Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
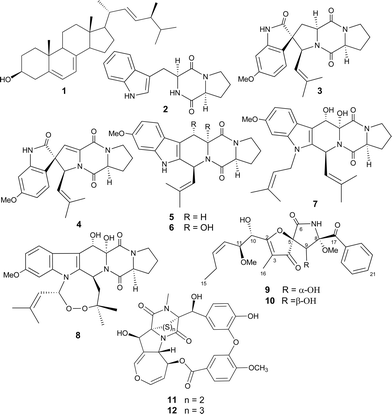
Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture†
Mostafa E.
Rateb
ab,
Irene
Hallyburton
c,
Wael E.
Houssen
ad,
Alan T.
Bull
e,
Michael
Goodfellow
f,
Rakesh
Santhanam
f,
Marcel
Jaspars
a and
Rainer
Ebel
*a
aMarine Biodiscovery Centre, Department of Chemistry, University of Aberdeen, Meston Walk, AB24 3UE, Scotland, UK. E-mail: r.ebel@abdn.ac.uk; Fax: +44 1224 272921; Tel: +44 1224 272930
bPharmacognosy Department, Faculty of Pharmacy, Beni-Suef University, Salah Salem St., 62111, Beni-Suef, Egypt
cDrug Discovery Unit, Division of Biological Chemistry and Drug Discovery, College of Life Sciences, University of Dundee, Sir James Black Centre, Dundee, DD1 5EH, UK
dInstitute of Medical Sciences, University of Aberdeen, Ashgrove Road West, Aberdeen AB25 2ZD, Scotland, UK
eSchool of Biosciences, University of Kent, Canterbury, Kent CT2 7NJ, UK
fSchool of Biology, University of Newcastle, Newcastle upon Tyne NE1 7RU, UK
First published on 10th June 2013
Abstract
An established culture of Aspergillus fumigatus MBC-F1-10 proved to be very receptive to external stimuli and reacted with the production of secondary metabolites which were undetectable when the fungus was grown under standard conditions. Firstly, co-cultivation with the type strain of Streptomyces bullii, an isolate from hyper-arid Atacama desert soil, led to the isolation of ergosterol 1, seven metabolites belonging to the diketopiperazine alkaloids; brevianamide F 2, spirotryprostatin A 3, 6-methoxy spirotryprostatin B 4, fumitremorgin C and its 12,13-dihydroxy derivative (5–6), fumitremorgin B 7, and verruculogen 8, in addition to 11-O-methylpseurotin A 9 and its new isomer 11-O-methylpseurotin A210. In an independent experiment, addition of N-butyryl-DL-homoserine lactone to the culture medium led to the production of emestrins A and B (11–12). Neither microbe produced these compounds when cultured alone. The structures of all compounds were elucidated using NMR spectroscopic techniques and mass spectrometric analysis. The isolated compounds were tested for their potential antibacterial and antiprotozoal activities.
Introduction
Microbial secondary metabolites have been considered one of the best resources for drug discovery.1 Up to early 2013, more than 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 natural compounds have been characterized from microorganisms and higher fungi.2 Parallel to this increase in the rate of isolation of new microbial natural products, the rate of re-isolation of known secondary metabolites has also increased significantly in the past few years.3 Given the fact that microorganisms naturally interact with each other, simulating microbial competition for nutrition and space is regarded as a major route for the induction of bioactive secondary metabolites.4 Thus, mimicking the natural microbial environment, cultivation of two different microbial strains together in one culture vessel (i.e., competing co-culture) allows direct interaction between these microbes which may lead to the induction of new secondary metabolites not previously observed in independent culture of the strains. This phenomenon might be due to microbial crosstalk or chemical defence mechanisms leading to the switching on of cryptic biosynthetic genes in a second microorganism as the result of a signal from the other microorganism.5,6
000 natural compounds have been characterized from microorganisms and higher fungi.2 Parallel to this increase in the rate of isolation of new microbial natural products, the rate of re-isolation of known secondary metabolites has also increased significantly in the past few years.3 Given the fact that microorganisms naturally interact with each other, simulating microbial competition for nutrition and space is regarded as a major route for the induction of bioactive secondary metabolites.4 Thus, mimicking the natural microbial environment, cultivation of two different microbial strains together in one culture vessel (i.e., competing co-culture) allows direct interaction between these microbes which may lead to the induction of new secondary metabolites not previously observed in independent culture of the strains. This phenomenon might be due to microbial crosstalk or chemical defence mechanisms leading to the switching on of cryptic biosynthetic genes in a second microorganism as the result of a signal from the other microorganism.5,6
Most previous co-culture experimental studies reported an increase in biological activity without the identification of the secondary metabolites responsible for this effect, confirmed the induction of antibiotic biosynthesis but not the induction of specific metabolic pathways or increased yields of previously described metabolites.7,8 However, the induction of a previously unexpressed biosynthetic pathway leading to the production of new secondary metabolites in response to co-culture is less often described in the literature. Fenical and co-workers reported that the competing co-culture of the marine-derived fungus Pestalotia sp. with a marine α-proteobacterium (strain CNJ-328) led to the production of the new antibiotic, pestalone9 and subsequently, culturing the same bacterium with the marine-derived fungus Libertella sp. resulted in the discovery of four potent cytotoxic pimarane diterpenoids, libertellenones A–D.4 They also isolated two new antibacterial cyclic depsipeptides, emericellamides A–B through the co-culture of the marine-derived fungus Emericella sp. and the obligate marine actinomycete Salinispora arenicola.10 More recently, a new antibiotic-antitumor metabolite, glionitrin A was isolated during the co-culture of a mine drainage-derived Aspergillus fumigatus and a mine drainage-derived Sphingomonas isolate strain KMK-001,11 while co-culturing A. fumigatus with Streptomyces peucetius led to the production of the new formyl xanthocillin analogue fumiformamide.12 Interaction of the two fungi Penicillium pinophilum and Trichoderma harzianum led to the discovery of a new prenylated polyketide, secopenicillide C,13 while co-cultivation of Pseudomonas aeruginosa and Candida albicans resulted in induction of a bacterial gene cluster required for phenazine biosynthesis.14 This listing does not aim at complete coverage, and the reader is referred to excellent recent review articles covering bacterial-fungal interactions15 as well as the control of secondary metabolite biosynthetic pathways in the genus Aspergillus.16
