 Open Access Article
Open Access ArticleA new approach to high resolution, high contrast electron microscopy of macromolecular block copolymer assemblies†
M. Adam
Dyson
a,
Ana M.
Sanchez
a,
Joseph P.
Patterson
b,
Rachel K.
O'Reilly
b,
Jeremy
Sloan
a and
Neil R.
Wilson
*a
aDepartment of Physics, University of Warwick, Coventry, UK. E-mail: neil.wilson@warwick.ac.uk
bDepartment of Chemistry, University of Warwick, Coventry, UK
First published on 20th February 2013
Abstract
Determining the structure of macromolecular samples is vital for understanding and adapting their function. Transmission electron microscopy (TEM) is widely used to achieve this, but, owing to the weak electron scattering cross-section of carbon, TEM images of macromolecular samples are generally low contrast and low resolution. Here we implement a fast and practically simple routine to achieve high-contrast imaging of macromolecular samples using exit wave reconstruction (EWR), revealing a new level of structural detail. This is only possible using ultra-low contrast supports such as the graphene oxide (GO) used here and as such represents a novel application of these substrates. We apply EWR on GO membranes to study self-assembled block copolymer structures, distinguishing not only the general morphology or nanostructure, but also evidence for the substructure (i.e. the polymer chains) which gives insight into their formation mechanisms and functional properties.
Introduction
Characterizing the structure of organic macromolecular materials at the sub-nanometer scale is a major challenge that is critical to advancing our understanding of biological processes and capitalizing on the remarkable opportunities afforded by self-assembly of amphiphilic macromolecules. A common approach is to use transmission electron microscopy (TEM), often with heavy metal staining.1 Although staining can be used to highlight functionality, it is often required merely to give sufficient contrast. Macromolecular samples are typically weak phase objects,2i.e. due to their low average atomic number they scatter the wave function of the electron beam changing its phase with little effect on the amplitude. The in-focus TEM image depends mainly on the intensity, i.e. on the amplitude, of the wave and hence the observed contrast for such samples is weak. Imaging them on conventional amorphous carbon supports is thus problematic, as contrast from the support usually dominates that from the object under investigation.Recent developments in ultra-low contrast TEM supports have now made it possible to routinely image organic macromolecular samples without staining. For example, graphene and its oxidised derivative graphene oxide (GO) are strong and almost transparent under the electron beam. For macromolecular samples, we have shown that using GO as a support it is possible to unambiguously resolve the morphology of block copolymer assemblies such as polymersomes without staining,3 whilst Pantelic et al. have demonstrated analysis of unstained vitrified biological macromolecules on GO with high signal to noise ratio (SNR).4 Pantelic et al. have also shown that due to their greater crystallinity and lower scattering cross-section graphene supports outperform even GO supports,5–7 and demonstrated their application through imaging the periodic structure of the tobacco mosaic virus.6 As that report exemplified, these supports decrease the noise (contrast in the background support) and thus increase the signal (scattering from the object) to noise ratio.
Coupled with advances in aberration corrected TEM (ac-TEM), which now routinely enables sub-Angstrom resolution imaging of crystalline inorganic materials, this should enable structural determination of macromolecular samples with unprecedented resolution. However, to get sufficient contrast it is typically necessary to image under defocus. This acts like Fresnel diffraction, interfering the scattered and unscattered waves to make phase contrast visible in the bright field TEM image, but in the process the resolution is reduced. This effect is captured in the contrast transfer function (CTF) which defines the information transferred from the object to the image and depends on the microscope and imaging parameters.8 The CTF oscillates from positive to negative as a function of spatial frequency, with the position of the oscillations dependent on focus. As the defocus is increased, the point resolution (defined as the first zero in the CTF) decreases. As a result, although the general morphology of the sample at the nanometre scale can be deduced by imaging under defocus, the higher spatial resolution information is generally lost. For a single image, contrast is typically only achieved by sacrificing resolution.
Approaches have been developed to overcome this problem. The use of a Zernike phase plate enables pure phase imaging, dramatically increasing image contrast for weak phase objects. Application of Zernike phase plates to TEM requires advances in instrumentation and additional hardware that have limited its uptake, but it has shown great promise for structural determination of biological specimens at subnanometer resolutions.9,10 To a limited extent, the CTF can be corrected for a single image, as is routinely applied for single particle analysis. However, this cannot recover the missing information from the length scales at which the CTF is zero and for noisy images CTF correction can even amplify the noise.11 A more sophisticated approach involves focal pair merging12 whereby information from two images, one near focus and one at large defocus, are combined to reconstruct a higher-contrast and higher-resolution image. In essence, this is similar to exit wave reconstruction (EWR), a technique developed to maximise the resolution of high resolution (HR)-TEM of inorganic materials.13–15 EWR works by acquiring a series of images whilst incrementally changing the focus (a focal series) and then recombining the information from each image to reconstruct the wave function as it left the object under investigation. In the process the image is corrected for aberrations, increasing the resolution that can be achieved. However, the crucial point here is that both the amplitude and phase of the wave function are recovered. EWR has previously been used for recovering phase information, for example as ‘inline electron holography’ for studying static magnetic and electric potentials,16 but has not been demonstrated for macromolecular samples such as the ones investigated here.
