled and re-inserted into the inner membrane. Retraction is supported by the hexameric retraction ATPase PilT.12,13 When a load is attached to the tip of the pilus, the retracting T4P exerts force on that load.12 Laser tweezers experiments have shown that during retraction of single T4P (Fig. 2a) considerable force is generated, in the range of 200 pN for Neisseria gonorrhoeae and Myxococcus xanthus.13–15 T4P can team up by forming bundles and generate forces in the range of 1 nN.16 For measuring forces in the nanonewton range, a micropillar assay can be used (Fig. 2b).17 These experiments demonstrate that the type IV pilus system is among the strongest molecular motors known, although the mechanism of high force generation remains elusive. It is worth mentioning that the term “molecular motor” will refer to the T4P system including the pilus polymer, the ATPases, and other proteins that most likely play a role in transmission of molecular movement from the cytoplasmic ATPases to the extracellular pilus polymer.
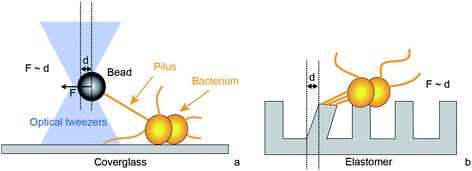 | ||
| Fig. 2 Experimental setups for characterization of force generation by type IV pili. (a) Laser tweezers. A micro-bead is trapped by laser tweezers. When a T4P binds and retracts it pulls the bead out of the center of the laser trap. By tracking the position of the bead, the length change of the T4P is measured with nanometer-resolution. The optical restoring force F increases with the distance d from the center enabling force measurement. This assay is useful for characterizing single T4P dynamics at high resolution. (b) Polyacrylamide micropillars. T4P adhere to adjacent pillars and deflect them by a distance d through retraction. By tracking the pillar position, the dynamics of T4P length change and the force F generated can be measured. This setup is useful for static force measurements and for force measurement by multiple T4P. | ||
Putative functions of force generation
The biological function of high force generation by type IV pilus retraction is currently unclear. For pathogenic Neisseria it has been suggested that force acts as a signal to its host cells and recent experiments support this hypothesis; during infection of epithelial cells, N. gonorrhoeae generate considerable force.18 Mechano-sensitive pathways are stimulated during infection of epithelial cells with force-generating N. gonorrhoeae.19 To demonstrate the T4P mediated mechanical force, which triggers cytoskeletal rearrangements within the host cells, force was applied to pilus-coated beads using laser tweezers. Cytoskeletal rearrangement occurred rapidly on a time scale of 2 min.20Application of force in the range of ∼1 nN can alter the structure of the pilus, exposing epitopes that are hidden in the absence of force.21 Furthermore, hidden epitopes become accessible during infection of endothelial cells, suggesting that T4P generated force may support exposure of these epitopes.22
Although there is increasing evidence that force generation by T4P affects the interaction between pathogenic Neisseria and their human host cells, it is unclear what function high force generation may have in other bacterial species that inhabit different niches.
Control of velocity
The type IV pilus motor works in distinct velocity modes, namely retraction at two different speeds and elongation (Fig. 3).23,24 Switching between retraction and elongation occurs at different time scales; in the range of milliseconds (corresponding to several pilins) and in the range of hundreds of milliseconds (corresponding to hundreds of pilins).24 The probability of switching from retraction to elongation increases with increasing force. Back-stepping at the level of single pilins is reminiscent of in vitro single molecule assays with processive molecular motors, but rigorous characterization will require higher spatial resolution of the laser tweezers experiments. Directional switching at the scale of several hundreds of pilins is more likely explained by different modes of the pilus systems, e.g. by binding and unbinding of the retraction ATPase PilT. Supporting this assumption, the probability for switching from the retraction mode to the elongation mode increases with decreasing concentration of PilT.24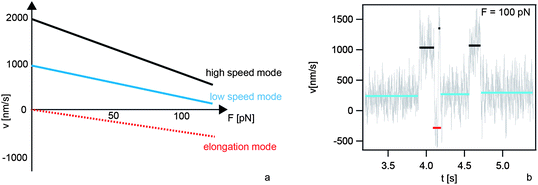 | ||
| Fig. 3 Velocity modes of single type IV pili. (a) Schematic relationship between the rate v of T4P length change and the force F applied to the T4P. (b) During a single T4P retraction event at 100 pN in N. gonorrhoeae the system switches between the high speed mode (black, 1000 nm s−1), the low speed mode (blue, 250 nm s−1), and elongation (red, −250 nm s−1). | ||
Furthermore, T4P retraction can occur in two distinct speed modes.24 This very unusual motor property is conserved between different bacterial species.14 For both speed modes the speed of pilus retraction decreases with increasing force (Fig. 3a). Furthermore, a retraction ATPase in N. gonorrhoeae, PilT2, increases the speed of both modes by a factor of ∼2.13 PilT2 is a paralog of PilT whose protein sequence has a high degree of similarity.