In this communication, we report on another example of the induction of a presumed fungal biosynthetic pathway in response to microbial co-culture. The addition of the type strain Streptomyces bullii,17 an isolate from hyper-arid Atacama Desert soil, to an established culture of A. fumigatus MBC-F1-10 has led to the isolation of ergosterol (1), seven metabolites belonging to the diketopiperazine (DKP) class of alkaloids; brevianamide F (2), spirotryprostatin A (3), 6-methoxy spirotryprostatin B (4), fumitremorgin C and its 12,13-dihydroxy derivative (5–6), fumitremorgin B (7), and verruculogen (8), in addition to two metabolites from the rare class of pseurotins, 11-O-methylpseurotin A (9) and its new isomer 11-O-methylpseurotin A2 (10). None of these compounds were observed in pure independent cultures of the fungus or the bacterium and all of them are considered to be of fungal origin due to their chemical nature.
Moreover, the addition of the known quorum sensing metabolite, N-butyryl-DL-homoserine lactone (HSL), to the culture media of the fungus led to the isolation of emestrins A–B (11–12) which were not traced in the pure fungal culture. The isolated compounds were tested for their potential antibacterial and antiprotozoal activities. Diseases caused by parasitic protozoa affect many people around the world, notably, Chagas' disease (American trypanosomiasis), sleeping sickness (human African trypanosomiasis), and leishmaniasis. The treatment for these diseases mainly depends on chemotherapy.18 Although several antiparasitic drugs are available, most have toxic side effects, and furthermore development of drug resistance is commonly observed. Therefore, searching for new and effective drugs against neglected tropical diseases (NTD) is needed. Since DKPs with diverse skeletons were reported to have efficient trypanocidal activity, especially disulfide-containing DKPs like gliotoxin which showed nanomolar activity against the whole cell parasite,19 DKPs isolated from Aspergillus fumigatus MBC-F1-10 were tested for their potential antiparasitic activity against Trypanosoma brucei brucei and Leishmania donovani. They were also tested against normal human MRC5 cells (diploid foetal lung fibroblasts) as a counter-screen for indications of toxicity and non-selective inhibition.
Results and discussion
Streptomyces sp. strain C2, which was recently proposed as the type strain of Streptomyces bullii,17 was one of several putatively novel Streptomyces strains from the Atacama Desert included in our chemical screening programme.20 Strain C2 was initially fermented in yeast-extract-malt extract broth21 on a rotary incubator for eight days at 28 °C. After extraction and fractionation, a significant amount of ergosterol22 (1), a strictly fungal metabolite, was isolated as pure crystals from the hexane fraction. Moreover, HPLC purification of the dichloromethane fraction resulted in the isolation of several DKPs which had previously been described from various fungal strains (vide infra). These results prompted us to re-cultivate the bacterial sterile ferment on ISP2 agar plates with incubation at 30 °C, which led to the isolation of a pure culture of strain C2 in addition to an unknown fungal strain, labelled MBC-F1-10. The fungus was purified by subculture on malt extract agar plates, and identified as Aspergillus fumigatus based on the comparison of the sequence of its ITS region with previously published sequences obtained from GenBank. From a chemotaxonomic point of view, the identification of the fungus was also reassuring, since strains of A. fumigatus have repeatedly been shown to be capable of producing secondary metabolites of both the DKP class of natural products including representatives of the fumitremorgin and the verruculogen families, and pseurotin congeners.23Quite unexpectedly, chemical analysis of the fungus A. fumigatus MBC-F1-10 cultivated on malt extract medium (Fig. 1a) resulted in a sparse metabolite profile, with hardly any dominating peaks visible in the LC-MS chromatogram. As a control, the S. bullii strain was also cultivated alone on ISP2 medium (Fig. 1b), resulting in a similar finding. However, deliberately co-cultivating both microorganisms together, i.e. addition of a fresh culture of S. bullii strain C2 to an established three day old culture of A. fumigatus MBC-F1-10, resulted in a very complex LC-MS profile, which was confirmed in two subsequent analogous experiments (Fig. 1c, only one representative HPLC chart depicted). Seven metabolites belonging to the DKP alkaloid family were isolated, specifically brevianamide F24 (2), spirotryprostatin A25 (3), 6-methoxyspirotryprostatin B26 (4), fumitremorgin C27 (5) and its 12,13-dihydroxy derivative28 (6), fumitremorgin B29 (7), and verruculogen30 (8), in addition to two metabolites from the rare class of pseurotins, 11-O-methylpseurotin A31 (9) and its new isomer 11-O-methylpseurotin A2 (10). All of these compounds were consistently detected in all three experiments, and based on their structures and their known fungal origin it is reasonable to assume that they were produced by the fungus and not by the bacterium during the fungal bacterial co-culture.