Here we show that through advances in support material and digital image acquisition and processing, it is now possible to use a fast and practically simple methodology to achieve high-contrast increased resolution imaging of macromolecular samples through exit wave reconstruction. The technique can be implemented without additional hardware on any conventional computer controlled TEM and results in significant improvements in image resolution and contrast for macromolecular objects. The information transferred is increased through the use of low-voltage aberration-corrected TEM. It is important to note that these advances are only possible through the use of ultra-low contrast supports, such as the graphene oxide sample support used here. We demonstrate the application of exit wave reconstruction through the analysis of two macromolecular structures formed by self-assembly of block copolymers, giving information beyond the basic nanostructure which is normally resolved. Imaging of an archetypal poly(acrylic acid)-b-poly(styrene) polymersome shows evidence for individual polymer chains that form a corona around it, while analysis of a poly(L-lactide) containing cylindrical micelle provides an indication of its formation mechanism.
Results and discussion
The effects of defocus on image contrast and resolution
Fig. 1 shows bright field TEM images of a polymersome from the solution assembly of the block copolymer poly(acrylic acid)11-b-poly(styrene)250 (at 0.65 mg mL−1) on a single layer GO support. The images were taken on a JEOL ARM200F operated at 80 kV with spherical aberration corrected to <1 μm. Note that GO is used rather than graphene due to the simplicity of preparation of the GO TEM supports (for the preparation method see for example the video abstract in ref. 17).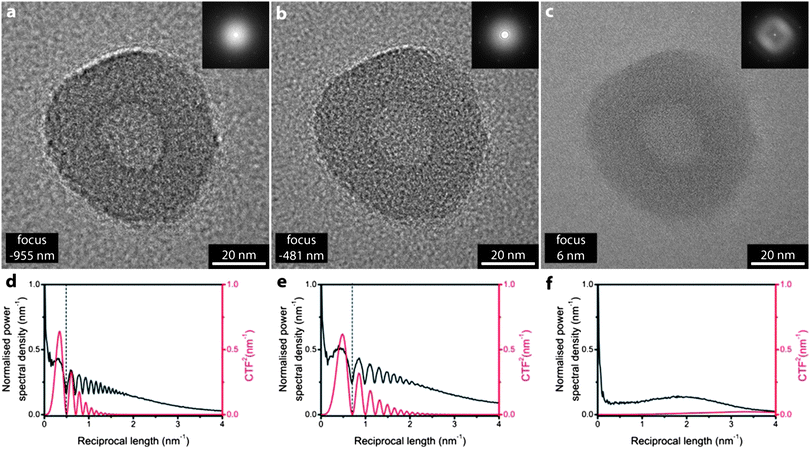 | ||
| Fig. 1 The effect of defocus on imaging. Bright field TEM images of a polyacrylic acid-b-polystyrene polymersome at (a) −955 nm focus, (b) −481 nm focus and (c) 6 nm focus. Inset are sections from their respective fast Fourier transforms. The normalized power spectral density (black) and square of the predicted contrast transfer function (red) for (a)–(c) are given in (d)–(f) respectively. | ||
The block copolymer was synthesized by the reversible addition fragmentation chain transfer (RAFT) polymerization, deprotected and then assembled using the solvent switch method as described previously.3 As shown in these images, and as expected from many previous investigations,18–20 this system self-assembles into well-defined polymersomes. The simplicity of the polymersome structure (a hollow sphere), its nanoscale dimensions and robustness of the morphology under irradiation in the electron beam make it an ideal test object. It also represents an important class of materials: self-assembled block copolymer systems in solution have applications in medicinal chemistry21–23 and catalysis.23–25 In both cases the primary function of the nanostructures is to provide a well-defined environment for the encapsulation of an active species (e.g. drugs, imaging agents, reactants, catalysts, etc.). The structure of the particles is thus particularly important as it defines their functional properties.
