 Open Access Article
Open Access ArticleMetallohelices with activity against cisplatin-resistant cancer cells; does the mechanism involve DNA binding?†
Viktor Brabec*b, Suzanne E. Howsona, Rebecca A. Kanera, Rianne M. Lordc, Jaroslav Malinab, Roger M. Phillipsd, Qasem M. A. Abdallahd, Patrick C. McGowanc, Alison Rodgera and Peter Scott*a
aDepartment of Chemistry, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL, UK. E-mail: peter.scott@warwick.ac.uk; Fax: +44 (0)24 7657 2710; Tel: +44 (0)24 7652 3238
bInstitute of Biophysics, Academy of Sciences of the Czech Republic, v.v.i., Královopolská 135, CZ-61265 Brno, Czech Republic. E-mail: brabec@ibp.cz
cSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK
dInstitute of Cancer Therapeutics, University of Bradford, Bradford, BD7 1DP, UK
First published on 10th September 2013
Abstract
Enantiomers of a relatively rigid DNA-binding metallo-helix are shown to have comparable activity to that of cisplatin against the cell lines MCF7 (human breast adenocarcinoma) and A2780 (human ovarian carcinoma) but are ca five times more active against the cisplatin-resistant A2780cis. The cell-line HCT116 p53+/+ (human colon carcinoma) is highly sensitive giving IC50 values in the nM range, far lower than the cisplatin control. The hypothesis that the biological target of such metallohelices is DNA is probed by various techniques. Tertiary structure changes in ct-DNA (formation of loops and intramolecular coiling) on exposure to the compounds are demonstrated by atomic force microscopy and supported by circular/linear dichroism in solution. Selectivity for 5′-CACATA and 5′-CACTAT segments is shown by DNase I footprinting. Various three- and four-way oligonucleotide junctions are stabilised, and remarkably only the Λ metallo-helix enantiomer stabilizes T-shaped 3WJs during gel electrophoresis; this is despite the lack of a known helix binding site. In studies with oligonucleotide duplexes with bulges it is also shown for the first time that the metallo-helix binding strength and the number of binding sites are dependent on the size of the bulge. In contrast to all the above, flexible metallo-helices show little propensity for structured or selective DNA binding, and while for A2780 the cancer cell line cytotoxicity is retained the A2780cis strain shows significant resistance. For all compounds in the study, H2AX FACS assays on HCT116 p53+/+ showed that no significant DNA damage occurs. In contrast, cell cycle analysis shows that the DNA binders arrest cells in the G2/mitosis phase, and while all compounds cause apoptosis, the DNA binders have the greater effect. Taken together these screening and mechanistic results are consistent with the more rigid helices acting via a DNA binding mechanism while the flexible assemblies do not.
Introduction
Clinical anticancer treatments commonly include the use of DNA-binding or -modifying drugs. Alkylators and DNA cleavage agents cause chemically irreversible reactions leading, in the absence of repair, to cell death,1,2 whereas DNA groove-binders and intercalators form non-covalent bonds, commonly affecting replication and transcription.3,4 Coordination chemistry plays an important role in both these areas;5,6 cisplatin chemotherapy is a resurgent field of research7–9 and metal scaffolds are being used to explore regions of biologically relevant chemical space inaccessible to organic chemistry alone.10–13One such architecture is provided by metal-templated helicates14–19 (Fig. 1, upper). These molecules commonly comprise three relatively rigid ditopic ligands such as the NN–NN system I20 wrapped in a helical array around two metal ions. The ligand (referred to as a helicand) mechanically couples21 the helicity at the two metal centres such that they have the same absolute configuration (Δ or Λ). Their structural novelty has encouraged testing in the healthcare arena e.g. towards diagnostic applications22–29 but there are rather few metallo-helix systems that lend themselves to projects in drug discovery.19
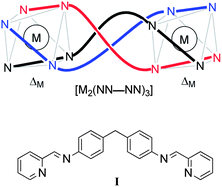 | ||
| Fig. 1 Schematic representation (upper) of one enantiomer of a bimetallic triple-stranded metallo-helicate incorporating a NN–NN bis(bidentate) helicand. | ||
Hannon's tetracations [M2(I)3]4+ (M = Fe, Ru) have been preeminent because of their simplicity and solubility,30 and despite their requirement for chromatographic resolution. They bind in the major groove of DNA with sequence-selectivity,31 induce intramolecular coiling32 and have some anticancer activity.33–35 Qu has discovered that the same Fe compound recognises human telomeric G-quadruplex DNA36–38 and targets the amyloid β peptide, reducing cytotoxicity and ameliorating memory deficits in a transgenic mouse model.39 Recently a derivative of [Fe2(I)3]4+ containing arginine units showed improved cytotoxicity against A2780 ovarian cancer cells.40
We recently developed optically and diastereochemically pure monometallic complexes containing functionalised pyridine/imine units41,42 and then used them to create via self-assembly processes water stable, optically pure bimetallic structures with flexible linkers [M2L3]2+ (Fig. 2).43 Since the stereoselectivity in these complexes does not rely on the helicate concept of mechanical coupling we describe them as flexicates,43,‡ although in other instances mechanical coupling is important.44 We also reported their promising antibiotic activity against MRSA (Methicillin-resistant Staphylococcus aureus) and Escherichia coli alongside modest toxicity towards the nematode worm Caenorhabditis elegans.43 In this work we describe the discovery of structure-dependent activity of flexicates against cancer cell-lines, including a cisplatin-resistant strain, and address the interactions of these compounds with potential drug targets in DNA and DNA-motifs via a range of biophysical techniques relevant to mechanism of action.45 In addition we probe the effects on cells via assay of DNA damage and cell-cycle analysis.
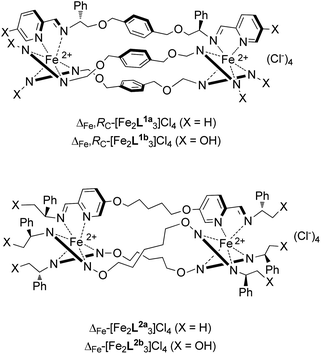 | ||
| Fig. 2 Diastereomerically pure metallo-helical “flexicate” complexes of chiral ligands L1 and L2. | ||
Results and discussion
Cancer cell-line activity
The absence of reactive metal centres in the helicates [M2(I)3]4+ (Fig. 1) described above means that a cisplatin-like lesion process is unlikely to be responsible for their cancer cell line cytotoxicity, yet it has been hypothesised that DNA binding is involved in the mechanism.33–35 The flexicate compounds [Fe2L3]4+ (Fig. 2) present an interesting series in this respect; linear dichroism studies indicate that while [Fe2L1a3]4+ binds to the major groove of calf thymus DNA, the system [Fe2L2a3]4+ does not. We thus set out to investigate the activity of the water soluble flexicates in cancer cell lines; MCF7 (human breast adenocarcinoma), A2780 (human ovarian carcinoma), the cisplatin resistant strain A2780cis and HCT116 p53+/+ (human colon carcinoma). In preliminary tests it was found that solutions of [Fe2L1b3]Cl4 deposited solid complex under assay conditions and so our studies were confined to the remaining six compounds (three pairs of enantiomers) containing L1a, L2a and L2b. The free ligands and major components were insufficiently soluble in DMSO/water for testing.The flexicates show promising anticancer properties with a range of IC50 values from ca 0.6–20 μM (Table 1). The enantiomers containing L1a were generally the most potent, and while a direct comparison with related helicates33,34 is not easy to make, reference to the cisplatin control experiments indicates that the activities here are at least comparable with the more recently reported arginine derivatives of Hannon.40 The most striking result however is that flexicates based on L1a exhibit potent cytotoxic effects on the cisplatin-resistant ovarian tumour cell line A2780cis. The effect is marked; there is an approximate 5-fold difference between the IC50 values for these flexicates and cisplatin towards A2780cis. Furthermore A2780cis is more sensitive to these L1a flexicates than is the parental line A2780.
