 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Lipid-insertion enables targeting functionalization of erythrocyte membrane-cloaked nanoparticles†
Ronnie H. Fangab,
Che-Ming J. Huab,
Kevin N. H. Chena,
Brian T. Lukbc,
Cody W. Carpentera,
Weiwei Gaoab,
Shulin Lid,
Dong-Er Zhangb,
Weiyue Lue and
Liangfang Zhang*ab
aDepartment of NanoEngineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA. E-mail: zhang@ucsd.edu; Tel: +1-858-246-0999
bMoores Cancer Center, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA
cDepartment of Bioengineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA
dDepartment of Pediatric Research, MD Anderson Cancer Center, Houston, TX 77030, USA
eKey Laboratory of Smart Drug Delivery, Ministry of Education & PLA, School of Pharmacy, Fudan University, Shanghai 201203, China
First published on 19th July 2013
Abstract
RBC membrane-cloaked polymeric nanoparticles represent an emerging nanocarrier platform with extended circulation in vivo. A lipid-insertion method is employed to functionalize these nanoparticles without the need for direct chemical conjugation. Insertion of both folate and the nucleolin-targeting aptamer AS1411 shows receptor-specific targeting against model cancer cell lines.
A major goal in engineering nanocarriers for systemic drug delivery is to achieve long circulation half-lives,1 as increased residence time in the bloodstream can improve the pharmacokinetic profile of therapeutic cargoes and allow a greater chance for the nanocarriers to reach the desired location through either passive2,3 or active4–6 targeting mechanisms. Ongoing search for new and effective ways to construct long-circulating nanoparticles has introduced numerous stealth functionalization strategies. While the use of synthetic polymers represents the current golden standard for prolonging particle residence time in circulation, recent efforts have drawn inspiration from nature to bestow immune-evasive properties on nanoparticles.7,8 Bridging the complex surface biochemistry of nature's carriers, RBCs, with the versatile cargo-carrying capacity of polymeric nanoparticles, an RBC membrane-cloaked nanoparticle (RBC-NP) platform represents a new class of bio-inspired nanocarriers with long-circulating capability.9,10 In this system, a top-down approach is used to coat polymeric nanoparticles with natural RBC membranes, which possess a litany of immunomodulatory proteins responsible for RBCs' extraordinarily long survival in circulation.11–14 This membrane cloak acts to camouflage the nanoparticle in vivo to evade immune attacks.9 Consisting entirely of biocompatible and biodegradable materials, the RBC-NPs have significant potential for drug delivery applications.
In applying RBC-NPs for disease treatments, particularly against cancers, target-selectivity is a desirable feature that promises minimization of off-target side effects.15–19 Cancer targeting has been made possible by carrier functionalization with ligands that target overexpressed tumor antigens, and numerous chemical conjugation techniques employing carboxyl-, amine-, or sulfhydryl-based chemistry have been used to decorate synthetic nanocarriers with targeting ligands.20–26 In the case of biologically derived carriers such as RBC-NPs, however, the presence of biological components on the particle surfaces demands a non-disruptive functionalization strategy since the immune evasion capabilities of cellular membranes are predicated upon having fully functional proteins. In order to address the issue of chemical denaturation, we report a lipid-insertion approach for functionalization of RBC-NPs that exploits the fluidity of bilayered RBC lipid membranes and precludes RBC membrane exposure to chemical reactions. As shown in Fig. 1, targeting moieties are incorporated onto RBC membranes through the aid of lipid tethers. The physical insertion of ligand–linker–lipid conjugates into the RBC membranes produces functionalized RBC membranes without damaging the existing surface proteins. Using two differently sized ligands, a small molecule folate (MW ∼ 441 Da) and a nucleolin-targeting aptamer AS1411 (MW ∼ 9000 Da), we demonstrate that the lipid-insertion technique can be applied to targeting ligands of different length scales. Following preparation of RBC-NPs with the functionalized RBC membranes, their receptor-specific targeting ability is verified in model cancer cell lines in vitro.