Speed switching between the different retraction speeds can be understood in terms of a two-state system.13 Interestingly, the probability of finding the T4P in one or the other mode is controlled differently in different species. In N. gonorrhoeae, speed switching from the high speed mode (2 μm s−1) to the low speed mode (1 μm s−1) can be triggered by depletion of oxygen. At time periods in which oxygen is nearly depleted, speed switching occurs during a single pilus retraction event, indicating bistability.13 The maximum force generated by a single T4P is lower under anaerobic conditions.13 In Myxococcus xanthus, oxygen depletion does not trigger speed switching; however, reversible switching occurs at forces exceeding 8 pN. In N. gonorrhoeae, reversible switching occurs only at forces exceeding 100 pN (Fig. 3b).
In summary, the biophysical properties of type IV pilus dynamics can be fine-tuned by mechanical force, concentration of ATPases, and oxygen. Although the mechanical properties are conserved between species, the speed regulation seems to have evolved differently. It will be exciting to assess the molecular mechanism of velocity switching and its biological function.
Translation of pilus retraction to bacterial movement
Cycles of type IV pilus elongation, adhesion, and retraction power bacterial surface motility as has been demonstrated using TIRF microscopy.25 Nevertheless, the question remains how individual T4P coordinate for generating movement. Recently, this question has been addressed with different bacterial species. N. gonorrhoeae employs T4P as their only molecular motor for movement while P. aeruginosa and M. xanthus use additional motility systems. Therefore, it is important to understand the coupling of T4P driven motility to these other motors.26There is recent evidence that different bacterial species use a two-dimensional tug-of-war mechanism for T4P-driven surface motility (Fig. 4a). N. gonorrhoeae are round bacteria whose pili are distributed all around the cell body (Fig. 1a) yet their movement exhibits directional persistence and the persistence time increases with increasing number of pili per cell.4 Their speed shows a mono-modal distribution around 1.5 μm s−1 under aerobic conditions and around 0.5 μm s−1 under anaerobic conditions. The movement can be modeled as a stochastic tug-of-war with directional memory (Marathe et al., unpublished), whereby directional memory most likely arises from pilus bundling or from the fact that the T4P complex is not disassembled after a full retraction. Thus in contrast to a one-dimensional tug-of-war found in cytoskeletal motors,27 directional memory is necessary to generate directional persistence in two dimensions.
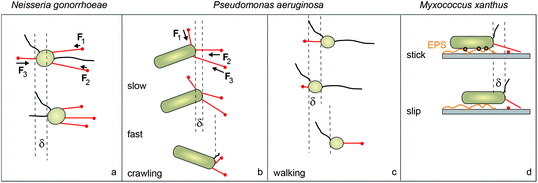 | ||
| Fig. 4 Coordination of type IV pili for surface movement. (a) In round N. gonorrhoeae T4P are distributed around the cell body. Their relatively smooth movement is consistent with a tug-of-war mechanism. (b) In rod-shaped P. aeruginosa T4P are polar. Their jerky crawling movement is consistent with a tug-of-war which is dominated by the rupture of T4P from the surface. (c) Walking of P. aeruginosa with the cell body oriented vertically shows low persistence in agreement with uncoordinated pilus retraction. (d) Rod-like M. xanthus show slip–stick movement resulting from interactions with secreted extracellular polysaccharides (EPS). Red: pili attached to the surface; black: pili detached from the surface. | ||
In rod-like P. aeruginosa, surface motility can switch between horizontal “crawling” and vertical “walking” (Fig. 4b and c).28 The transition from crawling to walking is facilitated by the second molecular motor used for movement by P. aeruginosa, namely flagella.29 The directional persistence of walking cells is lower than for crawling bacteria. Crawling has the characteristics of a sling-shot as it is characterized by a succession of slow (0.3 μm s−1) and fast (1 μm s−1) movements.30 The fast movement is accompanied by a rotation of the bacterium. Mechanistically, this type of movement can be understood as a tug-of-war between pili at the leading pole; movement is dominated by rupture of pili from the surface, leading to fast rotation and movement at the speed of individual pilus retraction (Fig. 4b). When fluid flow is applied, the rod-like P. aeruginosa align within the flow. As a consequence, the bacteria tend to move upstream.31