| Complex | IC50/μM (esd) | |||
|---|---|---|---|---|
| MCF7 | A2780 | A2780cis | HCT116 p53+/+ | |
| Cisplatin | 1.33 (0.23) | 0.93 (0.04) | 10.46 (0.15) | 3.51 (1.50) |
| Λ-[Fe2L1a3]Cl4 | 3.67 (0.14) | 4.80 (0.15) | 2.18 (0.07) | 1.66 (1.05) |
| Δ-[Fe2L1a3]Cl4 | 2.95 (0.77) | 3.75 (0.10) | 2.39 (0.12) | 0.61 (0.31) |
| Λ-[Fe2L2a3]Cl4 | 5.50 (0.52) | 3.29 (0.09) | 7.34 (0.32) | 0.62 (0.08) |
| Δ-[Fe2L2a3]Cl4 | 10.16 (0.18) | 3.48 (0.04) | 14.39 (0.39) | 0.87 (0.13) |
| Λ-[Fe2L2b3]Cl4 | 6.08 (0.21) | 4.29 (0.08) | 12.72 (0.03) | 0.64 (0.15) |
| Δ-[Fe2L2b3]Cl4 | 8.26 (0.16) | 3.10 (0.10) | 18.20 (0.15) | 0.61 (0.07) |
The flexicates based on L2a/2b are generally less cytotoxic but the differences in sensitivity between cell lines (i.e. selectivity) is marked. In particular A2780cis is resistant to these compounds, with IC50 values 2–6 times higher than those for the parent cell line. At the same time, some of the differences in IC50 between enantiomers are very significant with the Λ-[Fe2L2a3]Cl4 isomer being more potent. Again, this contrasts with the L1a complex enantiomers where only marginal differences were observed.
The HCT116 p53+/+ cell line is sensitive to all the flexicates and IC50 values in the nM range are observed, significantly lower than the cisplatin control.
Across the panel there are large differences in sensitivity to individual compounds e.g. a factor of ca 30 in sensitivity between the highest and lowest IC50 observations for Δ-[Fe2L2b3]Cl4.
Given the range of IC50 values in Table 1 and the ready availability and stability in water of enantiomers of the test compounds we have been able to address in more detail the relationship between the binding of various DNA motifs and cytotoxicity.
Atomic force microscopy
The influence of the flexicates on the tertiary structure of single DNA molecules was studied by direct visualisation of linearized plasmid pSP73 (2464 bp) mixed with increasing concentrations of the flexicates using atomic force microscopy (AFM) tapping mode in air. Samples for imaging were prepared by adsorption of the flexicates onto freshly cleaved mica in the presence of 5 mM Mg(II). The non-modified linearized plasmid molecules appeared as relaxed and well-separated strands; crossing strands were rarely observed [see ESI, Fig. S1(a and b)†]. The addition of [Fe2L1a3]Cl4 and [Fe2L2a3]Cl4 flexicates affected the conformation of linear plasmid DNA yielding typical images as shown in Fig. 3.![AFM images of linear plasmid pSP73 (2464 bp) mixed with flexicates at various DNA base : flexicate ratios. (A–C) DNA with ΛFe,SC-[Fe2L1a3]Cl4 at 5 : 1, 3 : 1 and 2 : 1 ratios, respectively. (D–F) DNA with ΛFe,RC-[Fe2L2a3]Cl4 at 1 : 2, 1 : 4 and 1 : 6 ratios, respectively.](/image/article/2013/SC/c3sc51731d/c3sc51731d-f3.gif) | ||
Fig. 3 AFM images of linear plasmid pSP73 (2464 bp) mixed with flexicates at various DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate ratios. (A–C) DNA with ΛFe,SC-[Fe2L1a3]Cl4 at 5 flexicate ratios. (A–C) DNA with ΛFe,SC-[Fe2L1a3]Cl4 at 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratios, respectively. (D–F) DNA with ΛFe,RC-[Fe2L2a3]Cl4 at 1 1 ratios, respectively. (D–F) DNA with ΛFe,RC-[Fe2L2a3]Cl4 at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1 2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and 1 4 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 ratios, respectively. 6 ratios, respectively. | ||
For [Fe2L1a3]Cl4, an increase in formation of loops and close strand contacts was seen for both enantiomers at lower DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate ratios up to ∼5
flexicate ratios up to ∼5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (shown for Λ-enantiomer in Fig. 3(A)). On increasing the concentration of flexicates (Fig. 3(B and C)) intramolecular coiling and intermolecular aggregation of DNA molecules was more pronounced. A level of cooperativity in the coiling process was apparent, since clusters and fully coiled DNA strands were observed in the presence of uncoiled DNA molecules. The extent of coiling and aggregation was slightly higher for the Λ enantiomer than the Δ enantiomer. The effects of the [Fe2L1a3]Cl4 flexicates on DNA coiling differ from that reported for the [Fe2(I)3]Cl4 helicate32 which induced almost exclusively intramolecular coiling of individual DNA molecules and did not exhibit a tendency to condense DNA into intermolecular clusters [see ESI, Fig. S1(c)†].
1 (shown for Λ-enantiomer in Fig. 3(A)). On increasing the concentration of flexicates (Fig. 3(B and C)) intramolecular coiling and intermolecular aggregation of DNA molecules was more pronounced. A level of cooperativity in the coiling process was apparent, since clusters and fully coiled DNA strands were observed in the presence of uncoiled DNA molecules. The extent of coiling and aggregation was slightly higher for the Λ enantiomer than the Δ enantiomer. The effects of the [Fe2L1a3]Cl4 flexicates on DNA coiling differ from that reported for the [Fe2(I)3]Cl4 helicate32 which induced almost exclusively intramolecular coiling of individual DNA molecules and did not exhibit a tendency to condense DNA into intermolecular clusters [see ESI, Fig. S1(c)†].
Typical AFM images of linear DNA mixed with [Fe2L2a3]Cl4 are shown in Fig. 3(D–F) and indicate different behaviour from [Fe2L1a3]Cl4. No intramolecular coiling was observed even at high loadings of [Fe2L2a3]Cl4 (Fig. 3(F)). Thick-stranded features made by tight coiling of two individual DNA strands and some intermolecular aggregates were predominantly seen (Fig. 3(E and F)). To observe a similar level of DNA condensation as seen with the [Fe2L1a3]Cl4 flexicates, the concentration of [Fe2L2a3]Cl4 was required to be approximately 10–20-fold higher. No difference between enantiomers was noticed. These observations are entirely consistent with the [Fe2L2a3]Cl4 flexicates only showing weak electrostatic binding to DNA.