The effect of defocus on image resolution and contrast can readily be seen in Fig. 1. As the defocus is decreased from Fig. 1a to c the most obvious change is the decrease in contrast. The characteristic structure of the polymersome, which has a radius of ∼30 nm with a roughly spherical hollow core of radius ∼10 nm, is readily apparent in the under focus images Fig. 1a and b. But in the near focus image, Fig. 1c, the lack of contrast makes it difficult to accurately discern the interface between the polymersome and the support or the boundary of its hollow core. Inset in the images are sections of their fast Fourier transforms (FFTs) which convey the information content in the image as a function of reciprocal length and hence give a clear visualization of the resolution in the image (detail further from the centre in the FFT corresponds to higher spatial resolution in the image). The FFTs have an essentially circular symmetry, as expected for this amorphous material, except for the hexagon of spots due to the single layer of graphene oxide. These GO spots give a useful inbuilt calibration for the images (corresponding to 4.69 nm−1)26 and it is remarkable that they are seen in the FFT despite no evident graphene-like lattice in the real space image. Plots of the power spectral density are given, Fig. 1d and e, with direct comparison to the square of the predicted phase contrast transfer function. The CTF2 shows a series of minima, often called Thon rings, which are also evident as dark circles in the FFT. The inner Thon ring defines the point resolution of the image. From Fig. 1a to c the defocus decreases (from −955 nm to −481 nm to 6 nm) with a corresponding increase in the point resolution (from 0.5 nm−1 to 0.7 nm−1 to around 4 nm−1) but with a concomitant decrease in contrast evident from the drop in magnitude of the CTF2. Note that this dependence of resolution and contrast on focus is a fundamental limit. Although advances in instrumentation reduce the aberrations and hence improve the CTF, the oscillations in the CTF are an inevitable consequence of defocus. Hence for individual images there is a compromise between contrast and resolution.
High resolution and high contrast through exit wave reconstruction
The solution to this problem, as implemented in exit-wave reconstruction, is to acquire a focal series. An algorithm is then used to combine the images to reconstruct the phase of the wave function as it leaves the object under investigation, combining the information from every image. Fig. 2 shows the result of such a reconstruction as operated on a focal series containing 40 images, with a nominal focal step between images of 26 nm. The images in Fig. 1 are examples extracted from this focal series (the focal series is given in ESI†, S1). The EWR algorithm used here is based on the focal and tilt series reconstruction (FTSR) algorithm developed by Kirkland and co-workers.14,15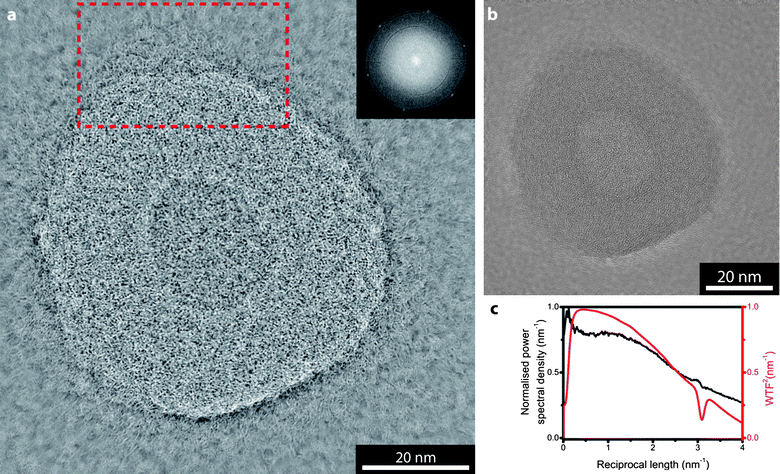 | ||
| Fig. 2 Exit wave reconstruction of the polymersome shown in Fig. 1. (a) phase image and (b) amplitude image. Inset in (a) is a section of its FFT, the red dashed box indicates the region shown in Fig. 3. In (c) the normalised power spectral density (black) for the phase image is compared with the square of the predicted wave transfer function (red). | ||
The EWR recreates the amplitude and phase of the exit wave as it leaves the object. The phase, Fig. 2a, shows both higher resolution and higher contrast than the original images while the amplitude, Fig. 2b, shows only weak contrast similar to the 6 nm focus image, Fig. 1c. This low amplitude contrast is as expected, and indeed as required to satisfy the weak phase object approximation used in the EWR algorithm.15 Inset in the phase image is a section of its FFT, again the hexagon of spots due to the graphene-like lattice is present and it is clear that the information content is uniform out to larger reciprocal distances than in the experimental images (i.e. they are higher resolution). The thin black arcs indicate that the reconstruction is not ideal, i.e. that information is missing. However, the amount of missing information is clearly small. Fig. 2c compares the power spectral density from the phase image with the predicted wave transfer function for this reconstruction (WTF, relates the exit wave reconstruction to the object in a similar way to how the CTF relates the image to the object27). The WTF2 illustrates that the EWR achieves both high contrast and high resolution, with a fairly flat curve out to beyond 4 nm−1 (2.5 Å).
