 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of a well-defined glycoconjugate vaccine by a tyrosine-selective conjugation strategy†
Qi-Ying
Hu
*a,
Martin
Allan
a,
Roberto
Adamo
b,
Doug
Quinn
a,
Huili
Zhai
a,
Guangxiang
Wu
a,
Kirk
Clark
a,
Jing
Zhou
a,
Sonia
Ortiz
a,
Bing
Wang
a,
Elisa
Danieli
b,
Stefano
Crotti
b,
Marta
Tontini
b,
Giulia
Brogioni
b and
Francesco
Berti
b
aNovartis Institutes for BioMedical Research, 100 Technology Square, Cambridge, MA 02139, USA. E-mail: qiying.hu@novartis.com; Tel: +1-617-871-7607
bNovartis Vaccine & Diagnostics, Via Fiorentina 1, 53100 Siena, Italy
First published on 1st July 2013
Abstract
An anti-candidiasis glycoconjugate vaccine was prepared via a tyrosine-selective alkynylation and a click chemistry mediated glycoconjugation sequence. It features a well-defined glycan, protein carrier, and connectivity. The construct, although with significantly lower carbohydrate loading and a shorter β-(1,3) glucan chain than the well-established anti-candidiasis vaccine derived from the random conjugation of laminarin at lysines, elicited a comparable level of specific IgG antibodies.
Introduction
Bioconjugation is of great importance to biotherapies and chemical biology.1–4 Glycoconjugate vaccines derived from the covalent coupling of microbial poly- or oligosaccharides to a protein carrier are crucial for the prevention of many life-threatening infectious diseases.5–7 To date, all licensed conjugate vaccines are still prepared and used as complex mixtures. Well-defined constructs are highly desirable to ensure consistent and optimal efficacy, and to facilitate studies of immunological mechanisms. Recently significant efforts have been applied to the development of chemical and enzymatic methods for the synthesis of structurally well-defined carbohydrates with high purity.8–12 However, the conjugation steps for almost all licensed vaccines employ the surface abundant lysines as the attachment points. The site-selectivity is difficult to control, and variability can be introduced.5–7 Furthermore, it has been recently reported that the carbohydrate epitope of a glycopeptide, generated in antigen-presenting cells upon endolysosomal processing from a glycoconjugate vaccine, binds major histocompatibility class II (MHCII) and stimulates T-cell dependent immunological responses.13 Therefore, not only the glycan antigen and protein carrier but also the connectivity may be crucial to the immunogenicity of the carbohydrate-based conjugate vaccines, and is a key aspect of the structure–activity relationships (SARs).14,15 Herein, we present a general strategy to a well-defined glycoconjugate vaccine against Candida albicans through tyrosine-selective conjugation.Results and discussion
Development of the selective tyrosine modification protocol
Our synthetic plan is outlined in Scheme 1. An azide–alkyne Huisgen cycloaddition is employed for the final coupling of the glycan and the carrier protein aiming for a broad substrate scope.16,17 We envisaged that a well-defined alkynyl-protein carrier with broad applicability would be essential to a general strategy, given the fact that various structurally-defined glycans can be obtained by total synthesis.8–12 The nontoxic mutant diphtheria toxin CRM19718 has been extensively used as the protein carrier in many licensed vaccines, and has well-proven safety and efficacy.5–7 Furthermore, unlike many other protein carriers, CRM197 has high purity and a well-defined structure.19 Thus, CRM197 was chosen as the carrier protein in our synthesis.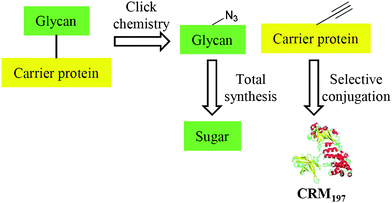 | ||
| Scheme 1 A general strategy to well-defined conjugate vaccines. | ||
First, we investigated the site-selective conjugation of CRM197. We predicted that the surface population of tyrosines on CRM197 is much lower than that of lysines.19 Therefore, targeting tyrosines on CRM197 could potentially offer good site-selectivity. Several tyrosine-targeting conjugation methods have been reported.20–24 The high reactivity of 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD) toward the phenol functionality in water is particularly attractive,24 but its application to CRM197 was found to be very challenging (Scheme 2). When CRM197 (30 μM in PBS, pH 7.4) was incubated with PTAD (10 eq.) at room temperature overnight, unexpected products were predominantly obtained, as indicated by LC-MS (ESI) analysis (Fig. 1). Theoretically, the desired conjugate derived from a PTAD addition to tyrosine should give a peak with a mass shift of +175 Da. In the analysis, a trace peak (C) showed the expected mass shift (+175 Da). In addition, a significant amount of unmodified CRM197 (A, 38%) was left, and a major peak (B, 33%) with an unexpected mass shift (+119 Da) was also found. Two additional peaks with higher mass shift were detected as D (19%) and E (8%). Apparently, they showed mass shifts of +119 Da and +175 Da from the peak B respectively. Subsequently, MS/MS analysis of proteolytic digests was performed to further characterize the reaction products. It was confirmed that the +175 Da labeling was on tyrosine as expected; however, the +119 Da labeling was detected on lysine.