DNA binding studies using spectroscopic methods
Circular dichroism titration experiments were used to investigate the binding of the water soluble [Fe2L3]Cl4 flexicate enantiomers to calf-thymus DNA (ct-DNA) as shown in Fig. 4 and in ESI.† Changes to peaks below 300 nm are predominantly due to the intrinsic ct-DNA CD signals that occur in this region. In the MLCT region, i.e. that of the intense bisignate curves between 450 and 650 nm, subtle changes in the intensity and wavelength are consistent with distortion of the complex in the region of the Fe(pyridine/imine)3 chromophore on binding to the DNA. The ΔFe,RC-[Fe2L1a3]Cl4 flexicate shows the largest change of the tested complexes. The changes in the CD of the corresponding Λ isomer of L1a (Fig. 4) as well as of the flexicates based on L2a and L2b (see ESI, Fig. S2 and S3†) are much smaller and therefore suggest smaller distortions to the geometries of the complexes on binding to DNA. Perturbations of charge-transfer absorptions were also observed in ct-DNA/[Fe2(I)3]4+ solutions.46![CD titration series for ΔFe/ΛFe-[Fe2L1a3]Cl4 at constant flexicate concentrations (15 μM) and increasing ct-DNA concentrations (indicated in legend). TRIZMA® base buffer (1 mM, pH 7.2). Path length 1.0 cm.](/image/article/2013/SC/c3sc51731d/c3sc51731d-f4.gif) | ||
| Fig. 4 CD titration series for ΔFe/ΛFe-[Fe2L1a3]Cl4 at constant flexicate concentrations (15 μM) and increasing ct-DNA concentrations (indicated in legend). TRIZMA® base buffer (1 mM, pH 7.2). Path length 1.0 cm. | ||
The extent to which these interactions cause disruption in the DNA structure in solution may be estimated from the reduction in the intensity of the DNA linear dichroism (LD) absorption at ca 260 nm. Suitable linear dichroism (LD) spectra (Fig. S4–S9) and details of calculation of the peak intensity are given in ESI.† The data are summarised in Fig. 5. This reduction in intensity almost certainly comes from bending or coiling, as we have seen in the solid state by AFM, that reduces alignment of the DNA with the laminar flow in the cell. It is rather less likely to result from an increase in the flexibility of the DNA. The percentage reduction in the DNA peak follows the trend L1a > L2a > L2b. The ΛFe compounds cause more disruption to the DNA structure, indicating stronger preferences for binding with the ΛFe enantiomer in each case. In comparison to the iron(II) flexicates reported here, [Fe2(I)3]4+ causes a higher degree of disruption to the DNA structure at the same DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) complex ratio32,47 and this correlates well with AFM data.
complex ratio32,47 and this correlates well with AFM data.
![Comparison of the reduction in the linear dichroism absorption of the ct-DNA 260 nm peak in the presence of various complexes. DNA base : complex ratio 7 : 1 throughout. Values for ΔFe/ΛFe-[Fe2L1a3]Cl4 flexicates are calculated using film LD and UV-Vis absorbance data. Data from Hannon and co-workers taken from previously reported work.32,47](/image/article/2013/SC/c3sc51731d/c3sc51731d-f5.gif) | ||
Fig. 5 Comparison of the reduction in the linear dichroism absorption of the ct-DNA 260 nm peak in the presence of various complexes. DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) complex ratio 7 complex ratio 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 throughout. Values for ΔFe/ΛFe-[Fe2L1a3]Cl4 flexicates are calculated using film LD and UV-Vis absorbance data. Data from Hannon and co-workers taken from previously reported work.32,47 1 throughout. Values for ΔFe/ΛFe-[Fe2L1a3]Cl4 flexicates are calculated using film LD and UV-Vis absorbance data. Data from Hannon and co-workers taken from previously reported work.32,47 | ||
DNase I footprinting
In order to obtain information on sequence-specificity in the binding events that appear to lead to the tertiary structure changes described above, footprinting methodology was used. Deoxyribonuclease I (DNase I) is the most commonly used nuclease for these experiments and the reaction conditions used were such that, on average, each DNA strand was cut once giving a mixture of different fragments. A compound bound to DNA protects the DNA from cleavage at their binding sites.Each flexicate was mixed with the 158 bp HindIII/NdeI restriction fragment of pSP73 followed by partial cleavage by DNase I. The autoradiograms of the DNA cleavage inhibition patterns are shown in Fig. 6. Comparing the patterns observed in the presence and absence of the flexicates shows some evidence of footprints in the gel particularly for the ΛFe,SC-[Fe2L1a3]Cl4 flexicate around positions 50 and 70 (Fig. 6(a), lane 2 and Fig. 6(b), lanes 2, 3) indicating specific sequences in DNA are recognised. There are only minor differences between the autoradiograms for the flexicates at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (DNA base
1 (DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate) ratios (Fig. 6(b)).
flexicate) ratios (Fig. 6(b)).
![Autoradiogram of DNase I footprint of 3′ end labeled top strand of the 158 bp HindIII/NdeI restriction fragment of the plasmid pSP73 in the presence of the flexicates. The nucleotide sequence of the fragment is shown on the right side of the gel and numbers refer to the sequence shown in the corresponding differential cleavage plots in Fig. 8. For (a) lane 1: DNA in the absence of flexicates, lanes 2–7: DNA mixed with ΛFe,SC-[Fe2L1a3]Cl4, ΔFe,RC-[Fe2L1a3]Cl4, ΛFe,RC-[Fe2L2a3]Cl4, ΔFe,SC-[Fe2L2a3]Cl4, ΛFe,SC-[Fe2L2b3]Cl4, ΔFe,RC-[Fe2L2b3]Cl4 respectively. All at 10 : 1 (DNA base : flexicate) ratios. For (b) lane 1: DNA in the absence of flexicates, lanes 2 and 3: DNA mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 20 : 1 and 10 : 1 (DNA base : flexicate) ratios respectively, lanes 4 and 5: DNA mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 20 : 1 and 10 : 1 ratios respectively, lanes G + A and G correspond to Maxam–Gilbert G + A and G ladders.](/image/article/2013/SC/c3sc51731d/c3sc51731d-f6.gif) | ||
Fig. 6 Autoradiogram of DNase I footprint of 3′ end labeled top strand of the 158 bp HindIII/NdeI restriction fragment of the plasmid pSP73 in the presence of the flexicates. The nucleotide sequence of the fragment is shown on the right side of the gel and numbers refer to the sequence shown in the corresponding differential cleavage plots in Fig. 8. For (a) lane 1: DNA in the absence of flexicates, lanes 2–7: DNA mixed with ΛFe,SC-[Fe2L1a3]Cl4, ΔFe,RC-[Fe2L1a3]Cl4, ΛFe,RC-[Fe2L2a3]Cl4, ΔFe,SC-[Fe2L2a3]Cl4, ΛFe,SC-[Fe2L2b3]Cl4, ΔFe,RC-[Fe2L2b3]Cl4 respectively. All at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (DNA base 1 (DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate) ratios. For (b) lane 1: DNA in the absence of flexicates, lanes 2 and 3: DNA mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 20 flexicate) ratios. For (b) lane 1: DNA in the absence of flexicates, lanes 2 and 3: DNA mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10 1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (DNA base 1 (DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate) ratios respectively, lanes 4 and 5: DNA mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 20 flexicate) ratios respectively, lanes 4 and 5: DNA mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10 1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratios respectively, lanes G + A and G correspond to Maxam–Gilbert G + A and G ladders. 1 ratios respectively, lanes G + A and G correspond to Maxam–Gilbert G + A and G ladders. | ||
Further information on the binding specificity of the [Fe2L1a3]Cl4 enantiomers was obtained from the intensities of the bands from the gel lanes in Fig. 6. For bands containing DNA mixed with the [Fe2L1a3]Cl4 enantiomers at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (DNA base
1 (DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate) ratios were measured by densitometry and the resulting differential cleavage plots are shown in Fig. 7. Negative values indicate inhibition of DNase I cleavage at that section whereas positive values indicate enhancement. It can be seen that ΔFe,RC-[Fe2L1a3]Cl4 does not display significant patterns of protection and enhancement. Conversely, the differential cleavage plot for ΛFe,SC-[Fe2L1a3]Cl4 contains two major regions where the flexicate has protected the DNA from DNase I cleavage. These sites are centred around positions 52 and 70 and extend over approximately 5–6 bp. Applying a shift of 2–3 bp in the 3′ direction is necessary to correct for the fact that DNase I binds across the minor groove.48,49 From this assumption, the two preferential binding sites of ΛFe,SC-[Fe2L1a3]Cl4 can be deduced as 5′-CACATA and 5′-CACTAT starting at positions 51 and 69, respectively. Hence there appear to be fewer preferential binding sites than has been observed31,50 for [Fe2(I)3]4+ and [Ru2(I)3]4+.
flexicate) ratios were measured by densitometry and the resulting differential cleavage plots are shown in Fig. 7. Negative values indicate inhibition of DNase I cleavage at that section whereas positive values indicate enhancement. It can be seen that ΔFe,RC-[Fe2L1a3]Cl4 does not display significant patterns of protection and enhancement. Conversely, the differential cleavage plot for ΛFe,SC-[Fe2L1a3]Cl4 contains two major regions where the flexicate has protected the DNA from DNase I cleavage. These sites are centred around positions 52 and 70 and extend over approximately 5–6 bp. Applying a shift of 2–3 bp in the 3′ direction is necessary to correct for the fact that DNase I binds across the minor groove.48,49 From this assumption, the two preferential binding sites of ΛFe,SC-[Fe2L1a3]Cl4 can be deduced as 5′-CACATA and 5′-CACTAT starting at positions 51 and 69, respectively. Hence there appear to be fewer preferential binding sites than has been observed31,50 for [Fe2(I)3]4+ and [Ru2(I)3]4+.