Resolving the substructure in polymersomes
As with a conventional TEM image, the EWR phase image is effectively a two-dimensional projection of a three-dimensional object and so interpreting the image is non-trivial. In addition, these images were taken without consideration of the electron dose applied to the sample and so it is inevitable that damage induced by the electron beam will have altered the fine structure that is now resolved. However, the result of this increase in resolution and contrast is that the morphology and some aspects of the structure of the polymersome can be investigated in greater detail. For example, in the reconstructed phase image a corona can be resolved around the outside of the polymersome.A magnified view of the marked section of Fig. 2a is shown in Fig. 3, along with the same section of the near focus image (Fig. 1c). In the near focus image, Fig. 3a, due to the low contrast it is difficult to discern where the edge of the polymersome is and, although some structure is apparent, the corona is not clearly resolved. From this image alone it would be challenging to draw conclusions about the fine structure of the polymersome. By comparison, the structure revealed in the EWR phase image, Fig. 3b, is startlingly clear. The edge of the nanostructure is resolved and shown to be a gradual transition with a corona of polymeric material surrounding the polymersome bilayer.
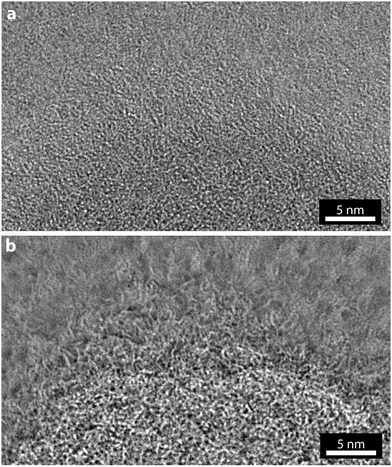 | ||
| Fig. 3 Comparison of conventional TEM imaging and EWR phase imaging. (a) Near focus TEM, and (b) EWR phase image, for the region of polymersome marked by the dashed box in Fig. 2. | ||
Three levels of hierarchy exist in the polymersomes; monomers, which are reacted together to form the polymers, which are assembled to form the nanostructures. In Fig. 1a–c only the highest level structure is observable (i.e. the size and shape of the nanostructure) whereas the EWR images give evidence for the structure at the next level down of information (i.e. the individual polymer chains) and consequently the method represents a significant step forward in imaging of self-assembled soft materials.
Practical considerations for EWR
The robustness of the reconstruction can readily be tested. Reconstructions using non-overlapping subsets of the focal series give the same result (ESI S3†), as do different EWR algorithms (ESI S5†). A test for consistency can be made by using the reconstructed exit wave to simulate the expected image as a function of focus and the results match the experimental images with a high degree of fidelity (ESI S6†).EWR does not require an aberration corrected TEM. On a conventional TEM operated under high resolution bright field imaging conditions EWR has an even greater effect, correcting some of the microscope aberrations and hence increasing the resolution further as well as restoring the high contrast of the phase image. The high magnification TEM image of a polyacrylic acid-b-polystyrene polymersome (as in Fig. 1–3) on graphene oxide shown in Fig. 4 was taken on a Jeol 2100 TEM with LaB6 filament operated at 200 kV. Fig. 4a shows a near focus image (−12 nm defocus), one of a focal series of 20 images with focal step of 34 nm. The amplitude and phase images from EWR of the focal series are given in Fig. 4b and c respectively. There are clear similarities in the fine structure of the polymersome resolved in the phase image here compared to that obtained from the aberration corrected TEM in Fig. 2c, although it is also readily apparent that the low voltage aberration corrected images result in greater information transfer to the EWR images.