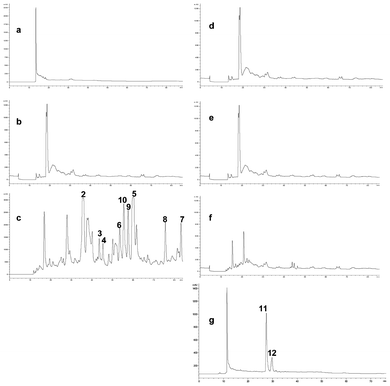 | ||
| Fig. 1 HPLC profiles of different bacterial/fungal co-culture approaches. (a) bacteria alone; (b) fungus alone; (c) fungal-bacterial co-cultivation; (d) fungus with bacteria-free ISP2 medium; (e) fungus with bacterial MeOH extract; (f) fungus with dead bacterial cells; (g) fungus with HSL. | ||
In order to gain insight into the possible mechanism of induction of the biosynthesis of these fungal metabolites by the streptomycete, an additional series of experiments was performed. To an established fungal culture cultured by itself and obtained as described above, was added a treated culture of the streptomycete cultured on its own, as follows: A bacteria-free medium only consisting of ISP2 medium (to rule out induction of media components) (Fig. 1d); an extract of the bacterial biomass obtained with MeOH (Fig. 1e), and a full bacterial culture broth, but inactivated by autoclaving (Fig. 1f). In all three cases, induction of the fungal metabolites 2–10 was not observed. However, addition of N-butyryl-DL-homoserine lactone, a known bacterial quorum sensing compound (Fig. 1g), to the established fungal culture led to induction of the known cyclic DKPs emestrin A32 (11) and B33 (12). It is worth noting that neither 11 nor 12 could be detected in the co-cultivation experiment represented in Fig. 1c, while none of the fungal metabolites 2–10 were observed in this experiment.
Compounds 1–9, 11, and 12 were identified based on their accurate mass analysis and comparison of their MS and NMR spectral data with those reported in the literature. The HRESIMS data of 10 established its molecular formula as C23H27NO8, the same as that of 11-O-methylpseurotin A (9). Both compounds displayed virtually identical UV and IR spectra. The analysis of the 1H and 13C NMR data of 10 (Table 1) indicated the presence of a monosubstituted benzene ring, a disubstituted cis double bond (J = 10.9 Hz), one aliphatic and one allylic methyl group, two O-methyl groups, and three carbonyls including one α,β-unsaturated ketone and one amide. The 1H- and 13C NMR data of both compounds were reminiscent of each other with the exception of slight variations in chemical shifts at four positions (Table 1, ESI†). The analysis of COSY and HMBC spectra (Table 1, Fig. 2) revealed that the planar structure of compound 10 was identical to that of 9, thus indicating that they were stereoisomers. Analysis of the 1D NMR data (Table S1, ESI†) suggested that both compounds should differ with regard to the relative configuration of the pyrrolidinone ring, and a comparison of the 13C NMR data of this substructure with that reported for the structurally related compounds, pseurotin A34 and its diastereomer pseurotin A2,35 revealed that 10 possessed the same relative stereochemistry as pseurotin A2, based on almost identical 13C chemical shift values (Table S1, ESI†). Similar optical rotation values for 10 and pseurotin A2, the absolute configuration of which was established on the basis of X-ray crystallographic analysis and the use of circular dichroism (CD) spectroscopy,35 indicated that both compounds should also have the same absolute configuration.