 | ||
| Fig. 1 Schematic of the preparation of RBC-NPs with targeting ability. Ligand–linker–lipid conjugates are synthesized and then inserted into RBC membrane ghosts. The resulting ligand-functionalized RBC membranes are used to coat polymeric cores to form targeted RBC-NPs. | ||
To demonstrate that lipid-tethered ligands can be spontaneously incorporated onto RBC membranes, a FITC–linker–lipid (excitation/emission = 495/519 nm) conjugate was used for a proof-of-concept test, where FITC was linked to a lipid molecule through a short polyethylene glycol chain (MW ∼ 2000 Da). In the study, 40 μg of FITC–PEG–lipid was first incubated with emptied red blood cells (RBC ghosts) collected from 1 mL of whole mouse blood for 30 min. The RBC ghosts were then centrifuged, washed with PBS, reconstituted, and examined using flow cytometric analysis (Fig. 2A). Compared to unmodified RBC ghosts, modified membrane ghosts had a significantly higher signal under the FITC channel. Visualization by fluorescence microscopy further confirmed the localization of the lipid-tethered FITC on the RBC membranes, as the microscopy image displayed strong FITC signals outlining the exterior of the RBC ghosts (Fig. 2B). To characterize the lipid-insertion efficiency and saturation level, varying amounts of FITC–PEG–lipid were incubated with the membrane ghosts followed by membrane purification through centrifugation. Fluorescence quantification showed that the retained FITC fluorescence increased with the initial FITC–PEG–lipid input (Fig. 2C). Saturation was observed as the retained FITC–PEG–lipid approached a plateau at approximately 40 μg per mL of RBCs. Based on the RBC concentration and the molecular weight of the ligand, it is estimated that each RBC ghost contains about 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 FITC–PEG–lipid. Fig. 2C also demonstrates the adjustability of ligand density on the membranes by controlling the lipid-tethered ligand input.
000 FITC–PEG–lipid. Fig. 2C also demonstrates the adjustability of ligand density on the membranes by controlling the lipid-tethered ligand input.
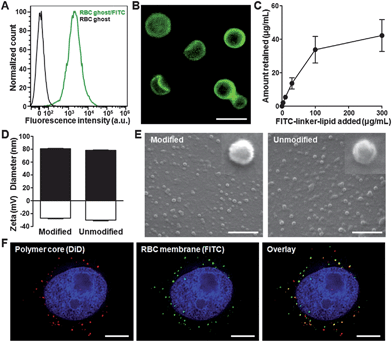 | ||
| Fig. 2 Lipid-insertion enables modification of RBC-NPs with FITC. (A) Flow cytometry histograms of plain RBC ghosts (black) and RBC ghosts incorporated with FITC–linker–lipid (green). (B) Fluorescence microscopy visualization of RBC ghosts modified with FITC (green). Scale bar = 8 μm. (C) FITC–linker–lipid was incubated with RBC ghosts derived from 1 mL of mouse blood. The amount of FITC–linker–lipid incorporated onto the RBC ghosts was then quantified after 30 min of incubation and plotted against the initial input. (D) Physicochemical characterizations (size and zeta potential) of both FITC-modified and unmodified RBC-NPs. (E) SEM images of FITC-modified and unmodified RBC-NPs. Insets represent a single particle with a size of ∼80 nm. Scale bars = 500 nm. (F) Colocalization of the polymeric core (red) and the FITC-modified RBC membrane shell (green) upon intracellular uptake by KB cells. Cellular nuclei were stained with DAPI (blue). Scale bars = 8 μm. | ||
Upon confirming the incorporation of FITC onto the RBC ghosts, the membrane materials were then used to prepare FITC-modified RBC-NPs. Following a previously reported protocol,9 the FITC-modified RBC membrane ghosts were extruded to form ∼100 nm vesicles, which were then mixed and extruded with 70 nm PLGA particles to generate FITC-modified RBC-NPs. For comparison, RBC-NPs coated with unmodified RBC membranes were also prepared. Characterization of the two particles by dynamic light scattering (DLS) showed similar physicochemical properties between the FITC-modified and unmodified RBC-NPs (Fig. 2D). Both particles were approximately 80 nm in mean diameter and with a zeta potential below −25 mV. Scanning electron microscopy (SEM) further demonstrated the similarity between the two particle types (Fig. 2E), both of which were spherical in morphology and exhibited monodisperse population distributions. To confirm the colocalization of lipid-tethered ligands with the polymeric cores, an in vitro fluorescence colocalization study was conducted by loading DiD dye (excitation/emission = 644/663 nm) into the polymeric cores. Following cellular uptake, significant overlap was observed between the DiD-specific red punctates and the FITC-specific green punctates (Fig. 2F). The colocalization pattern confirms the presence of lipid-tethered FITC on the surface of the polymeric cores, demonstrating successful preparation of ligand-modified RBC-NPs.