![Caption differential cleavage plots for the ΛFe- and ΔFe-enantiomers of [Fe2L1a3]Cl4 showing the induced differences in susceptibility to DNase I digestion on the 158 bp HindIII/NdeI restriction fragment of the plasmid pSP73 at 10 : 1 (DNA base : flexicate) ratio. The vertical scale is in units of ln(fc) − ln(f0), where fc is the fractional cleavage at any bond in the presence of flexicate and f0 is the fractional cleavage of the same bond in the control. Positive values indicate enhancement, negative values indicate inhibition.](/image/article/2013/SC/c3sc51731d/c3sc51731d-f7.gif) | ||
Fig. 7 Caption differential cleavage plots for the ΛFe- and ΔFe-enantiomers of [Fe2L1a3]Cl4 showing the induced differences in susceptibility to DNase I digestion on the 158 bp HindIII/NdeI restriction fragment of the plasmid pSP73 at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (DNA base 1 (DNA base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) flexicate) ratio. The vertical scale is in units of ln(fc) − ln(f0), where fc is the fractional cleavage at any bond in the presence of flexicate and f0 is the fractional cleavage of the same bond in the control. Positive values indicate enhancement, negative values indicate inhibition. flexicate) ratio. The vertical scale is in units of ln(fc) − ln(f0), where fc is the fractional cleavage at any bond in the presence of flexicate and f0 is the fractional cleavage of the same bond in the control. Positive values indicate enhancement, negative values indicate inhibition. | ||
Supramolecular interactions of flexicates with oligonucleotide motifs
In this section we investigate the possibility of flexicate interactions with other potential drug target DNA structures relevant to DNA replication and recombination.Three-way junctions
Three-way junctions (3WJs) consist of three double strands converging at one point (Fig. 8).51 Typically found in both DNA (e.g. replication forks)52 and RNA,53 they are potential drug targets. Fig. 8(a) shows a Y-shaped 3WJ (Tm = 35.0 °C) of the type shown to bind one particular helicate molecule in the central cavity.54 T-shaped 3WJs, which do not in their relaxed state have such a cavity are found where unpaired bases appear in the junction region;51 the systems of Fig. 8(b and c) have two unpaired adenosines (3WJ-AA) and two unpaired thymidines (3WJ-TT) and higher melting temperatures of 41.4 °C and 38.5 °C respectively. As shown in Table 2, addition of flexicates to solutions of all of these oligonucleotide assemblies increases the melting temperatures (Tm) i.e. increases thermal stability.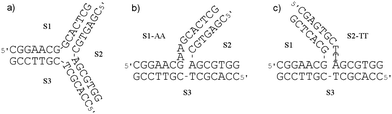 | ||
| Fig. 8 The sequences of the oligonucleotides used to form (a) Y-shaped 3WJ, and T-shaped 3WJs containing two unpaired (b) adenosines 3WJ-AA or (c) thymidines 3WJ-TT at the branch point of the junction. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate
1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios
3WJ) ratios
| Complex | Y-shaped 3WJ ΔTm (°C) | T-shaped 3WJ-AA ΔTm (°C) | T-shaped 3WJ-TT ΔTm (°C) | |||
|---|---|---|---|---|---|---|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | |
| Λ-[Fe2L1a3]Cl4 | 20.1 | 20.3 | 8.3 | 9.4 | 9.6 | 10.7 |
| Δ-[Fe2L1a3]Cl4 | 18.2 | 18.5 | 7.0 | 8.1 | 9.6 | 10.7 |
| Λ-[Fe2L2a3]Cl4 | 9.0 | 10.4 | 0.2 | 0.4 | 2.8 | 3.6 |
| Δ-[Fe2L2a3]3Cl4 | 13.1 | 12.6 | 1.7 | 2.5 | 5.4 | 6.2 |
| Λ-[Fe2L2b3]Cl4 | 7.1 | 8.9 | 0.5 | 0.8 | 2.6 | 3.5 |
| Δ-[Fe2L2b3]Cl4 | 11.0 | 11.8 | 1.6 | 2.4 | 5.1 | 6.2 |
The L1a flexicates are consistently more efficient at stabilising the 3WJs than are the L2 flexicates. In all cases, doubling the ratio from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2
1 to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate
1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) had almost no further effect on the stability suggesting all the 3WJs contain just one major binding site.
3WJ) had almost no further effect on the stability suggesting all the 3WJs contain just one major binding site.
Gel electrophoresis was used to gain more information about the stabilisation effect that the flexicates bring to DNA 3WJs.55 The three 14 base single strand oligonucleotides used in the Y-shaped 3WJ of Fig. 8(a) provide the minimum length required for the assembly to persist during electrophoresis at low temperatures (5 °C) in the presence of magnesium(II) ions.56 Without Mg2+ ions present, these 3WJs split into single strands [Fig. 9(a) lane C]. Fig. 9(a) shows that both [Fe2L1a3]4+ enantiomers (lanes 1–6) are able to stabilise the Y-shaped 3WJ during electrophoresis in the absence of Mg2+ at room temperature. The L2 flexicates, ΔFe/ΛFe-[Fe2L2a3]Cl4 (lanes 7, 8) and ΔFe/ΛFe-[Fe2L2b3]Cl4 (lanes 9, 10), had no or very little effect on the stability of this 3WJ under these conditions. Consistent with the melting experiments above, a plot of the percentage of 3WJs remaining after electrophoresis as a function of flexicate concentration [Fig. 9(b)] clearly shows that for [Fe2L1a3]Cl4, the ΛFe enantiomer stabilises the Y-shaped 3WJ during electrophoresis to a greater extent compared with the ΔFe enantiomer (Fig. 10).