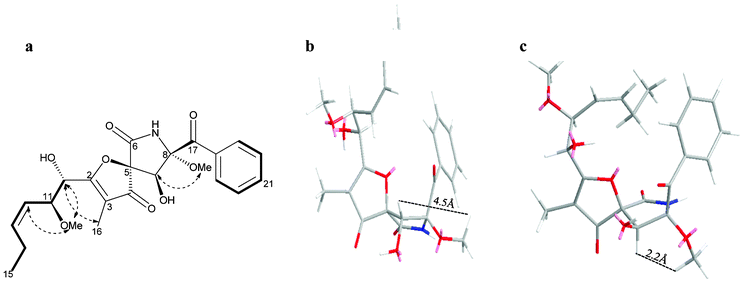 | ||
Fig. 2 Selected COSY ( ) and NOESY ( ) and NOESY ( ) correlations of 10 (a). Molecular mechanics models of the 9α-OH (in 9, b) and 9β-OH (in 10, c) indicated that the observed NOE of H-9 to 8-OMe was only consistent with the 9β-OH stereochemistry in 10. ) correlations of 10 (a). Molecular mechanics models of the 9α-OH (in 9, b) and 9β-OH (in 10, c) indicated that the observed NOE of H-9 to 8-OMe was only consistent with the 9β-OH stereochemistry in 10. | ||
| Position | δ H, mult. (J in Hz)a | δ C, multa | δ H, multb (J in Hz) | δ C, multb | HMBCc |
|---|---|---|---|---|---|
| a In CDCl3. b In DMSO-d6. c From H to C. | |||||
| 2 | 184.4, C | 186.8, C | |||
| 3 | 113.3, C | 111.4, C | |||
| 4 | 198.1, C | 199.8, C | |||
| 5 | 88.5, C | 88.2, C | |||
| 6 | 167.9, C | 167.5, C | |||
| 8 | 95.4, C | 97.0, C | |||
| 9 | 4.78, s | 76.5, CH | 4.62 (d, 6.2) | 75.6, CH | 4, 6, 8 |
| 10 | 4.56 (d, 6.4) | 70.3, CH | 4.48 (t, 5.4) | 70.4, CH | 2, 11, 12 |
| 11 | 4.28 (dd, 6.5, 9.1) | 77.6, CH | 4.17 (dd, 5.5, 9.3) | 77.1, CH | 2, 10, 13 |
| 12 | 5.27 (t, 10.7) | 124.7, CH | 5.31 (t, 10.9) | 125.6, CH | 14 |
| 13 | 5.82, m | 140.3, CH | 5.62, m | 136.9, CH | 11 |
| 14 | 2.10, m | 21.4, CH2 | 2.04, m | 20.7, CH2 | 12, 13, 15 |
| 1.97, m | |||||
| 15 | 0.98 (t, 7.5) | 14.3, CH3 | 0.87 (t, 7.6) | 14.1, CH3 | 13, 14 |
| 16 | 1.77, s | 5.8, CH3 | 1.64, s | 5.6, CH3 | 2, 3, 4 |
| 17 | 193.3, C | 194.7, C | |||
| 18 | 134.0, C | 134.8, C | |||
| 19/23 | 8.21 (d, 7.9) | 129.9, CH | 8.11 (d, 7.7) | 129.6, CH | 17, 21 |
| 20/22 | 7.47 (t, 7.8) | 128.7, CH | 7.52 (t, 7.8) | 128.3, CH | 18 |
| 21 | 7.60 (t, 7.2) | 134.0, CH | 7.63 (t, 7.6) | 133.1, CH | |
| 8-OMe | 3.29, s | 51.6, CH3 | 3.12, s | 51.4, CH3 | 8 |
| 11-OMe | 3.25, s | 56.7, CH3 | 3.15, s | 55.7, CH3 | 11 |
| 9-OH | 6.09 (d, 6.2) | ||||
| 10-OH | 5.75 (d, 5.8) | ||||
| NH-7 | 7.40, br s | 9.89, s | 5 | ||
Molecular mechanics simulations as well as a comparison of their NOESY spectra were carried out for both 9 (featuring a 9α-OH) and 10 (displaying a 9β-OH, Fig. 2). The proximity between H-9 and 8-OMe was confirmed by a strong NOE correlation from H-9 to 8-OMe in 10, indicating that the 9-OH was in the β-configuration in 10. This fact was also corroborated by the absence of an NOE between H-9 and 8-OMe in 9 when 9-OH was in the α-configuration. On this basis 10 was identified as the previously unreported 9-epimer of 11-O-methylpseurotin A for which the name 11-O-methylpseurotin A2 is suggested.
