Hierarchical multiscale modeling of macromolecules and their assemblies†
Abstract
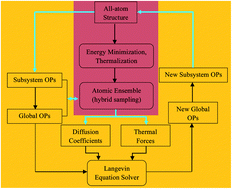
Soft materials (e.g., enveloped viruses, liposomes, membranes and supercooled liquids) simultaneously deform or display collective behaviors, while undergoing atomic scale vibrations and collisions. While the multiple space-time character of such systems often makes traditional molecular dynamics simulation impractical, a multiscale approach has been presented that allows for long-time simulation with atomic detail based on the co-evolution of slowly varying order parameters (OPs) with the quasi-equilibrium probability density of atomic configurations. However, this approach breaks down when the structural change is extreme, e.g., when nearest-neighbor connectivity between structural subsystems is not maintained. In the current study, a self-consistent approach is presented wherein OPs and a reference structure co-evolve slowly to yield long-time simulation for dynamical soft-matter phenomena such as structural transitions and

 Please wait while we load your content...
Please wait while we load your content...