 | ||
| Fig. 4 EWR of a polymersome on GO using a conventional TEM at 200 kV. (a) Near focus image (focus −12 nm), (b) EWR amplitude image and (c) EWR phase image. | ||
Advances in digital acquisition and processing now make EWR a fast and relatively simple technique. For this work, acquisition of the focal series has been automated by a script written for Gatan's Digital Micrograph (DMG) software, the most common TEM image acquisition software, and takes around a minute for a typical 20 image sequence. During this acquisition the sample exposure time need only be a few seconds, the majority of the time is the latency in taking the images from the camera. We have found that there is no need to go to large defocus; provided the SNR in the images is sufficient to align the images, focal steps of around 25 nm starting near focus give good results. We have increased the speed of the EWR algorithm and integrated a version into DMG so that a basic reconstruction can be performed in the microscope software in a few minutes immediately after image acquisition (ESI S5†).‡ This rapid acquisition and reconstruction allows EWR analysis to be used as a routine technique. The full reconstruction, including additional refinements of the image series alignment and determination of focal drift which are not accounted for in the rapid EWR algorithm, has also been accelerated by utilising general-purpose computing on graphics processing units so that it takes between 1 and 15 seconds for a full FTSR reconstruction depending on the number of images and pixels. The EWR algorithms require some prior information from the specific microscope being used (for example the modulation transfer function of the camera, the aberrations and the minimum focal increment27), but this information need only be acquired once and subsequently acquiring the focal series requires no more expertise than conventional TEM imaging. Most reconstruction algorithms are valid only in the linear imaging regime (i.e. thin samples with limited scattering). However, reconstructions with more complete algorithms such as the maximum likelihood method (MAL),28 which are valid even for significant non-linear contributions, provide similar results (ESI S5†). This indicates that EWR is valid for these comparatively large macromolecular samples, however, for these thicker amorphous samples interpreting the contrast is complicated. In a similar way, the sample support also plays a critical role.
The importance of an ultra-low contrast support
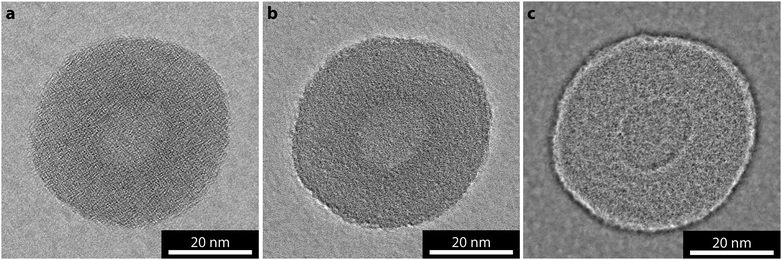
In recreating the phase image through EWR, the contrast and resolution at which the support is imaged is also increased. As a result, in order to resolve a weak phase object, EWR places strict requirements on the thickness and structure of the support. Fig. 5 compares images of a poly(lactide) containing cylindrical micelle, which was prepared as previously reported,29 on an amorphous carbon support Fig. 5a–c and a GO support Fig. 5d–f. In the near focus image of the cylindrical micelle on amorphous carbon, Fig. 5a, the cylindrical micelle shows little contrast with respect to the background, indicating a low SNR as expected. This is one of a focal series of 20 images with 26 nm focal step. The amplitude image from the EWR of this series, Fig. 5b, shows similarly low contrast to the near focus image. The structure and contrast in the amorphous carbon support dominates the phase image, Fig. 5c, making it difficult to discern where the cylindrical micelle is and almost impossible to separate the structure of the micelle from that of the support.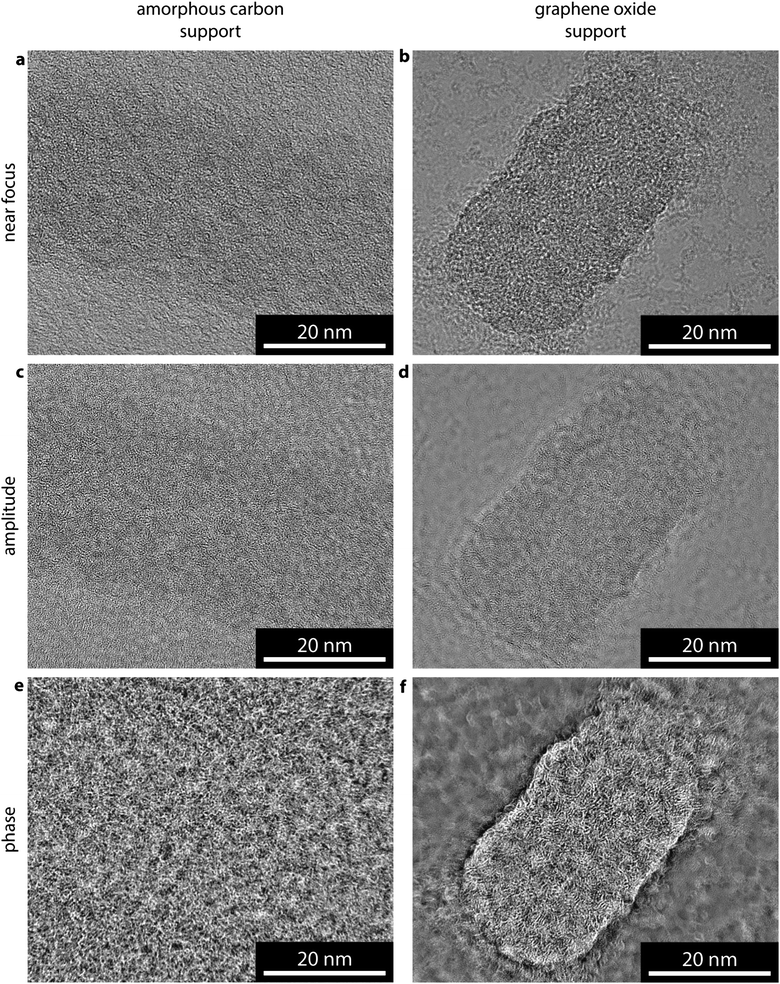 | ||
| Fig. 5 Comparison of EWR on amorphous carbon and graphene oxide supports. (a and d) Near focus, (b and e) EWR amplitude, and (c and f) EWR phase images of a poly(lactide) containing cylindrical micelle on amorphous carbon and graphene oxide supports respectively. The near focus images were taken with focus of −5 nm (a), and −30 nm (d). | ||
Insight into self-assembly of cylindrical micelles through EWR