Since DKP production was induced through a microbial co-culture, which can be considered as a simulated competitive environment, antibacterial activity was expected for these compounds. However, none of the isolated compounds showed significant antibacterial activity when screened against Staphylococcus aureus and Escherichia coli (data not shown). This result does not exclude that these compounds were produced by the fungus to combat the existing bacteria in the culture environment, but could also be due to the compounds acting as quorum sensing inhibitors which do not inevitably kill or inhibit the growth of a microbial strain but instead modulate its phenotype, for instance via virulence attenuation.36,37 Meanwhile, assays for trypanocidal and leishmanicidal activities were performed on the isolated compounds (Table 2). All tested compounds proved active against these two parasites except brevianamide F (2) and 11-O-methylpseurotin A (9). Fumitremorgin B (7) and verruculogen (8) were the most potent growth inhibitors on both parasites exhibiting EC50 values of 0.2 μM on Trypanosoma brucei brucei and 3–4 μM on Leishmania donovani, followed by 12,13-dihydroxyfumitremorgin C (6) which exhibited an EC50 value of 7.4 μM on Trypanosoma brucei brucei. Although these compounds proved moderate to potent inhibitors of these parasites, they were not selective and showed pronounced toxicity on MRC5 cells (normal human lung fibroblasts) which suppressed further investigation and precluded their value as a lead scaffold. These results were in agreement with the literature where the fused pentacyclic DKPs showed pronounced trypanocidal activity when compared with other DKPs with a different skeleton.18 It could be concluded that the 12,13-diol moiety in DKP derivatives plays an important role in potentiating the antiparasitic effect of the fused pentacyclic DKPs. 6-methoxyspirostatin B (4) showed a moderate leishmanicidal activity with an EC50 value of 24.7 μM, but displayed selectivity in not being toxic toward MRC5 cells.
Conclusions
In conclusion, microbial co-culture (also referred to as mixed fermentation) proved to be an efficient technique for the production of microbial secondary metabolites. Although in the last few years, various and diverse interactions between bacteria and fungi have been investigated, the current study describes an extreme example of induction of otherwise cryptic biogenetic pathways. Apart from the multitude of structurally diverse metabolites induced in A. fumigatus MBC-F1-10 by co-cultivation with S. bullii strain C2, it is remarkable that simulating bacterial presence by addition of well-established quorum sensing metabolite, N-butyryl-DL-homoserine lactone, resulted in inducing the biosynthesis of yet another two metabolites, and none of these compounds was detected when the fungus was cultivated alone. Application of mixed fermentation for the discovery of new microbial metabolites is still in its infancy, probably due to the fear of lack of reproducibility.5 Our current study demonstrates that this approach is capable of delivering reproducible metabolite patterns, provided relevant fermentation parameters are first established and then carefully maintained.Experimental
General experimental procedures
Optical rotations were recorded using a Perkin-Elmer 343 polarimeter. UV and IR spectra were measured on a Perkin-Elmer Lambda 25 UV-vis spectrometer and a Thermo Nicolet IR 100 FT-IR spectrometer, respectively. NMR data were acquired on a Varian Unity INOVA 600 MHz spectrometer. High resolution mass spectrometric data were obtained using a Thermo Instruments MS system (LTQ XL/LTQ Orbitrap Discovery) coupled to a Thermo Instruments HPLC system (Accela PDA detector, Accela PDA autosampler, and Accela pump). The following conditions were used: capillary voltage 45 V, capillary temperature 320 °C, auxiliary gas flow rate 10–20 arbitrary units, sheath gas flow rate 40–50 arbitrary units, spray voltage 4.5 kV, mass range 100–2000 amu (maximum resolution 30,000). HPLC separations were carried out using a Phenomenex reversed-phase (C18, 250 × 10 mm, L × i.d.) column connected to an Agilent 1200 series binary pump and monitored using an Agilent photodiode array detector. Detection was carried out at 220, 254, 280, 350, and 400 nm. N-butyryl-DL-homoserine lactone was obtained from Sigma-Aldrich. Diaion HP-20 was obtained from Resindion S.R.L., a subsidiary of Mitsubishi Chemical Co., Binasco, Italy.Microorganisms and fermentation
The S. bullii strain was isolated from the soil taken from the Laguna de Chaxa of the Salar de Atacama (in the northern part of the Atacama Desert, Chile) as described previously. The first-stage seed culture was prepared by inoculation of an agar grown culture into 100 mL of ISP2 broth (4.0 g yeast extract, 10.0 g malt extract, 4.0 g glucose, distilled water 1000 mL adjusted to pH 7.2) for 24 h.Aspergillus fumigatus strain MBC-F1-10 was isolated from the S. bullii culture, purified on malt extract agar (15.0 g malt extract, 15.0 g agar, distilled water 1000 mL adjusted to pH 7.2) and identified by sequence comparison of its amplified ITS region (GenBank accession number JN153038) with published sequences obtained from GenBank as described previously.38 The fungal strain was inoculated in 10 × 100 mL malt extract broth (15.0 g malt extract, distilled water 1000 mL adjusted to pH 7.2 in the presence of 50 g Diaion HP-20 resin) for 5 days in a rotary incubator at 28 °C with shaking at 150 rpm.