![(a) Autoradiogram of the gel run at room temperature. Lane ss: control containing a single strand. Lane C: control containing all three strands S1, S2 and S3. Lane 1–3: S1, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5 : 1, 1 : 1 and 2 : 1 (flexicate : 3WJ) ratios, respectively. Lanes 4–6: S1, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5 : 1, 1 : 1 and 2 : 1 (flexicate : 3WJ) ratios, respectively. Lanes 7–10: S1, S2 and S3 mixed with ΛFe,RC-[Fe2L2a3]Cl4 (7), ΔFe,SC-[Fe2L2a3]Cl4 (8), ΛFe,SC-[Fe2L2b3]Cl4 (9) and ΔFe,RC-[Fe2L2b3]Cl4 (10) at 1 : 1 (flexicate : 3WJ) ratio. (b) Plot of the % of 3WJs remaining after electrophoresis as a function of flexicate concentration.](/image/article/2013/SC/c3sc51731d/c3sc51731d-f9.gif) | ||
Fig. 9 (a) Autoradiogram of the gel run at room temperature. Lane ss: control containing a single strand. Lane C: control containing all three strands S1, S2 and S3. Lane 1–3: S1, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios, respectively. Lanes 4–6: S1, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5 3WJ) ratios, respectively. Lanes 4–6: S1, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios, respectively. Lanes 7–10: S1, S2 and S3 mixed with ΛFe,RC-[Fe2L2a3]Cl4 (7), ΔFe,SC-[Fe2L2a3]Cl4 (8), ΛFe,SC-[Fe2L2b3]Cl4 (9) and ΔFe,RC-[Fe2L2b3]Cl4 (10) at 1 3WJ) ratios, respectively. Lanes 7–10: S1, S2 and S3 mixed with ΛFe,RC-[Fe2L2a3]Cl4 (7), ΔFe,SC-[Fe2L2a3]Cl4 (8), ΛFe,SC-[Fe2L2b3]Cl4 (9) and ΔFe,RC-[Fe2L2b3]Cl4 (10) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratio. (b) Plot of the % of 3WJs remaining after electrophoresis as a function of flexicate concentration. 3WJ) ratio. (b) Plot of the % of 3WJs remaining after electrophoresis as a function of flexicate concentration. | ||
![Autoradiograms of the gels run at room temperature. Lane C1: control containing the three strands, S1-AA, S2 and S3. Lanes 1–3: S1-AA, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5 : 1, 1 : 1 and 2 : 1 (flexicate : 3WJ) ratios, respectively. Lanes 4–6: S1-AA, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5 : 1, 1 : 1 and 2 : 1 (flexicate : 3WJ) ratios, respectively. Lane C2: control containing the three strands, S1-TT, S2 and S3. Lanes 7–9: S1-TT, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5 : 1, 1 : 1 and 2 : 1 (flexicate : 3WJ) ratios, respectively. Lanes 4–6: S1-TT, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5 : 1, 1 : 1 and 2 : 1 (flexicate : 3WJ) ratios, respectively.](/image/article/2013/SC/c3sc51731d/c3sc51731d-f10.gif) | ||
Fig. 10 Autoradiograms of the gels run at room temperature. Lane C1: control containing the three strands, S1-AA, S2 and S3. Lanes 1–3: S1-AA, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios, respectively. Lanes 4–6: S1-AA, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5 3WJ) ratios, respectively. Lanes 4–6: S1-AA, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios, respectively. Lane C2: control containing the three strands, S1-TT, S2 and S3. Lanes 7–9: S1-TT, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5 3WJ) ratios, respectively. Lane C2: control containing the three strands, S1-TT, S2 and S3. Lanes 7–9: S1-TT, S2 and S3 mixed with ΛFe,SC-[Fe2L1a3]Cl4 at 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios, respectively. Lanes 4–6: S1-TT, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5 3WJ) ratios, respectively. Lanes 4–6: S1-TT, S2 and S3 mixed with ΔFe,RC-[Fe2L1a3]Cl4 at 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2 1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate 1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3WJ) ratios, respectively. 3WJ) ratios, respectively. | ||
The related gel electrophoresis experiments on the T-shaped 3WJs of Fig. 8(b and c)] show an extraordinary difference in the behaviour of enantiomers ΔFe/ΛFe-[Fe2L1a3]Cl4; with both 3WJ-AA and 3WJ-TT, stabilisation was only observed with the ΛFe enantiomer (lanes 1–3 and 7–9).
The mechanisms of binding to Y- and T-shaped 3WJ must thus differ. With the Y-shaped 3WJ, it is likely that the flexicates bind to the central hollow cavity at the branch point of the junction;54 notably the ΔTm are consistently higher for these “preorganised” systems. In contrast for the T-shaped 3WJs we propose the structure of the junction may be being changed on binding to accommodate the flexicate; something that may explain the enhanced enantioselectivity.
We note that while only Λ isomer of [Fe2L1a3]Cl4 stabilizes T-shaped 3WJ-TT during gel electrophoresis, both Λ and Δ compounds increase the melting temperatures equally. A plausible explanation might be while the gel electrophoresis in the absence of Mg2+ was carried out at room temperature (22 °C) and observes the proportion of 3WJs remaining after electrophoresis, the melting temperatures (∼40 °C) rather reflect the properties of the 3WJs at the elevated temperatures when 3WJs start to melt. Thus, melting temperatures may not reflect relatively small differences in the capability of the two isomers to stabilize T-shaped 3WJs-TT whereas the gel electrophoresis apparently can.
Four-way junctions
Four-way junctions (4WJs), e.g. the Holliday junction,57 consist of four double strands converging at one point and are also potential DNA drug targets.58 The thermal stability of the 4WJ shown in Fig. 11 (Tm = 42.4 °C) in the presence of the flexicates was investigated. The L2 systems (ΔFe/ΛFe-[Fe2L2a3]Cl4 and ΔFe/ΛFe-[Fe2L2b3]Cl4) had small or even a negative effect on the stability of this 4WJ (Table 3). The melting temperature was, however, found to increase in the presence of both enantiomers of the [Fe2L1a3]Cl4 flexicates with the ΛFe enantiomer providing some additional stability in comparison to the ΔFe enantiomer. Additionally it was found that the increase in melting temperature of this 4WJ as the concentration of the L1a flexicates increases above the stoichiometric ratio suggests that, unlike the 3WJs, the 4WJ contains more than one binding site for these complexes.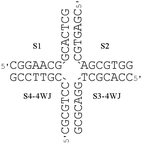 | ||
| Fig. 11 The sequences of the oligonucleotides used to form the 4WJ. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate
1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4WJ) ratios
4WJ) ratios
| Flexicate | ΔTm (°C) of 4WJ at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | ΔTm (°C) of 4WJ at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
|---|---|---|
| Λ-[Fe2L1a3]Cl4 | 6.7 | 9.7 |
| Δ-[Fe2L1a3]Cl4 | 5.8 | 6.5 |
| Λ-[Fe2L2a3]Cl4 | −1.5 | −1.5 |
| Δ-[Fe2L2a3]Cl4 | −1.4 | −1.4 |
| Λ-[Fe2L2b3]Cl4 | 0.0 | 0.2 |
| Δ-[Fe2L2b3]Cl4 | 0.5 | 1.0 |
Oligonucleotide duplexes with bulges
Oligonucleotide duplexes with bulges occur when one or more of the bases on one of the strands have no base(s) on the complementary strand to form a base-pair with. Bulge sites in DNA have been shown to bind some proteins more strongly than standard duplex DNA59 and therefore are of significant interest as targets for novel drugs,60,61 particularly in the context of peptidomimetic helices. The thermal stabilities of the oligonucleotide duplexes containing three-, two- and one-adenine bulges (A3–A1) [Fig. 12(a–c)] in the presence of the flexicates were analysed. The melting temperature of the duplexes depends on the size of the bulge, increasing from 35.2 °C (A3) to 38.7 °C (A2) to 45.0 °C (A1) as the number of unpaired adenines in the bulge decreases. The same duplex without a bulge was also investigated as a control and has a melting temperature 54.9 °C. The results are listed in Table 4 and show that only the ΔFe/ΛFe-[Fe2L1a3]Cl4 flexicates have positive impacts on the thermal stability.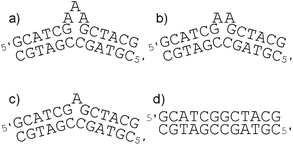 | ||
| Fig. 12 (a–c) The sequences of the oligonucleotide duplexes containing a three-, two- and one-adenine bulge, respectively. (d) The corresponding duplex used as a control. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flexicate
1 (flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) duplex) ratiosa
duplex) ratiosa
| Complex | A3 bulge ΔTm (°C) | A2 bulge ΔTm (°C) | A1 bulge ΔTm (°C) | No bulge ΔTm (°C) | ||||
|---|---|---|---|---|---|---|---|---|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 | |
| a Tm for only one set of exo-imine flexicates with A2 and Al bulges were measured due to their similarly low stabilising effects with the A3 bulge. As stabilisation was only observed with ΔFe/ΛFe-[Fe2L1a3]Cl4, Tm for the no-bulge control was measured for these flexicates only. | ||||||||
| Λ-[Fe2L1a3]Cl4 | 8.2 | 10.2 | 4.6 | 5.5 | 3.6 | 6.9 | 1.2 | 2.4 |
| Δ-[Fe2L1a3]Cl4 | 8.8 | 10.3 | 4.2 | 5.5 | 1.4 | 2.5 | 0.0 | 0.0 |
| Λ-[Fe2L2a3]Cl4 | 0.2 | 0.1 | −0.1 | −0.1 | −0.3 | −0.7 | — | — |
| Δ-[Fe2L2a3]3Cl4 | 0.1 | 0.2 | −0.1 | −0.1 | −0.1 | −0.1 | — | — |
| Λ-[Fe2L2b3]Cl4 | 0.0 | 0.1 | — | — | — | — | — | — |
| Δ-[Fe2L2b3]Cl4 | 0.2 | 0.1 | — | — | — | — | — | — |
With the duplexes containing A3 and A2 bulges, the stabilising effect of the flexicates decreases with the size of the bulge. The thermal stability of these duplexes is further increased when the flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) duplex ratio is increased from 1
duplex ratio is increased from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2
1 to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, but the increase is only relatively small (1–2°). This is consistent with a single dominant binding site for the flexicates on these bulges with up-take slightly higher at the increased 2
1, but the increase is only relatively small (1–2°). This is consistent with a single dominant binding site for the flexicates on these bulges with up-take slightly higher at the increased 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentration. In addition, there are no significant differences between enantiomers.