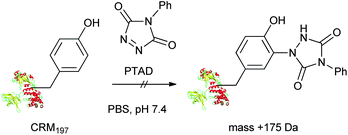 | ||
| Scheme 2 Reaction of PTAD with CRM197 in PBS. | ||
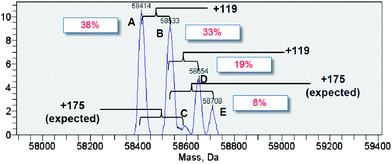 | ||
| Fig. 1 Major unexpected conjugates indicated by the LC-MS (ESI) spectrum of the reaction of PTAD with CRM197 in phosphate buffer. | ||
It is not trivial to assign the unexpected protein conjugates, therefore we employed a penta-peptide thymopentin (1), bearing lysine and tyrosine, as a surrogate substrate in the study to enable the use of NMR. Thymopentin was treated with PTAD (1.1 eq.) in phosphate buffer (pH 7.4) at room temperature overnight. The tyrosine-mediated conjugate (+175 Da, 2a) and the lysine-mediated conjugate (+119 Da, 2b) were obtained in 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio as indicated by LC-MS (Scheme 3). The mixture was separated by HPLC, and two conjugates were assigned by 2D NMR. 2a was confirmed as the expected tyrosine-mediated conjugate, and 2b was assigned as a urea formed at lysine. At this point, we recalled that the cranberry red solution of PTAD always turned to light yellow immediately after dropping into buffer, indicating a rapid decomposition. It is worth noting that isocyanate has been suggested as a degradation product of PTAD in previous studies.25 Thus, we postulated that the +119 Da lysine conjugate was derived from the reaction of lysine with the isocyanate generated from the degradation of PTAD (Scheme 4). To verify the hypothesis, myoglobin without exposed tyrosines was treated with PTAD in phosphate buffer. A significant amount of +119 Da conjugate was obtained, and the MS/MS analysis of proteolytic digests confirmed lysines as the conjugation sites.
1 ratio as indicated by LC-MS (Scheme 3). The mixture was separated by HPLC, and two conjugates were assigned by 2D NMR. 2a was confirmed as the expected tyrosine-mediated conjugate, and 2b was assigned as a urea formed at lysine. At this point, we recalled that the cranberry red solution of PTAD always turned to light yellow immediately after dropping into buffer, indicating a rapid decomposition. It is worth noting that isocyanate has been suggested as a degradation product of PTAD in previous studies.25 Thus, we postulated that the +119 Da lysine conjugate was derived from the reaction of lysine with the isocyanate generated from the degradation of PTAD (Scheme 4). To verify the hypothesis, myoglobin without exposed tyrosines was treated with PTAD in phosphate buffer. A significant amount of +119 Da conjugate was obtained, and the MS/MS analysis of proteolytic digests confirmed lysines as the conjugation sites.