After validating the lipid-insertion method for RBC-NP functionalization using lipid-tethered fluorescent probes, particle modification with cancer-targeting ligands was explored. A small molecule ligand, folate (MW ∼ 441 Da), which has a similar molecular weight to FITC, was first examined. Folate-functionalized nanocarriers have broad applicability as folate receptors are overexpressed on several types of cancers.27 Upon receptor-mediated binding, folate-functionalized nanocarriers can deliver their cargoes intracellularly through an endocytic uptake pathway. The benefit of folate-induced cancer targeting has been demonstrated on several nanocarrier platforms,28,29 and thus its incorporation onto RBC-NPs would improve the particles' utility in cancer drug delivery.
To prepare folate-functionalized RBC-NPs, a commercially available folate–PEG–lipid conjugate was used (Fig. 3A). Folate-functionalized RBC-NPs were prepared using RBC ghosts inserted with folate–PEG–lipid. Since the targeting ability of folate-functionalized nanoparticles has already been well-established, the KB cell line, a model cancer cell line overexpressing the folate receptor that is commonly used to evaluate folate targeting,30–32 was used to confirm successful functionalization of the RBC-NPs. To assess for folate-mediated differential uptake, the cells were cultured in folate-free media and incubated with folate-functionalized RBC-NPs, non-targeted RBC-NPs, or folate-functionalized RBC-NPs together with 1 mM of free folate. The cells from each sample were then detached, washed, and analyzed using flow cytometry (Fig. 3B and C). Compared to the non-targeted RBC-NPs, incubation of folate-functionalized RBC-NPs resulted in an 8-fold increase in cellular uptake. Conjoint incubation with folate-functionalized RBC-NPs and 1 mM of free folate yielded a similar level of cellular uptake as compared to non-targeted RBC-NPs, which indicated that the increased uptake of folate-functionalized RBC-NPs was receptor-specific. Fluorescence microscopy visualization of particle uptake further confirmed the results observed from flow cytometry. As shown in Fig. 3D, fluorescence from the DiD dye encapsulated inside the particles was only observed in cells incubated with folate-functionalized RBC-NPs in the absence of free folate molecules. To demonstrate that the targeting effects were exclusive to cells overexpressing the folate receptor, a negative cancer cell line, A549,33 that does not overexpress the folate receptor was incubated with either unmodified or folate-functionalized RBC-NPs. No increased uptake was observed for the targeted nanoparticles compared to the unmodified nanoparticles using both flow cytometry and fluorescence imaging (ESI Fig. S1†). It is also important to note that no cytotoxicity was observed for the RBC-NPs when incubated with human umbilical vein endothelial cells (HUVECs), a normal cell line, at the concentrations used in these studies (ESI Fig. S2†). Overall, the results confirm the receptor-specific targeting capability of folate-functionalized RBC-NPs.