A set of experiments (a–g) were performed on these 2 cultures, each experiment was performed in triplicate on small scale cultures (1 L) and monitored by HPLC while experiments c and g were repeated on a larger scale basis with 3 L ISP2 medium. The time frame for experiment (c) was established during 11 independent experiments, in which the addition of the bacterial to the fungal culture was changed by 12 h each, ranging from 0 h to 144 h, and maximum induction of metabolites was observed after 72 h (data not shown). (a) Bacteria alone: 1 mL S. bullii culture was transferred aseptically to 100 mL fresh ISP2 broth and re-incubated using the same conditions for 5 days. (b) Fungus alone: 1 mL fungal culture was transferred aseptically to 100 mL fresh malt extract broth and re-incubated using the same conditions for 5 days. (c) Bacterial fungal interaction: 1 mL S. bullii culture was transferred aseptically to 100 mL three day-established fungal culture in ISP2 broth and re-incubated using the same conditions for 5 days. (d) Fungus with bacteria-free ISP2 broth: 1 mL fresh ISP2 broth was transferred aseptically to 100 mL fungal culture in malt extract broth and re-incubated using the same conditions for 5 days. (e) Fungus with bacterial extract: 0.5 mL biomass extract [(bacterial biomass obtained from 100 mL bacterial culture for 3 days was extracted with MeOH; the resulting extract was concentrated and the residue dissolved in 1 mL MeOH:H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)] was transferred aseptically to 100 mL fungal culture in ISP2 broth and re-incubated using the same conditions for 5 days. (f) Fungus with dead bacterial cells: 1 mL autoclaved bacterial broth was transferred aseptically to 100 mL fungal culture in ISP2 broth and re-incubated using the same conditions for 5 days. (g) Fungus with HSL: 1 mg L−1N-butyryl-DL-homoserine lactone was added aseptically to the fresh fungal culture in ISP2 broth and incubated using the same conditions for 7 days.
1)] was transferred aseptically to 100 mL fungal culture in ISP2 broth and re-incubated using the same conditions for 5 days. (f) Fungus with dead bacterial cells: 1 mL autoclaved bacterial broth was transferred aseptically to 100 mL fungal culture in ISP2 broth and re-incubated using the same conditions for 5 days. (g) Fungus with HSL: 1 mg L−1N-butyryl-DL-homoserine lactone was added aseptically to the fresh fungal culture in ISP2 broth and incubated using the same conditions for 7 days.
Extraction and isolation
The same procedure was performed for small and large scale screening experiments. The harvested fermentation broth from the fungal strain (ISP2, 3 L) was centrifuged at 3000 rpm for 20 min, and the HP20 resin together with the biomass was washed with distilled water and then extracted with methanol (5 × 200 mL). The successive MeOH extracts were combined and concentrated in vacuo yielding 250 mL of materials that were fractionated with n-hexane (3 × 200 mL), CH2Cl2 (4 × 200 mL) and EtOAc (3 × 200 mL). The CH2Cl2 fraction was loaded on a Sephadex LH-20 column equilibrated with MeOH-CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and two fractions were collected. The second fraction was purified by RP-HPLC using a gradient of MeOH in H2O as eluent (50–100% over 70 min, 100% for 20 min) at a flow rate of 1.25 mL min−1. HPLC of the large scale of experiments (c and g) afforded the pure compounds 2 (14 mg), 3 (5 mg), 4 (6 mg), 5 (23 mg), 6 (6 mg), 7 (3 mg), 8 (2 mg), 9 (10 mg), 10 (7 mg), 11 (13 mg), and 12 (1.5 mg). 11-O-methylpseurotin A2 (10): colorless powder [α]20D −13.8 (c 0.1, CH3OH); UV (MeCN) λmax (log ∈) 202 (3.92), 251 (3.22), 281 (2.80) nm; IR (film) υmax 3244, 1710, 1680, 1605, 1122 cm−1; 1H and 13C NMR, see Table 1; HRESIMS m/z 446.1809 [M+H]+ (calcd for C23H27NO8, 446.1809).
1) and two fractions were collected. The second fraction was purified by RP-HPLC using a gradient of MeOH in H2O as eluent (50–100% over 70 min, 100% for 20 min) at a flow rate of 1.25 mL min−1. HPLC of the large scale of experiments (c and g) afforded the pure compounds 2 (14 mg), 3 (5 mg), 4 (6 mg), 5 (23 mg), 6 (6 mg), 7 (3 mg), 8 (2 mg), 9 (10 mg), 10 (7 mg), 11 (13 mg), and 12 (1.5 mg). 11-O-methylpseurotin A2 (10): colorless powder [α]20D −13.8 (c 0.1, CH3OH); UV (MeCN) λmax (log ∈) 202 (3.92), 251 (3.22), 281 (2.80) nm; IR (film) υmax 3244, 1710, 1680, 1605, 1122 cm−1; 1H and 13C NMR, see Table 1; HRESIMS m/z 446.1809 [M+H]+ (calcd for C23H27NO8, 446.1809).