The near focus image of a similar micelle on GO, Fig. 5d, shows a significantly higher SNR with the micelle clearly resolved. This is from a focal series of 20 images with 26 nm focal step. The resultant EWR amplitude image, Fig. 5e, is again similar to the near focus image. However, the phase image gives high contrast structural detail about the cylindrical micelle. The image represents a two-dimensional projection through a three-dimensional structure making it difficult to interpret the three-dimensional arrangement of the block copolymers inside the micelle and, as for the polymersome, the images were taken without consideration of electron dose so that some beam damage is inevitable. However, new structural insight can be gained from this image. Comparison with Fig. 2a shows that this structure has more order, i.e. more parallel, aligned features, consistent with these micelles being semi-crystalline.30 In addition the sides of the cylindrical micelle appear more structurally confined (closed) than the ends which appear less densely packed (open), this supports a unimer assembly process as suggested recently.29 This assembly mechanism proposes that the ends of the cylinder are ‘living’ and therefore structurally different from the sides; in the near focus image on GO, Fig. 5d, this is difficult to discern but becomes clear in the EWR image, Fig. 5f. Being able to probe small structural differences within individual self-assembled structures, promises far greater mechanistic insight into the synthesis of such systems.31,32 Furthermore, the closed sides of the cylinders confirm the confined hydrophobic environment within these structures although the open ends may affect the ability of the nanostructures to retain hydrophobic guests within the core for extended periods, which would have implications for their application as encapsulation or delivery vessels.Conclusions
The information extracted here from EWR analysis demonstrates the capability of this technique. The polymersome and cylindrical micelle represent important classes of self-assembled structures, particularly for biomedical applications. The potential to resolve structural detail at the polymer chain level promises to give greater insight into the function of these nanomaterials. For example, the corona length, spacing and cross-linking density are critical parameters which dictate circulation times and stability.21 These parameters can be probed by bulk scattering techniques, however, as is demonstrated in the analysis of the cylindrical micelles, in some cases there are structural differences within individual self-assembled structures. Therefore, the ability to analyze individual particles is of paramount importance.We also expect that EWR utilising ultra-low background supports will be applicable to many other macromolecular samples, including biological ones. To advance this technique it will be critical to image under low-dose conditions and to demonstrate its compatibility with vitrified samples. The reconstruction in Fig. 2 used a focal series consisting of 40 images, but as shown in ESI S4† most of the information can be recovered with far fewer images, nominally down to three. Sufficient SNR has to be achieved in each image to enable their alignment, but the latest (second) generation of direct detection cameras that feature electron counting and high speed read out will enable the electron dose to be minimised, making it feasible to rapidly acquire a focal series with the same dose currently required for one low dose image. Here, again, the importance of the support is paramount. As Pantelic et al. have shown, graphene and GO supports offer significantly increased SNR over conventional supports even for hydrated vitrified samples.4,5
We have shown that the new generation of ultra-low contrast supports enable the application of imaging techniques for macromolecular samples that were not practical before. Exit wave reconstruction significantly increases both the contrast and the resolution with which macromolecular samples can be investigated in the TEM. Conventional TEM images resolve only the general morphology or nanostructure of these samples; here we have shown that EWR has the capability of distinguishing the next level of structural detail with the potential to reveal insights into formation mechanisms and functional properties.