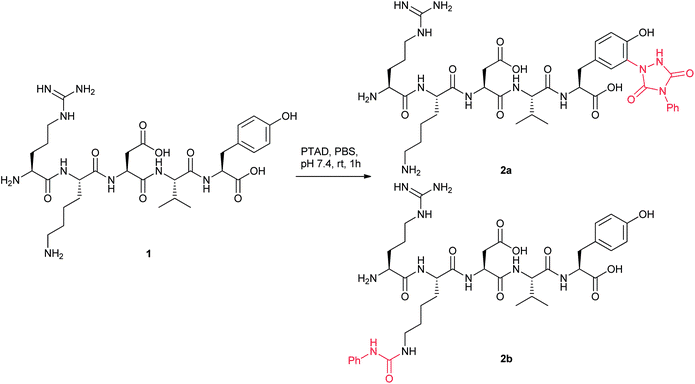 | ||
| Scheme 3 Reaction of thymopentin 1 with PTAD in PBS. | ||
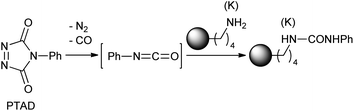 | ||
| Scheme 4 Postulated pathway to the lysine conjugate. | ||
After we successfully characterized the unexpected conjugates, we explored conditions to favor the selective formation of the tyrosine conjugation. First, we investigated the stability of PTAD. 1H NMR spectra of PTAD in deuterated acetonitrile, DMF and DMSO were collected at different time points. No detectable decomposition of PTAD was observed in acetonitrile after 16 h. Partial degradation of PTAD was observed in DMF and DMSO. Interestingly, the cranberry red PTAD solution rapidly turned light yellow, with the emission of gas after the addition of D2O. In the immediately collected 1H NMR spectra, only a trace amount of PTAD could be found.
This observation suggested the rapidly accelerated decomposition of PTAD by water. Therefore, a nucleophilic amine was employed in the reaction to trap isocyanate and prevent the undesired lysine labeling, in addition to minimizing the speed of PTAD addition. Tris(hydroxymethyl)aminomethane (Tris) buffer was selected to function as both the isocyanate scavenger and buffer. Interestingly, only tyrosine-mediated conjugate was obtained without any detectable lysine-mediated conjugate in the reaction of thymopentin with PTAD in Tris buffer (pH 7.4, 1.0 M). LC-MS indicated a peak corresponding to the urea derivative of Tris, which supported the proposed function of Tris as the isocyanate scavenger. Several substrates were investigated with the new protocol (Table 1). Tyrosine-mediated conjugates were obtained as the major product in the presence of Tris buffer in all cases. It is worth noting that the concentration of Tris buffer is important for the full elimination of the undesired lysine-mediated conjugation.
|
|
|||||
|---|---|---|---|---|---|
| Substrate | Conditionsa | Conjugatesb (%) | |||
| T/°C | Tris (M) | Unmod. | K | Yc | |
| a 1.1 eq. of PTAD was used for thymopentin, and 30 eq. of PTAD was used for other cases. b Determined according to peak intensity of the ESI-MS spectra. c Not excluding potential trace amount of W labeling. | |||||
| Thymopentin | 20 | 1.0 | 22 | 0 | 78 |
| Myoglobin | 20 | 0.2 | 90 | 10 | 0 |
| 4 | 1.0 | 87 | 0 | 13 | |
| α-Chymotrypsinogen A | 20 | 0.2 | 45 | 10 | 45 |
| 4 | 0.2 | 16 | 7 | 77 | |
| 4 | 1.0 | 25 | 0 | 75 | |
| CRM197 | 20 | 0.2 | 0 | 0 | 100 |
With the establishment of the tyrosine-selective conjugation method, we began to selectively functionalize CRM197 for the synthesis of well-defined glycoconjugate vaccines (Scheme 5). Reagent 3 was prepared according to the literature protocol.26 The reaction of 3 with CRM197 gave a modified CRM1974 bearing terminal alkynes at tyrosines. The product was purified by single size exclusion desalting chromatography. LC-MS (ESI) indicated the generation of the desired product in good homogeneity with an average labeling ratio of 3.8 alkyne functionalities. The recovery of purified protein was determined as 82% according to its UV absorption at 210 nm. MS/MS analysis of proteolytic digests indicated that Y27, 46, 358 and 380 were the major labeling sites out of total 18 tyrosines. The reaction gave consistent results over several repeats. Other reactive amino acid residues including S, T, W, H, K, C and M have also been searched for potential modification, and only a trace amount of modification was found on W50.