Antibacterial screening
The antibacterial activity of the isolated compounds was evaluated against S. aureus ATCC25923 and E. coli ATCC25922 using the agar diffusion method.39Antiprotozoal activity
Trypanosoma brucei brucei (bloodstream form 427, VSG 221) and Leishmania donovani axenic amastigotes, (WHO design, MHOM/SD/62/1S-CL2D) were used for this assay. Normal human MRC5 cells (diploid foetal lung fibroblasts) were used as a counter-screen for toxicity and non-selective inhibitors. The full method was performed using the 384 well plate adaptation of an assay described in the literature.40 In brief, bloodstream-form T. brucei brucei cells were grown at 37 °C and 5% CO2 in a modified HMI9 medium, L. donovani amastigotes were grown axenically at 37 °C and 5% CO2 in medium-199, while MRC5 cells were grown at 37 °C and 5% CO2 in a humidified incubator in Eagle's Minimal Essential Medium.3 Different concentrations of the tested compounds were examined in duplicate. Tested compounds were solubilised to 10 mM in DMSO. 10-point, 1 in 3 dilution curves were made from the compounds, and then 250 nL of each compound was stamped into 384-well assay plate so that the top concentration for the test compounds was 50 μM. 50 μL of either T. brucei brucei or L. donovani culture was added to each well at a density of 5 × 103 mL−1. MRC5 cells were plated at 2 × 104 mL−1 in 80 μL medium, allowed to adhere overnight, before the compound, which had been pre-diluted in media, was added to achieve a final top concentration of 50 μM. All plates were incubated for 69 h at 37 °C in 5% CO2. Then, resazurin (Sigma-Aldrich) dissolved in water was added to a final concentration of 45.5 μM in each well and incubated for a further 3–4 h. Plates were read for fluorescence on a Biotec Instruments FLX 800 fluorescent plate reader at an excitation/emission of 528/590 nm, with a filter cutoff at 550 nm. EC50 values were analysed using IDBS activity base XLFit 4.2 Model 205.Acknowledgements
We thank DNA Sequencing & Services (MRCPPU, College of Life Sciences, University of Dundee, Scotland, www.dnaseq.co.uk) for DNA sequencing, A. Crossman, Department of Chemistry, University of Dundee, for determination of the optical activity, A. Raab, Department of Chemistry, University of Aberdeen, for the accurate mass analysis, and the Egyptian Government for a Ph.D. scholarship to MER. WEH is the recipient of a SULSA post-doctoral fellowship. We thank the College of Physical Sciences, University of Aberdeen for provision of infrastructure and facilities in the Marine Biodiscovery Centre.Notes and references
- R. K. Pettit, Mar. Biotechnol., 2011, 13, 1–11 CrossRef CAS.
- H. Laatsch, AntiBase 2012 The Natural Compound Identifier, Wiley VCH, Weinheim, Germany, ( 2013) Search PubMed.
- K. Hong, A.-H. Gao, Q.-Y. Xie, H. G. Gao, L. Zhuang, H.-P. Lin, H.-P. Yu, J. Li, X.-S. Yao, M. Goodfellow and J.-S. Ruan, Mar. Drugs, 2009, 7, 24–44 CrossRef CAS.
- D.-C. Oh, P. R. Jensen, C. A. Kauffman and W. Fenical, Bioorg. Med. Chem., 2005, 13, 5267–5273 CrossRef CAS.
- R. K. Pettit, Appl. Microbiol. Biotechnol., 2009, 83, 19–25 CrossRef CAS.
- V. Schroeckh, K. Scherlach, H.-W. Nützmann, E. Shelest, W. Schmidt-Heck, J. Schuemann, K. Martin, C. Hertweck and A. A. Brakhage, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14558–14563 CrossRef CAS.
- J. G. Burgess, E. M. Jordan, M. Bregu, A. Mearns-Spragg and K. G. Boyd, J. Biotechnol., 1999, 70, 27–32 CrossRef CAS.
- J. Sonnenbichler, J. Dietrich and H. Peipp, Biol. Chem. Hoppe-Seyler, 1994, 375, 71 CrossRef CAS.
- M. Cueto, P. R. Jensen, C. Kauffman, W. Fenical, E. Lobkovsky and J. Clardy, J. Nat. Prod., 2001, 64, 1444–1446 CrossRef CAS.
- D.-C. Oh, C. A. Kauffman, P. R. Jensen and W. Fenical, J. Nat. Prod., 2007, 70, 515–520 CrossRef CAS.
- H. B. Park, H. C. Kwon, C.-H. Lee and H. O. Yang, J. Nat. Prod., 2009, 72, 248–252 CrossRef CAS.
- K. M. Zuck, S. Shipley and D. J. Newman, J. Nat. Prod., 2011, 74, 1653–1657 CrossRef CAS.
- K. Nonaka, T. Abe, M. Iwatsuki, M. Mori, T. Yamamoto, K. Shiomi, S. Omura and R. Masuma, J. Antibiot., 2011, 64, 769–774 CrossRef CAS.
- J. Gibson, A. Sood and D. A. Hogan, Appl. Environ. Microbiol., 2009, 75, 504–513 CrossRef CAS.
- P. Frey-Klett, P. Burlinson, A. Deveau, M. Barret, M. Tarkka and A. Sarniguet, Microbiol. Mol. Biol. Rev., 2011, 75, 583–609 CrossRef CAS.
- J. F. Sanchez, A. D. Somoza, N. P. Keller and C. C. C. Wang, Nat. Prod. Rep., 2012, 29, 351–371 RSC.