The rod-like M. xanthus tightly controls the location of its T4P through a molecular clock that triggers the process of switching the location of the T4P between both poles every 6–8 min.32 Interestingly, the PilT retraction ATPase resides mostly at the lagging pole.33 A few PilT molecules are located at the leading pole, triggering pilus retraction and movement. T4P motility in M. xanthus shows different biophysical characteristics when compared to P. aeruginosa and N. gonorrhoeae. During movement, M. xanthus deposits extracellular polysaccharides (EPS) and other slime components on the surface.34 Apparently, the interaction of the cell body with the EPS impairs pilus-driven movement.35 Eventually the cell body yields to the pilus-generated force and the bacterium starts sliding. Thus the movement has an aperiodic slip–stick character (Fig. 4d). Slip duration and slip displacement follow scaling laws known from earthquake dynamics.35 The different behavior of M. xanthus may be connected to the second motor for surface motility that seems to be specific to myxobacteria, termed A-motility.36 A-motility involves transient adhesion complexes that remained at fixed positions relative to the substratum as cells move forward.37 This movement is mediated by membrane-bound cytoplasmic complexes consisting of motor and regulatory proteins that are directionally transported down the axis of a cell at constant velocity.38
In conclusion, the translation of single T4P retractions to bacterial surface movement can be understood in terms of force-balance. Importantly, the shape of the bacterium, surface interaction, and the coupling to other molecular motors determine the dynamics of bacterial movement.
Bacterial motility in micro-structured environments
Type IV pilus driven surface motility depends on the physico-chemical properties of the surface. So far no unique receptor has been found, but it has been shown that T4P mediate adhesion to very different surfaces including glass, polystyrene, BSA, lipids, and eukaryotic cells. On the other hand, the biophysical properties of surface motility strongly depend on the surface. The characteristic slip–stick motion of M. xanthus depends on their ability to secrete extracellular polysaccharides.35N. gonorrhoeae can adhere to solid-supported lipid membranes. The speed of surface movement increases with decreasing membrane fluidity.39With these results in mind, structured surfaces have been designed where the physico-chemical properties control bacterial movement. For example, bacterial motility can be confined to stripes or to islands (Fig. 5a).39 In particular, on BSA-islands surrounded by fluid lipid membrane, motile N. gonorrhoeae are trapped on the islands. Mechanistically, bacteria cannot translate T4P retraction into movement on fluid lipid membranes and therefore, they are unlikely to move from the rigid BSA-surface into the fluid membrane. Furthermore, the location of bacterial clusters can be controlled by microcontact printing.39 The transition from single cell motility to microcolonies occurs when the surface coverage of the islands exceeds ∼50%. In a different approach, motile bacteria were guided by elevations with dimension and depth corresponding to the size of the bacteria.40 Fences of 1 μm height tend to reflect N. gonorrhoeae. Grooves with micrometer-dimensions attract motile N. gonorrhoeae and M. xanthus and force them to move preferentially within the grooves (Fig. 5b). Most likely, the reason why bacteria are trapped in grooves is that they encounter a larger adhesive surface within the groove than on the ridge.
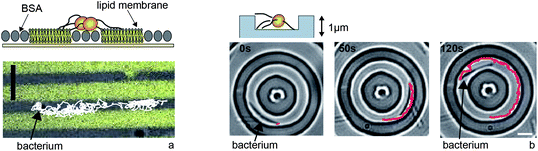 | ||
| Fig. 5 Type IV pilus driven motility can be guided by microstructures. (a) Physico-chemical surface properties control motility of N. gonorrhoeae. Bacteria move along BSA stripes but hardly enter the fluid lipid membrane. (b) M. xanthus is trapped in grooves of 1 μm height (adapted from Meel et al.40). Full lines: tracks of individual bacterium. Scale bars: 5 μm. | ||
In their natural habitat, bacteria encounter spatially inhomogeneous environments. The experimental techniques discussed above allow us to control the spatial residence of bacteria. In particular, the formation of bacterial clusters as controlled by microcontact printing39 can be considered the first step to biofilm formation. It would thus be interesting to rationally design surfaces that can control the formation and architecture of bacterial clusters and biofilms.