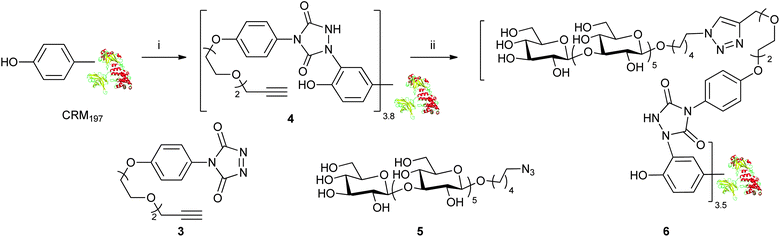 | ||
| Scheme 5 Tyrosine-selective labeling of CRM197 to an anti-candidiasis vaccine. (i) 3, Tris, 0.5 M, pH 7.4; (ii) 5, CuSO4, sodium ascorbate, THPTA, sodium phosphate, 0.1 M, pH 7.0. | ||
To rationalize the selectivity, the solvent exposure of residues in the crystal structure, 4AE0, was examined.19 The buried surface area of each residue was calculated using the software package, ICM.27 All highly labeled residues have high surface exposure. Interestingly, we noted that Y478 showed no detectable labeling, although it has high surface exposure area according to the crystal structure.
CRM197 is a homodimer in solution; however, the dimer's 2-fold axis is coincident with a crystallographic 2-fold symmetry axis in the crystal, and the deposited structure includes only the unique monomer. The surface area calculations were repeated with the reconstructed homodimer structure. Based on the homodimer, there is good correlation between a residue's surface exposure area and the extent of its labeling. The unlabeled Y478 is fully buried in the dimer interface, and not solvent exposed. The predicted labeling conditions were able to preserve the dimer structure, and selectively label the exposed residues of the dimer.
Conjugation of the carbohydrate antigen
Next, we investigated the coupling of 4 with a synthetic glycan antigen bearing an azide group. Candidiasis is the fourth leading cause of nosocomial blood stream infections, and has attracted our interest for years.28–30 In our previous studies, CRM197 conjugates prepared by random coupling to lysines of synthetic β-(1,3)-glucan hexasaccharides conferred murine IgG titres comparable to the structurally more complex laminarin–CRM197 conjugate, a well-established candidate for an anti-candidiasis vaccine.30 Hexasaccharide 5 was prepared by a modified route (see ESI†), and the glycoconjugation of the tyrosine modified CRM1974 by click chemistry was explored. A mixture of the alkyne functionalized CRM1974 and hexasaccharide 5 (3.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, mol ratio) was incubated with copper sulphate, sodium ascorbate and tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) in phosphate buffer (pH 7.0) at room temperature for 5 h.31
20, mol ratio) was incubated with copper sulphate, sodium ascorbate and tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) in phosphate buffer (pH 7.0) at room temperature for 5 h.31
The desired glycoconjugate was obtained, as confirmed by mass spectra and SDS-page analysis. An average loading of 3.5 carbohydrate residues, which was consistent with the number of alkyne moieties of 4, was estimated by MALDI-TOF MS (see ESI†). Therefore, conjugate 6 was obtained with practically complete coupling of all the alkynes of 4 and in 85% yield from 4 based on the recovery of the purified glycoprotein.
Immunization study
The conjugate 6 was tested in CD1 mice for its ability to induce specific antibodies. The laminarin–CRM197 conjugate (7), a well-established candidate for an anti-candidiasis vaccine, prepared by random conjugation to the lysines of CRM197 of heterogeneous 15–20mer β-(1,3)-glucans with sporadic β-(1,6) arms and presenting an average glycosylation degree of 7.3 oligosaccharides,30 was used as the positive control. Groups of 8 CD1 mice were immunized with 2 μg of carbohydrate-based dose of conjugates formulated with the adjuvant MF59.32 Sera collected two weeks after the third dose were analyzed by ELISA for their content of anti-laminarin IgG. It has been traditionally believed that lower glycosylation degree and shorter oligosaccharides correspond to lower specific IgG titres.33 In contrast, vaccine 6, with significantly lower carbohydrate loading and a shorter glycan chain than 7, elicited comparable IgG titres (Fig. 2). Recently, it has been reported that glycoconjugate vaccines after endolysosomal processing in antigen-presenting cells generate glycopeptides, whose carbohydrate epitope binds the major histocompatibility class II (MHC-II), and stimulates T-cell dependent immunological responses.13 Thus, we postulated that the well-defined glycoconjugate 6 might have generated more homogenous glycopeptides, and evoked a stronger immune response. Furthermore, the attachment point may also have impacted the antigen presentation and the efficacy of conjugate vaccines.![Immune response induced in CD1 mice by the conjugate 6 and a laminarin conjugate vaccine 7 prepared by active ester chemistry on lysine30 at 2 μg saccharide dose and formulated with adjuvant MF59. Table shows GMT ELISA 95% CI. [** P = 0.0021 7vs. PBS. *** P = 0.0008 6vs. PBS].](/image/article/2013/SC/c3sc51694f/c3sc51694f-f2.gif) | ||
| Fig. 2 Immune response induced in CD1 mice by the conjugate 6 and a laminarin conjugate vaccine 7 prepared by active ester chemistry on lysine30 at 2 μg saccharide dose and formulated with adjuvant MF59. Table shows GMT ELISA 95% CI. [** P = 0.0021 7vs. PBS. *** P = 0.0008 6vs. PBS]. | ||
Some literature suggests that rigid and constrained spacers elicit a significant amount of undesirable antibodies, and potentially drive the immune response away from the targeted epitope on the hapten.5 In our experiments, there is no noticeable shift of the antibody response from the carbohydrate antigens to the linker. This is in agreement with previous reports on conjugates bearing a rigid triazole moiety on the linker.34,35 Further studies along this line are ongoing in our laboratory.