 | ||
| Fig. 3 Lipid-insertion enables targeting functionalization of RBC-NPs with folate. (A) Schematic representation of folate–linker–lipid. (B) Flow cytometry histograms of KB cells alone (black) and the cells incubated with folate-functionalized RBC-NPs (red), non-targeted RBC-NPs (blue), and folate-functionalized RBC-NPs together with free folate (orange). (C) Quantification of the mean fluorescence intensity of the histograms in (B). (D) Fluorescence microscopy image of KB cells incubated with folate-functionalized RBC-NPs, non-targeted RBC-NPs, and folate-functionalized RBC-NPs together with free folate. A fluorescent probe DiD was loaded inside the RBC-NPs for visualization (red) and cellular nuclei were stained with DAPI (blue). Scale bars = 25 μm. | ||
To demonstrate that the lipid-insertion method can be applied to targeting ligands of different length scales, particle functionalization with a nucleolin-targeting oligonucleotide, AS1411 aptamer (MW ∼ 9000 Da), was also tested. Oligonucleotide-based targeting agents, or aptamers, are a versatile class of ligands that can be customized against specific receptors through affinity screening.34 AS1411, a 26-mer DNA aptamer with the sequence GGT GGT GGT GGT TGT GGT GGT GGT GG, has shown targeting capability against several cancer cell types owing to frequent overexpression of surface nucleolin on cancerous cells.35–37 The ligand has also been applied for the preparation of cancer-targeted nanoparticles,38,39 thus its integration onto the RBC-NPs would greatly benefit the utility of the platform.
To incorporate aptamers onto RBC-NPs via lipid-insertion, a lipid–tethered AS1411 was first prepared. AS1411 aptamers containing a 3′ thiol modifier was reduced using tris[2-carboxyethyl]phosphine (TCEP) and conjugated to lipid–PEG–maleimide via maleimide–sulfhydryl chemistry (Fig. 4A). After purification, the AS1411–PEG–lipid conjugates were used to prepare RBC-NPs following the aforementioned procedures. In a cellular uptake study using a surface nucleolin expressing breast cancer cell line, MCF-7,35 differential targeting was observed. Flow cytometry analysis revealed that the targeted nanoparticles induced a two-fold increase in cellular uptake as compared to the non-targeted RBC-NPs (Fig. 4B and C). The uptake enhancement by the targeted nanoparticles was also confirmed to be receptor-specific, as blocking by free AS1411 reduced the particle uptake to the level of the non-targeted nanoparticles. Fluorescence imaging corroborated the flow cytometry results with the AS1411-fuctionalized RBC-NPs showing much greater uptake than the non-targeted and blocked samples (Fig. 4D). The results demonstrate that the lipid-insertion method can be applied to relatively large targeting ligands (e.g., MW ∼ 9000 Da) with a molecular weight larger than that of the lipid anchor (MW ∼ 748 Da).