Conclusion
The single motor properties including velocity and force generation are very well conserved between different bacterial species. It will be a future challenge to understand the molecular basis of high force generation and of velocity control. The coordination of multiple motors and the response to different environmental inputs has evolved very differently. In future studies it would be interesting to investigate the biophysical response of the type IV pilus system to environmental inputs including cell–cell communication and tactic signals, such as lipid chemotaxis41,42 or phototaxis.43Acknowledgements
I gratefully thank the Maier lab, Stefan Klumpp, Gerard Wong, and Mike Koomey for helpful discussions. This work has been supported by the Deutsche Forschungsgemeinschaft through MA 3898.References
- I. Le Trong, P. Aprikian, B. A. Kidd, M. Forero-Shelton, V. Tchesnokova, P. Rajagopal, V. Rodriguez, G. Interlandi, R. Klevit, V. Vogel, R. E. Stenkamp, E. V. Sokurenko and W. E. Thomas, Cell, 2010, 141, 645–655 CrossRef CAS.
- M. Forero, O. Yakovenko, E. V. Sokurenko, W. E. Thomas and V. Vogel, PLoS Biol., 2006, 4, e298 Search PubMed.
- L. Craig and J. Li, Curr. Opin. Struct. Biol., 2008, 18, 267–277 CrossRef CAS.
- C. Holz, D. Opitz, L. Greune, R. Kurre, M. Koomey, M. A. Schmidt and B. Maier, Phys. Rev. Lett., 2010, 104, 178104 CrossRef.
- L. Craig, R. K. Taylor, M. E. Pique, B. D. Adair, A. S. Arvai, M. Singh, S. J. Lloyd, D. S. Shin, E. D. Getzoff, M. Yeager, K. T. Forest and J. A. Tainer, Mol. Cell, 2003, 11, 1139–1150 CrossRef CAS.
- P. C. Morand, E. Bille, S. Morelle, E. Eugene, J. L. Beretti, M. Wolfgang, T. F. Meyer, M. Koomey and X. Nassif, EMBO J., 2004, 23, 2009–2017 CrossRef CAS.
- N. E. Freitag, H. S. Seifert and M. Koomey, Mol. Microbiol., 1995, 16, 575–586 CrossRef CAS.
- H. C. Winther-Larsen, M. Wolfgang, S. Dunham, J. P. van Putten, D. Dorward, C. Lovold, F. E. Aas and M. Koomey, Mol. Microbiol., 2005, 56, 903–917 CrossRef CAS.
- A. Cehovin, P. J. Simpson, M. A. McDowell, D. R. Brown, R. Noschese, M. Pallett, J. Brady, G. S. Baldwin, S. M. Lea, S. J. Matthews and V. Pelicic, Proc. Natl. Acad. Sci. U. S. A., 2013, 3065–3070 CrossRef CAS.
- J. Chamot-Rooke, G. Mikaty, C. Malosse, M. Soyer, A. Dumont, J. Gault, A. F. Imhaus, P. Martin, M. Trellet, G. Clary, P. Chafey, L. Camoin, M. Nilges, X. Nassif and G. Dumenil, Science, 2011, 331, 778–782 CrossRef CAS.
- C. L. Giltner, Y. Nguyen and L. L. Burrows, Microbiol. Mol. Biol. Rev., 2012, 76, 740–772 CrossRef CAS.
- A. J. Merz, M. So and M. P. Sheetz, Nature, 2000, 407, 98–102 CrossRef CAS.
- R. Kurre, A. Hone, M. Clausen, C. Meel and B. Maier, Mol. Microbiol., 2012, 857–865 CAS.
- M. Clausen, V. Jakovljevic, L. Sogaard-Andersen and B. Maier, J. Bacteriol., 2009, 191, 4633–4638 CrossRef CAS.
- B. Maier, L. Potter, M. So, C. D. Long, H. S. Seifert and M. P. Sheetz, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16012–16017 CrossRef CAS.
- N. Biais, B. Ladoux, D. Higashi, M. So and M. Sheetz, PLoS Biol., 2008, 6, e87 Search PubMed.