Experimental
Materials
Chemicals were used as received from Aldrich, Fluka and Acros. L-Lactide was donated by Purac and further purified/dried over 4 Å molecular sieves in dichloromethane before being dried under vacuum and sublimed. Lactides were stored in a nitrogen-filled glove box. Tetrahydropyran acrylate (THPA) was prepared as described previously and stored below 4 °C.33 Lacy carbon and ultrathin (6 nm) amorphous carbon TEM grids were purchased from Agar Scientific.Synthesis and self-assembly of nanomaterials
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 water–tetrahydrofuran at 65 °C were used to deprotect the tetrahydropyran groups and self-assemble the resulting P(AA)333-b-P(L-LA)37 amphiphile into cylindrical micelles.
1 water–tetrahydrofuran at 65 °C were used to deprotect the tetrahydropyran groups and self-assemble the resulting P(AA)333-b-P(L-LA)37 amphiphile into cylindrical micelles.
Graphene oxide was prepared using a modified Hummers' technique as reported previously.26
Preparation of GO-TEM grids
Dried GO powder was re-dispersed in water at 0.1 mg mL−1 by stirring overnight. Solutions were then sonicated for approximately 5 minutes to exfoliate the sheets. Prior to deposition of the GO, lacy carbon TEM grids were cleaned and made hydrophilic by exposure to an air plasma (otherwise known as glow discharge or ‘plasma ash’ treatment) to aid the dispersion and adhesion of the GO sheets on the grids. The TEM grids were placed on a piece of filter paper and one drop (from a glass pipette, ca. 0.08 mL) of GO solution was dropped onto the grids. Most of the GO solution was wicked away by the filter paper and the grids were left to dry for ca. 30 minutes.Application of aqueous assemblies to GO-TEM grids
4 μL of aqueous sample was added to the GO-TEM grids and left for 2 minutes before being blotted away using filter paper. The grids were then left to dry in air for at least 30 minutes.Application of aqueous samples to 6 nm thick carbon TEM grids
The TEM grids were glow discharge treated. 4 μL of aqueous sample was then added and the grids left for 2 minutes before being blotted away using filter paper. The grids were left to dry in air for at least 30 minutes.Electron microscopy
The focal steps in the microscopes were calibrated by taking pairs of images with varying focal differences, and the focal difference between the pairs of images determined via the phase compensated phase correlation function (PCPCF) method.15 Focal series of 20 to 40 images were recorded via a custom EWR plugin for Digital Micrograph with focal increments of approximately 20 to 30 nm between images. The aberration corrected JEOL JEM-ARM200F was operated at 80 kV with spherical aberration (Cs) tuned to approximately +1 μm and images were recorded on a Gatan SC-1000 Orius CCD camera. The JEOL JEM-2100 LaB6 was operated at 200 kV (estimated Cs = 1 mm) and images were recorded on a Gatan SC-600 Orius CCD camera.Acknowledgements
We thank Nikos Petzetakis for help with cylindrical micelle sample preparation. We thank the EPSRC for funding through a studentship for M. A. D. and support through the Warwick Centre for Analytical Science (EP/F034210/1). A. M. S. thanks the Science City Research Alliance and the HEFCE Strategic Development Fund for funding support. Some items of equipment that were used in this research were funded by Birmingham Science City, with support from Advantage West Midlands and part funded by the European Regional Development Fund.Notes and references
- J. R. Harris and D. Scheffler, Micron, 2002, 33, 461–480 CrossRef
.
- R. H. Wade, Ultramicroscopy, 1992, 46, 145–156 CrossRef CAS
.
- J. P. Patterson, A. M. Sanchez, N. Petzetakis, T. P. Smart, T. H. Epps III, I. Portman, N. R. Wilson and R. K. O'Reilly, Soft Matter, 2012, 8, 3322–3328 RSC
.
- R. S. Pantelic, J. C. Meyer, U. Kaiser, W. Baumeister and J. M. Plitzko, J. Struct. Biol., 2010, 170, 152–156 CrossRef CAS
.
- R. S. Pantelic, J. W. Suk, Y. Hao, R. S. Ruoff and H. Stahlberg, Nano Lett., 2011, 11, 4319–4323 CrossRef CAS
.
- R. S. Pantelic, J. C. Meyer, U. Kaiser and H. Stahlberg, Solid State Commun., 2012, 152, 1375–1382 CrossRef CAS
.
- R. S. Pantelic, J. W. Suk, C. W. Magnuson, J. C. Meyer, P. Wachsmuth, U. Kaiser, R. S. Ruoff and H. Stahlberg, J. Struct. Biol., 2011, 174, 234–238 CrossRef CAS
.