- R. Santhanam, X. Y. Rong, Y. Huang, B. A. Andrews, J. A. Asenjo and M. Goodfellow, Antonie van Leeuwenhoek, 2013, 103, 367–373 CrossRef.
- L. Gros, V. M. Castillo-Acosta, C. J. Jiménez, M. Sealey-Cardona, S. Vargas, A. Manuel Estévez, V. Yardley, L. Rattray, S. L. Croft, L. M. Ruiz-Perez, J. A. Urbina, I. H. Gilbert and D. G. Pacanowska, Antimicrob. Agents Chemother., 2006, 50, 2595–2601 CrossRef CAS.
- K. R. Watts, J. Ratnam, K.-H. Ang, K. Tenney, J. E. Compton, J. McKerrow and P. Crews, Bioorg. Med. Chem., 2010, 18, 2566–2574 CrossRef CAS.
- C. Okoro, R. Brown, A. Jones, B. Andrews, J. Asenjo, M. Goodfellow and A. Bull, Antonie van Leeuwenhoek, 2009, 95, 121–133 CrossRef.
- E. B. Shirling and D. Gottlieb, Int. J. Syst. Bacteriol., 1966, 16, 313–340 CrossRef.
- X. Xiang-qian, H. Wen-rong and C. Dai-jie, Zhongguo Kangshengsu Zazhi (Chin. J. Antibiot.), 2006, 31, 379–381 Search PubMed.
- J. C. Frisvad, C. Rank, K. F. Nielsen and T. O. Larsen, Med. Mycol., 2009, 47, S53–S71 CrossRef CAS.
- M. Kobayashi, S. Aoki, K. Gato, K. Matsunami, M. Kurosu and I. Kitagawa, Chem. Pharm. Bull., 1994, 42, 2449–2451 CrossRef CAS.
- C.-B. Cui, H. Kakeya and H. Osada, Tetrahedron, 1996, 52, 12651–12666 CrossRef CAS.
- M. Zhang, W.-L. Wang, Y.-C. Fang, T.-J. Zhu, Q.-Q. Gu and W.-M. Zhu, J. Nat. Prod., 2008, 71, 985–989 CrossRef CAS.
- C. B. Cui, H. Kakeya and H. Osada, J. Antibiot., 1996, 49, 534–540 CrossRef CAS.
- W.-R. Abraham and H.-A. Arfmann, Phytochemistry, 1990, 29, 1025–1026 CrossRef CAS.
- M. Yamazaki, K. Suzuki, H. Fujimoto, T. Akiyama, U. Sankawa and Y. Iitaka, Chem. Pharm. Bull., 1980, 28, 861–865 CrossRef CAS.
- J. Fayos, D. Lokensgard, J. Clardy, R. J. Cole and J. W. Kirksey, J. Am. Chem. Soc., 1974, 96, 6785–6787 CrossRef CAS.
- C. M. Boot, N. C. Gassner, J. E. Compton, K. Tenney, C. M. Tamble, R. S. Lokey, T. R. Holman and P. Crews, J. Nat. Prod., 2007, 70, 1672–1675 CrossRef CAS.
- H. Seya, K. Nozawa, S. Nakajima, K.-i. Kawai and S.-i. Udagawa, J. Chem. Soc., Perkin Trans. 1, 1986, 109–116 RSC.
- K. Nozawa, S. I. Udagawa, S. Nakajima and K. I. Kawai, Chem. Pharm. Bull., 1987, 35, 3460–3463 CrossRef CAS.
- O. Ando, H. Satake, M. Nakajima, A. Sato, T. Nakamura, T. Kinoshita, K. Furuya and T. Haneishi, J. Antibiot., 1991, 44, 382–389 CrossRef CAS.
- F. Z. Wang, D. H. Li, T. J. Zhu, M. Zhang and Q. Q. Gu, Can. J. Chem., 2011, 89, 72–76 CrossRef CAS.
- M. Cámara, P. Williams and A. Hardman, Lancet Infect. Dis., 2002, 2, 667–676 CrossRef.
- M. Mansson, A. Nielsen, L. Kjærulff, C. H. Gotfredsen, M. Wietz, H. Ingmer, L. Gram and T. O. Larsen, Mar. Drugs, 2011, 9, 2537–2552 CrossRef CAS.
- M. E. Rateb, W. E. Houssen, N. M. Legrave, C. Clements, M. Jaspars and R. Ebel, Bot. Mar., 2010, 53, 499 CrossRef CAS.
- G. Kronvall, J. Clin. Microbiol., 1983, 17, 975–980 CAS.
- D. C. Jones, I. Hallyburton, L. Stojanovski, K. D. Read, J. A. Frearson and A. H. Fairlamb, Biochem. Pharmacol., 2010, 80, 1478–1486 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: NMR spectra of 9–10 including 1H and 13C in CDCl3 and DMSO-d6, COSY, HSQC, HMBC and NOESY in CDCl3. See DOI: 10.1039/c3ra42378f |
| This journal is © The Royal Society of Chemistry 2013 |