Conclusion
In summary, an anti-candidiasis glycoconjugate vaccine with well-defined glycan, protein carrier and attachment points has been successfully prepared. To the best of our knowledge, this represents the first example of a well-defined conjugate vaccine glycosylated at tyrosines. Furthermore, this well-defined construct, although with lower carbohydrate loading and a shorter glycan chain length than the corresponding lysine-glycosylated conjugate, elicited comparable specific IgG titres in an immunization study in mice. This clearly underpins the crucial impact of vaccine structure on immunogenicity. The further investigation of various glycans, linkers5–7 and attachment at different amino acids1 is ongoing, and will be presented in due course. Finally, the tyrosine-selective conjugation method described here36 has a well-characterized reaction pathway, high selectivity and reproducibility.Experimental section
Procedure for CRM197 labeling
To a solution of CRM197 (2.92 mg, 0.05 μmol) in Tris·HCl (2.0 mL, 0.2 M, pH 7.4) 10 additions of freshly prepared reagent 3 (2.5 μL, 100 mM in CH3CN, 0.25 μmol) was added every minute (50 eq. in total). The mixture was agitated at 20 °C for 30 min. The mixture was passed through a Zeba 7K MWCO spin column with PBS (pH 7.4) as the eluting buffer three times. The modified CRM1974 (2 mL, 1.2 mg mL−1) was obtained (82%, the recovery was determined according to the UV absorption at 210 nm in comparison with standard). An average loading of 3.8 modified tyrosines per protein was derived from the peak intensity in the ESI-MS spectra.Procedure for glycosylation by copper catalyzed click chemistry
To a solution of 4 (300 μg, 0.005 μmol) in 100 mM sodium phosphate pH 7.0 (70 μL) and 5 (0.10 μmol), a premixed solution of 5 mM CuSO4·5H2O (5 μL) and 25 mM THPTA (5 μL) was added under a nitrogen atmosphere, followed by 5 mM aminoguanidine hydrochloride (5 μL) and 10 mM sodium ascorbate (5 μL). The mixture was stirred at ambient temperature for 1.5 h, at which time the glycoprotein was washed on a 30 kDa Amicon centrifugal filter with 10 mM EDTA/10 mM sodium phosphate pH 7.0 (2 × 100 μL) and 10 mM sodium phosphate pH 7.0 (8 × 100 μL), and subsequently reconstituted with 10 mM sodium phosphate (pH 7.0). Yield (recovered glycoprotein): 85%.Procedure for mice immunization
Animal experimental guidelines set forth by the Novartis Animal Care Department were followed in all animal studies performed. Groups of eight female CD1 mice were immunized at days 1, 14, and 28 with glycoconjugate 6, 7 containing 2 μg of carbohydrate antigens (laminarin or glucan hexasaccharide) or negative control (PBS). 6 and 7 were formulated with MF59 and delivered in a volume of 150 μL by subcutaneous injection. Bleedings were performed at days 0, 27, and 42.Procedure for ELISA analysis
Ninety-six-well Maxisorp plates (Nunc, Thermo Fisher Scientific) were coated overnight at 4 °C with laminarin 5 μg per well in 0.05 M bicarbonate buffer at pH 9.6. After coating, the plates were washed three times with 300 μL per well of TPBS (PBS with 0.05% Tween 20, pH 7.4) and blocked with 100 μL per well of 3% BSA (Sigma-Aldrich) for 1 h at 37 °C. Subsequently, each incubation step was followed by a triple TPBS wash. Sera, prediluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25–1
25–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in TPBS, were transferred into coated-plates (200 μL) and then serially two-fold diluted. After 2 h incubation at 37 °C, 100 μL per well of 1
100 in TPBS, were transferred into coated-plates (200 μL) and then serially two-fold diluted. After 2 h incubation at 37 °C, 100 μL per well of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in TPBS antimouse IgG alkaline phosphatase conjugated (Sigma-Aldrich) were added, and plates were incubated for 1 h at 37 °C. Plates were then developed for 30 min at room temperature with 100 μL per well of 1 mg mL−1p-nitrophenyl phosphate disodium (Sigma-Aldrich) in 1 M diethanolamine (pH 9.8) and read at 405 nm with a microplate spectrophotometer (Bio-Rad).