-
D. B. Williams and C. B. Carter, Transmission Electron Microscopy: Imaging, Plenum Press, 1996 Search PubMed
.
- W.-H. Chang, M. T. K. Chiu, C.-Y. Chen, C.-F. Yen, Y.-C. Lin, Y.-P. Weng, J.-C. Chang, Y.-M. Wu, H. Cheng, J. Fu and I. P. Tu, Structure, 2010, 18, 17–27 CrossRef CAS
.
- K. Murata, X. Liu, R. Danev, J. Jakana, M. F. Schmid, J. King, K. Nagayama and W. Chiu, Structure, 2010, 18, 903–912 CrossRef CAS
.
- K. H. Downing and R. M. Glaeser, Ultramicroscopy, 2008, 108, 921–928 CrossRef CAS
.
- S. J. Ludtke and W. Chiu, J. Struct. Biol., 2003, 144, 73–78 CrossRef
.
- L. J. Allen, W. McBride, N. L. O'Leary and M. P. Oxley, Ultramicroscopy, 2004, 100, 91–104 CrossRef CAS
.
- A. I. Kirkland and R. R. Meyer, Microsc. Microanal., 2004, 10, 401–413 CrossRef CAS
.
- R. R. Meyer, A. I. Kirkland and W. O. Saxton, Ultramicroscopy, 2002, 92, 89–109 CrossRef CAS
.
- C. T. Koch and A. Lubk, Ultramicroscopy, 2010, 110, 460–471 CrossRef CAS
.
- N. R. Wilson, P. A. Pandey, R. Beanland, J. P. Rourke, U. Lupo, G. Rowlands and R. A. Römer, New J. Phys., 2010, 12, 125010 CrossRef
.
- T. Azzam and A. Eisenberg, Angew. Chem., Int. Ed., 2006, 45, 7443–7447 CrossRef
.
- P. Lim Soo and A. Eisenberg, J. Polym. Sci., Part B: Polym. Phys., 2004, 42, 923–938 CrossRef
.
- L. F. Zhang and A. Eisenberg, Science, 1995, 268, 1728–1731 CAS
.
- M. Elsabahy and K. L. Wooley, Chem. Soc. Rev., 2012, 41, 2545–2561 RSC
.
- J. Z. Du and R. K. O'Reilly, Soft Matter, 2009, 5, 3544–3561 RSC
.
- F. H. Schacher, P. A. Rupar and I. Manners, Angew. Chem., Int. Ed., 2012, 51, 7898–7921 CrossRef CAS
.
- K. T. Kim, S. A. Meeuwissen, R. J. Nolte and J. C. van Hest, Nanoscale, 2010, 2, 844–858 RSC
.
- P. Cotanda, A. Lu, J. P. Patterson, N. Petzetakis and R. K. O'Reilly, Macromolecules, 2012, 45, 2377–2384 CrossRef CAS
.
- N. R. Wilson, P. A. Pandey, R. Beanland, R. J. Young, I. A. Kinloch, L. Gong, Z. Liu, K. Suenaga, J. P. Rourke, S. J. York and J. Sloan, ACS Nano, 2009, 3, 2547–2556 CrossRef CAS
.
-
R. R. Meyer, Ph.D. thesis, Technische Universität Dresden, 2002
.
- W. M. J. Coene, A. Thust, M. Op de Beeck and D. Van Dyck, Ultramicroscopy, 1996, 64, 109–135 CrossRef CAS
.
- N. Petzetakis, D. Walker, A. P. Dove and R. K. O'Reilly, Soft Matter, 2012, 8, 7408–7414 RSC
.
- N. Petzetakis, A. P. Dove and R. K. O'Reilly, Chem. Sci., 2011, 2, 955–960 RSC
.
- X. Wang, G. Guerin, H. Wang, Y. Wang, I. Manners and M. A. Winnik, Science, 2007, 317, 644–647 CrossRef CAS
.
- J. B. Gilroy, T. Gaedt, G. R. Whittell, L. Chabanne, J. M. Mitchels, R. M. Richardson, M. A. Winnik and I. Manners, Nat. Chem., 2010, 2, 566–570 CrossRef CAS
.
- K. L. Wooley, R. K. O'Reilly, M. J. Joralemon and C. J. Hawker, Chem.–Eur. J., 2006, 12, 6776–6786 CrossRef
.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional information on EWR and full original image series. See DOI: 10.1039/c3sm27787a |
| ‡ Plugin available on request from E-mail: m.a.dyson@warwick.ac.uk |
| This journal is © The Royal Society of Chemistry 2013 |