1 concentration. In addition, there are no significant differences between enantiomers.
The effect of ΔFe/ΛFe-[Fe2L1a3]Cl4 on the thermal stability of the duplex containing the A1 bulge shows different trends. At a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 flexicate
1 flexicate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) duplex ratio the increase in melting temperature is relatively small, however on increasing this ratio to 2
duplex ratio the increase in melting temperature is relatively small, however on increasing this ratio to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 the ΔTm approximately doubles, suggesting at least two binding sites are present per duplex. This indicates a different binding mode of ΔFe/ΛFe-[Fe2L1a3]Cl4 to the A1 bulge than to the A2 and A3 bulges. Furthermore, enantiomeric differences are observed with the A1 bulge. The ΛFe enantiomer has the greater stabilising effect on the A1 bulge, with ΔTm more than twice that observed for the ΔFe enantiomer.
1 the ΔTm approximately doubles, suggesting at least two binding sites are present per duplex. This indicates a different binding mode of ΔFe/ΛFe-[Fe2L1a3]Cl4 to the A1 bulge than to the A2 and A3 bulges. Furthermore, enantiomeric differences are observed with the A1 bulge. The ΛFe enantiomer has the greater stabilising effect on the A1 bulge, with ΔTm more than twice that observed for the ΔFe enantiomer.
H2AX expression analysis
We noted earlier that in contrast to DNA cleavage agents and “alkylators” (including cisplatin), groove-binders reversibly form non-covalent bonds such as those indicated in the above studies, so it is of interest in terms of the mechanism of action in cells to study the extent of DNA damage. H2AX is a histone which plays a key role in the repair of damaged DNA62–65 and its expression is a useful marker for several types of DNA damage.66 We selected the cell line HCT116 p53+/+ for this study since it is sensitive to all the flexicates.HCT116 p53+/+ cells (5 × 105 cells in 10 ml RPMI medium) were incubated with 10 μM of each flexicate for 24 h (a dose that kills 55 to 72% of cells) under appropriate conditions (see ESI†). These, along with a control of untreated HCT116 p53+/+ cells were analysed using fluorescence-activated cell sorting (FACS). Fig. 13 shows that there are no significant alterations in the production of H2AX, indicating that no DNA damage (increase in expression) or interruption of the H2AX pathway (decrease) has occurred.
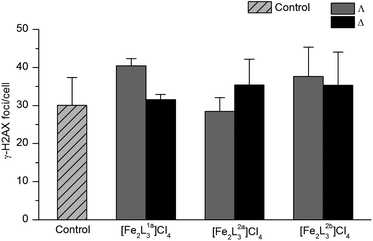 | ||
| Fig. 13 H2AX expression of HCT116 p53+/+ cells after treatment with flexicates (10 μM) for 24 h, and untreated cells (control). | ||
Cell cycle analysis
We further investigated mechanism of action of flexicates by assessing how they affect the population of phases of the cell cycle for HCT116 p53+/+. Using standard methods the cells were permeabilised and treated with the fluorescent dye propidium iodide (PI) which stains DNA quantitatively. The proportion of cells in the various phases – sub G1 (apoptotic67), G1 (increase in size in readiness for DNA replication), S (DNA synthesis), G2 (preparation for mitosis) and M (mitosis) – were determined by fluorescence via FACS analysis as a result of the differing amounts of DNA in the cells (see ESI†).As can be seen in Fig. 14, [Fe2L1a3]Cl4 flexicates show a dramatic increase in the proportion of cells in G2/M phase compared to the control (from ca 20 to 40% of cells); such arresting of cell growth at this phase is likely to be a significant factor in the mechanism of action. As in the cell-free DNA binding studies, the ΛFe enantiomer has the strongest effect of all compounds tested. Interestingly [Fe2L2a–b3]Cl4 flexicates did not show a significant increase in the G2/M phase, indicating a different mechanism. Fig. 14 also shows a very pronounced increase in population of sub G1 cells, from 4% in the control to between 13 and 30%, the latter being again for ΛFe-[Fe2L1a3]Cl4. Cells in the sub G1 phase are considered apoptotic67,68 and this data is thus consistent with flexicates inducing programmed cell death. Apoptosis induced by [Fe2L2a–b3]Cl4 flexicates varies little between enantiomers; this is consistent with the chemosensitivity data reported in this cell line.
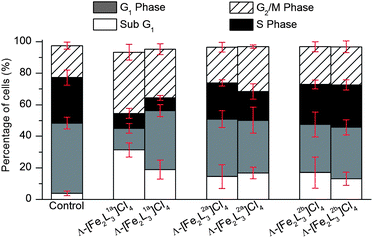 | ||
| Fig. 14 Cell cycle analysis of HCT116 p53+/+ cells after treatment with flexicates (10 μM) for 24 h, and untreated cells (control). | ||
Conclusions
We have shown that while the flexicate enantiomers based on L1a have comparable activity to cisplatin against MCF7 (human breast adenocarcinoma) and A2780 (human ovarian carcinoma) they are ca five times more potent than cisplatin against A2780cis. For L2a/b complexes the trend is reversed and A2780cis is rather resistant to L2a/b. The human colon carcinoma cell line HCT116 p53+/+ is very sensitive to all the compounds, giving IC50 values in the nM range, substantially more potent than cisplatin. For some flexicates the range of sensitivity is an order of magnitude or greater, indicating very promising selectively.The L1a flexicates are very good DNA binders and induce tertiary structural changes of the DNA as seen by AFM and correlated with solution spectroscopic techniques. The location of binding in the major groove is supported by the observation of nucleotide sequence selectivity according to DNase I footprinting. The overall picture is that the degree of DNA distortion falls in the order L1a > L2a > L2b. Given the very similar dimensions of all the complexes we suggest that this may be a function of flexibility; the linker units in L1a are much more rigid than those in L2a/b. Also, while the L1a complexes cause formation of loops and close strand contacts, with intramolecular coiling and intermolecular aggregation at higher loading the L2a complexes require substantially higher concentrations before tight coiling of two individual DNA strands was observed.