000 in TPBS antimouse IgG alkaline phosphatase conjugated (Sigma-Aldrich) were added, and plates were incubated for 1 h at 37 °C. Plates were then developed for 30 min at room temperature with 100 μL per well of 1 mg mL−1p-nitrophenyl phosphate disodium (Sigma-Aldrich) in 1 M diethanolamine (pH 9.8) and read at 405 nm with a microplate spectrophotometer (Bio-Rad).
Antibody titres were defined as reciprocals of sera dilution using an optical density (OD) of 0.2 as cutoff. Statistical and graphical analysis was performed using GraphPad Prism 5 software by applying the Mann–Whitney as statistical analysis.
Notes and references
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974 CrossRef CAS
.
- Y.-X. Chen, G. Triola and H. Waldmann, Acc. Chem. Res., 2011, 44, 762 CrossRef CAS
.
- N. Stephanopoulos and M. B. Francis, Nat. Chem. Biol., 2011, 7, 876 CrossRef CAS
.
- J. M. Chalker, G. J. L. Bernardes and B. G. Davis, Acc. Chem. Res., 2011, 44, 730 CrossRef CAS
.
- P. Costantino, R. Rappuoli and F. Berti, Expert Opin. Drug Discovery, 2011, 6, 1045 CrossRef CAS
.
- D. Pace, Expert Opin. Biol. Ther., 2013, 13, 11 CrossRef CAS
.
-
D. R. Bundle, Vaccine Design, ed. R. Rappuoli and F. Bagnoli, Caister Academic Press, Norfolk, UK, 2011, p. 69 Search PubMed
.
- V. Verez-Bencomo, V. Fernández-Santana, E. Hardy, M. E. Toledo, M. C. Rodríguez, L. Heynngnezz, A. Rodriguez, A. Baly, L. Herrera, M. Izquierdo, A. Villar, Y. Valdés, K. Cosme, M. L. Deler, M. Montane, E. Garcia, A. Ramos, A. Aguilar, E. Medina, G. Toraño, I. Sosa, I. Hernandez, R. Martínez, A. Muzachio, A. Carmenates, L. Costa, F. Cardoso, C. Campa, M. Diaz and R. Roy, Science, 2004, 305, 522 CrossRef CAS
.
- L. Morelli, L. Poletti and L. Lay, Eur. J. Org. Chem., 2011, 5723 CrossRef CAS
.
- M. L. Hecht, P. Stallforth, D. Varon Silva, A. Adibekiany and P. H. Seeberger, Curr. Opin. Chem. Biol., 2009, 13, 354 CrossRef CAS
.
- C.-H. Hsu, S.-C. Hung, C.-Y. Wu and C.-H. Wong, Angew. Chem., Int. Ed., 2011, 50, 11872 CrossRef CAS
.
- E. Danieli, L. Lay, D. Proietti, F. Berti, P. Costantino and R. Adamo, Org. Lett., 2011, 13, 378 CrossRef CAS
.
- F. I. Avci, X. Li, M. Tsu and D. L. Kasper, Nat. Med., 2011, 17, 1602 CrossRef CAS
.