 | ||
| Fig. 4 Lipid-insertion enables targeting functionalization of RBC-NPs with AS1411 aptamer. (A) Schematic representation of AS1411–linker–lipid. (B) Flow cytometry histograms of MCF-7 cells alone (black) and the cells incubated with AS1411-functionalized RBC-NPs (red), non-targeted RBC-NPs (blue), and AS1411-functionalized RBC-NPs together with the free AS1411 aptamer (orange). (C) Quantification of the mean fluorescence intensity of the histograms in (B). (D) Fluorescence microscopy of MCF-7 cells incubated with AS1411-functionalized RBC-NPs, non-targeted RBC-NPs, and AS1411-functionalized RBC-NPs together with free AS1411. A fluorescent probe DiD was loaded inside the RBC-NPs for visualization (red) and cellular nuclei were stained with DAPI (blue). Scale bars = 25 μm. | ||
Conclusions
In summary, by employing a lipid-insertion technique for the functionalization of biological membranes, targeted RBC-NPs were successfully prepared with two different types of targeting ligands. Through the aid of lipid tethers and the dynamic conformation of membrane bilayers, targeting ligands can be spontaneously incorporated onto the RBC-NP platform without exposing the biological membranes to chemical reactions. The robustness and simplicity of this functionalization scheme can enable a wide array of functionalized RBC-NPs for specific disease treatments. In addition, the technique can be generalized to help improve the applicability of emerging biologically inspired nanocarriers possessing complex surface chemistry. The capability to control and adjust ligand density through the lipid-insertion technique also provides versatility for platform optimization. Future studies are warranted to examine the in vivo implications of ligand functionalization on RBC-NPs. The targeted RBC-NPs reported in the present work possess significant potential for cancer treatments as they integrate nature's immune-evasive moieties with cancer-binding ligands.Acknowledgements
This work is supported by the National Science Foundation Grant DMR-1216461. R.F. is supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program. B.L. is supported by a National Institutes of Health R25CA153915 training grant from the National Cancer Institute.Notes and references
- S. D. Li and L. Huang, Mol. Pharmaceutics, 2008, 5, 496–504 CrossRef CAS.
- H. Maeda, J. Wu, T. Sawa, Y. Matsumura and K. Hori, J. Controlled Release, 2000, 65, 271–284 CrossRef CAS.
- A. K. Iyer, G. Khaled, J. Fang and H. Maeda, Drug Discovery Today, 2006, 11, 812–818 CrossRef CAS.
- R. Singh and J. W. Lillard, Jr, Exp. Mol. Pathol., 2009, 86, 215–223 CrossRef CAS.
- D. F. Emerich and C. G. Thanos, J. Drug Targeting, 2007, 15, 163–183 CrossRef CAS.
- L. Brannon-Peppas and J. O. Blanchette, Adv. Drug Delivery Rev., 2004, 56, 1649–1659 CrossRef CAS.
- R. H. Fang, C. M. Hu and L. Zhang, Expert Opin. Biol. Ther., 2012, 12, 385–389 CrossRef CAS.
- C. M. Hu, R. H. Fang and L. Zhang, Adv. Healthcare Mater., 2012, 1, 537–547 CrossRef CAS.
- C. M. Hu, L. Zhang, S. Aryal, C. Cheung, R. H. Fang and L. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10980–10985 CrossRef CAS.
- C. M. Hu, R. H. Fang, J. Copp, B. T. Luk and L. Zhang, Nat. Nanotechnol., 2013, 8, 336–340 CrossRef CAS.
- D. D. Kim and W. C. Song, Clin. Immunol., 2006, 118, 127–136 CrossRef CAS.
- P. A. Oldenborg, A. Zheleznyak, Y. F. Fang, C. F. Lagenaur, H. D. Gresham and F. P. Lindberg, Science, 2000, 288, 2051–2054 CrossRef CAS.
- R. K. Tsai and D. E. Discher, J. Cell Biol., 2008, 180, 989–1003 CrossRef CAS.
- R. K. Tsai, P. L. Rodriguez and D. E. Discher, Blood Cells, Mol., Dis., 2010, 45, 67–74 CrossRef CAS.
- O. C. Farokhzad and R. Langer, Adv. Drug Delivery Rev., 2006, 58, 1456–1459 CrossRef CAS.
- M. E. Davis, Z. Chen and D. M. Shin, Nat. Rev. Drug Discovery, 2008, 7, 771–782 CrossRef CAS.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS.
- V. Wagner, A. Dullaart, A. K. Bock and A. Zweck, Nat. Biotechnol., 2006, 24, 1211–1217 CrossRef CAS.
- L. Zhang, F. X. Gu, J. M. Chan, A. Z. Wang, R. S. Langer and O. C. Farokhzad, Clin. Pharmacol. Ther., 2008, 83, 761–769 CrossRef CAS.