Binding of the flexicates to various other chemotherapeutic drug targets (3WJs, 4WJs and oligonucleotide duplexes with bulges) also follows the trend L1a ≫ L2a > L2b. Fascinatingly, only the Λ isomer of [Fe2L1a3]Cl4 stabilised T-shaped 3WJs during gel electrophoresis indicating the importance of our approach to creating optically pure and non-racemising systems. We have also shown for the first time that the interactions (binding strength and number of binding sites) of metallo-helices with oligonucleotide duplexes with bulges are dependent on the size of the bulge. This suggests that different sized bulges may be targeted by changing flexicate dimensions; something that is almost uniquely available from our approach to metallo-helix design. It is possible for example to design more or less flexible, longer or fatter, or differently functionalised analogues; this will feature in future reports aimed at further probing the existing correlation between metallohelix–DNA interactions and cytotoxic action.
The H2AX expression data for all compounds in the study indicate that if reversible DNA major groove binding is involved in the mode of action, it does not cause DNA lesions; the chemosensitivity is not caused by DNA damage. The cell cycle analysis in HCT116 p53+/+ cells nevertheless shows dramatic changes in cell-cycle population and that the compounds, particularly the strongest DNA binders, induce apoptosis.
Taken together, the trends in the cytotoxicity data, the dramatically different abilities to bind DNA motifs and the cell cycle analysis suggest that the biological targets of [Fe2L1a3]Cl4 and [Fe2L2a/b3]Cl4 are different. The potency of [Fe2L1a3]Cl4 against cisplatin resistant A2780cis and its ability to strongly and selectively bind DNA suggests the involvement of such an event in the mechanism. We also note that the flexible systems L2a/b are based on a pyridine analogue of the antimicrobial/antiparasitic agent pentamidine,69 which is a potent minor groove binder70 and has been proposed as an antitumor drug.71 The data here do not exclude the possibility that the assemblies [Fe2L2a/b3]4+ are acting as vectors for delivery of a ligand L2 or ligand fragments that otherwise have insufficient solubility for cancer cell-line testing. Nevertheless, we know that the mechanism of action of [Fe2L2a/b3]4+ does not involve DNA binding or significant DNA damage, and the observed differences in chemosensitivity for particularly the L2a compounds probably does not arise from differential DNA binding ability.
These anticancer and preliminary mechanistic results provide a case for further investigating derivatives, both as α-helix mimetic groove binders and pro-drugs, depending on the detail of the metallo-helix design.
Acknowledgements
This work was supported by: EPSRC; the University of Warwick; the Czech Science Foundation (Grant P205/11/0856) and the Academy of Sciences of the Czech Republic (Grant M200041201). The authors also acknowledge that their participation in the EU COST Action CM1105 enabled them to exchange regularly the most recent ideas in the field of metallodrugs with several European colleagues.Notes and references
- D. Fu, J. A. Calvo and L. D. Samson, Nat. Rev. Cancer, 2012, 12, 104–120 CAS.
- Z. H. Siddik, Oncogene, 2003, 22, 7265–7279 CrossRef CAS PubMed.
- P. L. Hamilton and D. P. Arya, Nat. Prod. Rep., 2012, 29, 134–143 RSC.
- G. S. Khan, A. Shah, R. Zia-ur-Rehman and D. Barker, J. Photochem. Photobiol., B, 2012, 115, 105–118 CrossRef CAS PubMed.
- T. W. Hambley, Dalton Trans., 2007, 4929–4937 RSC.
- T. W. Hambley, Science, 2007, 318, 1392–1393 CrossRef CAS PubMed.
- Y. Jung and S. J. Lippard, Chem. Rev., 2007, 107, 1387–1407 CrossRef CAS PubMed.
- D. Wang and S. J. Lippard, Nat. Rev. Drug Discovery, 2005, 4, 307–320 CrossRef CAS PubMed.
- L. Kelland, Nat. Rev. Cancer, 2007, 7, 573–584 CrossRef CAS PubMed.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179–3192 RSC.
- J. K. Barton, E. D. Olmon and P. A. Sontz, Coord. Chem. Rev., 2011, 255, 619–634 CrossRef CAS PubMed.
- S. Blanck, J. Maksimoska, J. Baumeister, K. Harms, R. Marmorstein and E. Meggers, Angew. Chem., Int. Ed., 2012, 51, 5244–5246 CrossRef CAS PubMed.
- K. J. Kilpin and P. J. Dyson, Chem. Sci., 2013, 4, 1410–1419 RSC.
- J. M. Lehn, A. Rigault, J. Siegel, J. Harrowfield, B. Chevrier and D. Moras, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 2565–2569 CrossRef CAS.
- C. Piguet, M. Borkovec, J. Hamacek and K. Zeckert, Coord. Chem. Rev., 2005, 249, 705–726 CrossRef CAS.
- M. Albrecht, Chem. Rev., 2001, 101, 3457–3498 CrossRef CAS PubMed.
- M. J. Hannon and L. J. Childs, Supramol. Chem., 2004, 16, 7–22 CrossRef CAS.
- D. L. Caulder and K. N. Raymond, Acc. Chem. Res., 1999, 32, 975–982 CrossRef CAS.
- S. E. Howson and P. Scott, Dalton Trans., 2011, 40, 10268–10277 RSC.
- M. J. Hannon, C. L. Painting, A. Jackson, J. Hamblin and W. Errington, Chem. Commun., 1997, 1807–1808 RSC.
- J. Xu, T. N. Parac and K. N. Raymond, Angew. Chem., Int. Ed., 1999, 38, 2878–2882 CrossRef CAS.
- M. Elhabiri, R. Scopelliti, J.-C. G. Bünzli and C. Piguet, J. Am. Chem. Soc., 1999, 121, 10747–10762 CrossRef CAS.
- N. André, T. B. Jensen, R. Scopelliti, D. Imbert, M. Elhabiri, G. Hopfgartner, C. Piguet and J.-C. G. Bünzli, Inorg. Chem., 2004, 43, 515–529 CrossRef PubMed.
- J.-C. G. Bünzli, Acc. Chem. Res., 2006, 39, 53–61 CrossRef PubMed.
- A.-S. Chauvin, S. Comby, B. Song, C. D. B. Vandevyver, F. Thomas and J.-C. G. Bünzli, Chem.–Eur. J., 2007, 13, 9515–9526 CrossRef CAS PubMed.
- C. D. B. Vandevyver, A.-S. Chauvin, S. Comby and J.-C. G. Bünzli, Chem. Commun., 2007, 1716–1718 RSC.
- J.-C. G. Bünzli, A.-S. Chauvin, C. D. B. Vandevyver, S. Bo and S. Comby, Ann. N.Y. Acad. Sci., 2008, 1130, 97–105 CrossRef PubMed.
- V. Fernández-Moreira, B. Song, V. Sivagnanam, A.-S. Chauvin, C. D. B. Vandevyver, M. Gijs, I. Hemmilä, H.-A. Lehr and J.-C. G. Bünzli, Analyst, 2010, 135, 42–52 RSC.
- J.-C. G. Bünzli, C. D. B. Vandevyver, A.-S. Chauvin, M. Gijs and A.-A. Lehr, CHIMIA International Journal for Chemistry, 2011, 65, 361 CrossRef.
- J. M. C. A. Kerckhoffs, J. C. Peberdy, I. Meistermann, L. J. Childs, C. J. Isaac, C. R. Pearmund, V. Reudegger, S. Khalid, N. W. Alcock, M. J. Hannon and A. Rodger, Dalton Trans., 2007, 734–742 RSC.
- J. Malina, M. J. Hannon and V. Brabec, Nucleic Acids Res., 2008, 36, 3630–3638 CrossRef CAS PubMed.
- M. J. Hannon, V. Moreno, M. J. Prieto, E. Moldrheim, E. Sletten, I. Meistermann, C. J. Isaac, K. J. Sanders and A. Rodger, Angew. Chem., Int. Ed., 2001, 40, 879–884 CrossRef.
- G. I. Pascu, A. C. G. Hotze, C. Sanchez-Cano, B. M. Kariuki and M. J. Hannon, Angew. Chem., Int. Ed., 2007, 46, 4374–4378 CrossRef CAS PubMed.