- E. J. Grayson, G. J. L. Bernardes, J. M. Chalker, O. Boutureira, J. R. Koeppe and B. G. Davis, Angew. Chem., Int. Ed., 2011, 50, 4127 CrossRef CAS
.
- R. Adamo, A. Nilo, B. Castagner, O. Boutureira, F. Berti and G. J. L. Bernardes, Chem. Sci., 2013, 4, 2995 RSC
.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004 CrossRef CAS
.
- C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. S. Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu and P. Wu, Angew. Chem., Int. Ed., 2011, 50, 8051 CrossRef CAS
.
- G. Giannini and R. Rappuoli, Nucleic Acids Res., 1984, 25, 4063 CrossRef
.
- E. Malito, B. Bursulaya, C. Chen, P. Lo Surdo, M. Picchianti, E. Balducci, M. Biancucci, A. Brock, F. Berti, M. J. Bottomley, M. Nissum, P. Costantino, R. Rappuoli and G. Spraggon, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 5229 CrossRef CAS
.
- N. S. Joshi, L. R. Whitaker and M. B. Francis, J. Am. Chem. Soc., 2004, 126, 15942 CrossRef CAS
.
- S. D. Tilley and M. B. Francis, J. Am. Chem. Soc., 2006, 128, 1080 CrossRef CAS
.
- J. M. McFarland, N. S. Joshi and M. B. Francis, J. Am. Chem. Soc., 2008, 130, 7639 CrossRef CAS
.
- K. L. Seim, A. C. Obermeyer and M. B. Francis, J. Am. Chem. Soc., 2011, 133, 16970 CrossRef CAS
.
- H. Ban, J. Gavrilyuk and C. F. Barbas, J. Am. Chem. Soc., 2010, 132, 1523 CrossRef CAS
.
- H. Wamhofp and K. Wald, Chem. Ber., 1977, 110, 1699 CrossRef
.
-
R. C. Cookson, S. S. Gupte, I. D. R. Stevens and C. T. Watts, Organic Syntheses, Wiley, New York, 1988, Coll. vol. 6, p. 936 Search PubMed
.
- R. A. Abagyan, M. M. Totrov and D. N. Kuznetsov, J. Comput. Chem., 1994, 15, 488 CrossRef CAS
.
- C. Bromuro, M. Romano, P. Chiani, F. Berti, M. Tontini, D. Proietti, E. Mori, A. Torosantucci, P. Costantino, R. Rappuoli and A. Cassone, Vaccine, 2010, 28, 2615 CrossRef CAS
.
- A. Torosantucci, C. Bromuro, P. Chiani, F. De Bernardis, F. Berti, C. Galli, F. Norelli, C. Bellucci, L. Polonelli, P. Costantino, R. Rappuoli and A. Cassone, J. Exp. Med., 2005, 202, 597 CrossRef CAS
.
- R. Adamo, M. Tontini, G. Brogioni, M. R. Romano, G. Costantini, E. Danieli, D. Proietti, F. Berti and P. Costantino, J. Carbohydr. Chem., 2011, 30, 249 CrossRef CAS
.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 4879 Search PubMed
.
- D. T. O'Hagan, R. Rappuoli, E. D. Gregorio, T. Tsai and G. D. Giudice, Expert Rev. Vaccines, 2011, 10, 447 CrossRef CAS
.
- V. Poszgay, C. Chu, L. Pannell, J. Wolfe, J. B. Robbins and R. Schneerson, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 5194 CrossRef
.
- T. Lipinski, T. Luu, P. I. Kitov, A. Szpacenko and D. R. Bundle, Glycoconjugate J., 2011, 28, 149 CrossRef CAS
.
- Z. Yin, H. G. Nguyen, S. Chowdhury, P. Bentley, M. A. Bruckman, A. Miermont, J. C. Gildersleeve, Q. Wang and X. Huang, Bioconjugate Chem., 2012, 23, 1694 CrossRef CAS
.
-
R. Adamo, M. Allan, F. Berti, E. Danieli and Q.-Y. Hu, PCT Int. Appl., WO 2013009564 A1, Jan 17, 2013 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Data for new compounds, experimental procedures, copies of spectra. See DOI: 10.1039/c3sc51694f |
| This journal is © The Royal Society of Chemistry 2013 |