- K. C. R. Bahadur, B. Thapa and P. S. Xu, Mol. Pharmaceutics, 2012, 9, 2719–2729 CrossRef.
- D. Bartczak and A. G. Kanaras, Langmuir, 2011, 27, 10119–10123 CrossRef CAS.
- T. Chen, I. Ocsoy, Q. Yuan, R. W. Wang, M. X. You, Z. L. Zhao, E. Q. Song, X. B. Zhang and W. H. Tan, J. Am. Chem. Soc., 2012, 134, 13164–13167 CrossRef CAS.
- C. M. Hu, S. Kaushal, H. S. T. Cao, S. Aryal, M. Sartor, S. Esener, M. Bouvet and L. Zhang, Mol. Pharmaceutics, 2010, 7, 914–920 CrossRef CAS.
- E. L. Jin, B. Zhang, X. R. Sun, Z. X. Zhou, X. P. Ma, Q. H. Sun, J. B. Tang, Y. Q. Shen, E. Van Kirk, W. J. Murdoch and M. Radosz, J. Am. Chem. Soc., 2013, 135, 933–940 CrossRef CAS.
- M. M. J. Kamphuis, A. P. R. Johnston, G. K. Such, H. H. Dam, R. A. Evans, A. M. Scott, E. C. Nice, J. K. Heath and F. Caruso, J. Am. Chem. Soc., 2010, 132, 15881–15883 CrossRef CAS.
- J. Wang, S. M. Tian, R. A. Petros, M. E. Napier and J. M. DeSimone, J. Am. Chem. Soc., 2010, 132, 11306–11313 CrossRef CAS.
- J. Sudimack and R. J. Lee, Adv. Drug Delivery Rev., 2000, 41, 147–162 CrossRef CAS.
- X. Pan and R. J. Lee, Expert Opin. Drug Delivery, 2004, 1, 7–17 CrossRef CAS.
- J. D. Byrne, T. Betancourt and L. Brannon-Peppas, Adv. Drug Delivery Rev., 2008, 60, 1615–1626 CrossRef CAS.
- R. J. Lee and P. S. Low, J. Biol. Chem., 1994, 269, 3198–3204 CAS.
- R. J. Lee and P. S. Low, Biochim. Biophys. Acta, Biomembr., 1995, 1233, 134–144 CrossRef.
- S. Wang, R. J. Lee, G. Cauchon, D. G. Gorenstein and P. S. Low, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 3318–3322 CrossRef CAS.
- H. S. Yoo and T. G. Park, J. Controlled Release, 2004, 100, 247–256 CrossRef CAS.
- G. Mayer, M. S. L. Ahmed, A. Dolf, E. Endl, P. A. Knolle and M. Famulok, Nat. Protoc., 2010, 5, 1993–2004 CrossRef CAS.
- S. Soundararajan, W. W. Chen, E. K. Spicer, N. Courtenay-Luck and D. J. Fernandes, Cancer Res., 2008, 68, 2358–2365 CrossRef CAS.
- F. Mongelard and P. Bouvet, Curr. Opin. Mol. Ther., 2010, 12, 107–114 CAS.
- S. Soundararajan, L. Wang, V. Sridharan, W. W. Chen, N. Courtenay-Luck, D. Jones, E. K. Spicer and D. J. Fernandes, Mol. Pharmaceutics, 2009, 76, 984–991 CrossRef CAS.
- A. Aravind, P. Jeyamohan, R. Nair, S. Veeranarayanan, Y. Nagaoka, Y. Yoshida, T. Maekawa and D. S. Kumar, Biotechnol. Bioeng., 2012, 109, 2920–2931 CrossRef CAS.
- D. W. Hwang, H. Y. Ko, J. H. Lee, H. Kang, S. H. Ryu, I. C. Song, D. S. Lee and S. Kim, J. Nucl. Med., 2010, 51, 98–105 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3nr03064d |
| This journal is © The Royal Society of Chemistry 2013 |