- N. Biais, D. Higashi, M. So and B. Ladoux, Methods Mol. Biol., 2012, 799, 197–216 CAS.
- D. Opitz, M. Clausen and B. Maier, ChemPhysChem, 2009, 10, 1614–1618 CrossRef CAS.
- H. L. Howie, M. Glogauer and M. So, PLoS Biol., 2005, 3, e100 Search PubMed.
- D. Opitz and B. Maier, PLoS One, 2011, 6, e17088 CAS.
- N. Biais, D. L. Higashi, J. Brujic, M. So and M. P. Sheetz, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11358–11363 CrossRef CAS.
- T. Brissac, G. Mikaty, G. Dumenil, M. Coureuil and X. Nassif, Infect. Immun., 2012, 80, 3297–3306 CrossRef CAS.
- B. Maier, M. Koomey and M. P. Sheetz, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10961–10966 CrossRef CAS.
- M. Clausen, M. Koomey and B. Maier, Biophys. J., 2009, 96, 1169–1177 CrossRef CAS.
- J. M. Skerker and H. C. Berg, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 6901–6904 CrossRef CAS.
- J. C. Conrad, Res. Microbiol., 2012, 163, 619–629 CrossRef CAS.
- M. J. Muller, S. Klumpp and R. Lipowsky, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4609–4614 CrossRef.
- M. L. Gibiansky, J. C. Conrad, F. Jin, V. D. Gordon, D. A. Motto, M. A. Mathewson, W. G. Stopka, D. C. Zelasko, J. D. Shrout and G. C. Wong, Science, 2010, 330, 197 CrossRef CAS.
- J. C. Conrad, M. L. Gibiansky, F. Jin, V. D. Gordon, D. A. Motto, M. A. Mathewson, W. G. Stopka, D. C. Zelasko, J. D. Shrout and G. C. Wong, Biophys. J., 2011, 100, 1608–1616 CrossRef CAS.
- F. Jin, J. C. Conrad, M. L. Gibiansky and G. C. Wong, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12617–12622 CrossRef CAS.
- Y. Shen, A. Siryaporn, S. Lecuyer, Z. Gitai and H. A. Stone, Biophys. J., 2012, 103, 146–151 CrossRef CAS.
- P. Lenz and L. Sogaard-Andersen, Nat. Rev. Microbiol., 2011, 9, 565–577 CrossRef CAS.
- I. Bulyha, C. Schmidt, P. Lenz, V. Jakovljevic, A. Hone, B. Maier, M. Hoppert and L. Sogaard-Andersen, Mol. Microbiol., 2009, 74, 691–706 CrossRef CAS.
- A. Ducret, M. P. Valignat, F. Mouhamar, T. Mignot and O. Theodoly, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 10036–10041 CrossRef CAS.
- M. L. Gibiansky, W. Hu, K. A. Dahmen, W. Shi and G. C. Wong, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2330–2335 CrossRef CAS.
- J. Luciano, R. Agrebi, A. V. Le Gall, M. Wartel, F. Fiegna, A. Ducret, C. Brochier-Armanet and T. Mignot, PLoS Genet., 2011, 7, e1002268 CAS.
- T. Mignot, J. W. Shaevitz, P. L. Hartzell and D. R. Zusman, Science, 2007, 315, 853–856 CrossRef CAS.
- M. Sun, M. Wartel, E. Cascales, J. W. Shaevitz and T. Mignot, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 7559–7564 CrossRef CAS.
- C. Holz, D. Opitz, J. Mehlich, B. J. Ravoo and B. Maier, Nano Lett., 2009, 9, 4553–4557 CrossRef CAS.
- C. Meel, N. Kouzel, E. R. Oldewurtel and B. Maier, Small, 2012, 8, 530–534 CrossRef CAS.
- R. M. Miller, A. P. Tomaras, A. P. Barker, D. R. Voelker, E. D. Chan, A. I. Vasil and M. L. Vasil, J. Bacteriol., 2008, 190, 4038–4049 CrossRef CAS.
- P. J. Bonner, Q. Xu, W. P. Black, Z. Li, Z. Yang and L. J. Shimkets, Mol. Microbiol., 2005, 57, 1499–1508 CrossRef CAS.
- D. Bhaya, Mol. Microbiol., 2004, 53, 745–754 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