- A. C. G. Hotze, N. J. Hodges, R. E. Hayden, C. Sanchez-Cano, C. Paines, N. Male, M.-K. Tse, C. M. Bunce, J. K. Chipman and M. J. Hannon, Chem. Biol., 2008, 15, 1258–1267 CrossRef CAS PubMed.
- A. D. Richards, A. Rodger, M. J. Hannon and A. Bolhuis, Int. J. Antimicrob. Agents, 2009, 33, 469–472 CrossRef CAS PubMed.
- H. Yu, X. Wang, M. Fu, J. Ren and X. Qu, Nucleic Acids Res., 2008, 36, 5695–5703 CrossRef CAS PubMed.
- H. Yu, C. Zhao, Y. Chen, M. Fu, J. Ren and X. Qu, J. Med. Chem., 2009, 53, 492–498 CrossRef PubMed.
- C. Zhao, J. Geng, L. Feng, J. Ren and X. Qu, Chem.–Eur. J., 2011, 17, 8209–8215 CrossRef CAS PubMed.
- H. Yu, M. Li, G. Liu, J. Geng, J. Wang, J. Ren, C. Zhao and X. Qu, Chem. Sci., 2012, 3, 3145–3153 RSC.
- L. Cardo, V. Sadovnikova, S. Phongtongpasuk, N. J. Hodges and M. J. Hannon, Chem. Commun., 2011, 47, 6575–6577 RSC.
- S. E. Howson, L. E. N. Allan, N. P. Chmel, G. J. Clarkson, R. van Gorkum and P. Scott, Chem. Commun., 2009, 1727–1729 RSC.
- S. E. Howson, L. E. N. Allan, N. P. Chmel, G. J. Clarkson, R. J. Deeth, A. D. Faulkner, D. H. Simpson and P. Scott, Dalton Trans., 2011, 40, 10416–10433 RSC.
- S. E. Howson, A. Bolhuis, V. Brabec, G. J. Clarkson, J. Malina, A. Rodger and P. Scott, Nat. Chem., 2012, 4, 31–36 CrossRef CAS PubMed.
- S. E. Howson, G. J. Clarkson, A. D. Faulkner, R. A. Kaner, M. J. Whitmore and P. Scott, Dalton Trans., 2013 10.1039/c3dt51725j.
- R. Palchaudhuri and P. J. Hergenrother, Curr. Opin. Biotechnol, 2007, 18, 497–503 CrossRef CAS PubMed.
- I. Meistermann, V. Moreno, M. J. Prieto, E. Moldrheim, E. Sletten, S. Khalid, P. M. Rodger, J. C. Peberdy, C. J. Isaac, A. Rodger and M. J. Hannon, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5069–5074 CrossRef CAS PubMed.
- A. Rodger, K. J. Sanders, M. J. Hannon, I. Meistermann, A. Parkinson, D. S. Vidler and I. S. Haworth, Chirality, 2000, 12, 221–236 CrossRef CAS PubMed.
- L. Fairall and D. Rhodes, Nucleic Acids Res., 1992, 20, 4727–4731 CrossRef CAS PubMed.
- in Methods in molecular biology – Drug–DNA interaction protocols, ed. K. R. Fox, Humana Press, Totowa, New Jersey, 1997 Search PubMed.
- J. Malina, M. J. Hannon and V. Brabec, Chem.–Eur. J., 2008, 14, 10408–10414 CrossRef CAS PubMed.
- N. B. Leontis, W. Kwok and J. S. Newman, Nucleic Acids Res., 1991, 19, 759–766 CrossRef CAS PubMed.
- M. R. Singleton, S. Scaife and D. B. Wigley, Cell, 2001, 107, 79–89 CrossRef CAS PubMed.
- A. Nikulin, A. Serganov, E. Ennifar, S. Tishchenko, N. Nevskaya, W. Shepard, C. Portier, M. Garber, B. Ehresmann, C. Ehresmann, S. Nikonov and P. Dumas, Nat. Struct. Biol., 2000, 7, 273–277 CrossRef CAS PubMed.
- A. Oleksi, A. G. Blanco, R. Boer, I. Usón, J. Aymamí, A. Rodger, M. J. Hannon and M. Coll, Angew. Chem., Int. Ed., 2006, 45, 1227–1231 CrossRef CAS PubMed.
- J. Malina, M. J. Hannon and V. Brabec, Chem.–Eur. J., 2007, 13, 3871–3877 CrossRef CAS PubMed.
- J. L. Kadrmas, A. J. Ravin and N. B. Leontis, Nucleic Acids Res., 1995, 23, 2212–2222 CrossRef CAS PubMed.
- R. Holliday, Genet. Res., 1964, 5, 282–304 CrossRef.
- M. Culyba, Y. Hwang, S. Attar, P. B. Madrid, J. Bupp, D. Huryn, L. Sanchez, J. Grobler, M. D. Miller and F. D. Bushman, Nucleic Acids Res., 2012, 40, e124 CrossRef CAS PubMed.
- K. Nakatani, S. Sando and I. Saito, J. Am. Chem. Soc., 2000, 122, 2172–2177 CrossRef CAS.
- L. Guan and M. D. Disney, ACS Chem. Biol., 2011, 7, 73–86 CrossRef PubMed.
- A. Davidson, T. C. Leeper, Z. Athanassiou, K. Patora-Komisarska, J. Karn, J. A. Robinson and G. Varani, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 11931–11936 CrossRef CAS PubMed.
- J. Kobayashi, J. Radiat. Res., 2004, 45, 473–478 CrossRef CAS PubMed.
- in Molecular Determinants of Radiation Response, ed. T. L. DeWeese and M. Laiho, Springer New York, New York, 2011 Search PubMed.
- W. M. Bonner, C. E. Redon, J. S. Dickey, A. J. Nakamura, O. A. Sedelnikova, S. Solier and Y. Pommier, Nat. Rev. Cancer, 2008, 8, 957–967 CrossRef CAS PubMed.
- A. Celeste, S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig and A. Nussenzweig, Science, 2002, 296, 922–927 CrossRef CAS PubMed.
- L. J. Kuo and L.-X. YANG, In Vivo, 2008, 22, 305–309 CAS.
- M. Kajstura, H. D. Halicka, J. Pryjma and Z. Darzynkiewicz, Cytometry, Part A, 2007, 71, 125–131 CrossRef PubMed.
- I. Nicoletti, G. Migliorati, M. C. Pagliacci, F. Grignani and C. Riccardi, J. Immunol. Methods, 1991, 139, 271–279 CrossRef CAS PubMed.
- S. M. Bakunova, S. A. Bakunov, T. Wenzler, T. Barszcz, K. A. Werbovetz, R. Brun and R. R. Tidwell, J. Med. Chem., 2009, 52, 4657–4667 CrossRef CAS PubMed.
- P. G. Baraldi, A. Bovero, F. Fruttarolo, D. Preti, M. A. Tabrizi, M. G. Pavani and R. Romagnoli, Med. Res. Rev., 2004, 24, 475–528 CrossRef CAS PubMed.
- M. S. Lee, L. Johansen, Y. Zhang, A. Wilson, M. Keegan, W. Avery, P. Elliott, A. A. Borisy and C. T. Keith, Cancer Res., 2007, 67, 11359–11367 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details for AFM imaging, DNA binding studies using CD and LD, DNase I footprinting, UV melting experiments and anticancer testing. See DOI: 10.1039/c3sc51731d |
| ‡ Correspondingly, and because some flexicates are optically and stereochemically pure but heterohelical (the sense of helicity is not the same throughout the molecule), we use the absolute configuration descriptors at each metal (Δ or Λ) rather than the helicity (P or M). The Δ and Λ flexicates have local P and M helicity respectively. |
| This journal is © The Royal Society of Chemistry 2013 |
