 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Three dimensional quantitative characterization of magnetite nanoparticles embedded in mesoporous silicon: local curvature, demagnetizing factors and magnetic Monte Carlo simulations†
Toni
Uusimäki
*ab,
Georgios
Margaris
c,
Kalliopi
Trohidou
c,
Petra
Granitzer
d,
Klemens
Rumpf
d,
Meltem
Sezen
be and
Gerald
Kothleitner
ab
aGraz University of Technology, Institute for Electron Microscopy and Nanoanalysis (FELMI), Steyrergasse 17, 8010, Graz, Austria. E-mail: toni.uusimaki@felmi-zfe.at; Tel: +43 316 8738320
bCenter for Electron Microscopy (ZFE), Steyrergasse 17, 8010, Graz, Austria
cInstitute of Materials Science, NCSR Demokritos, Ag. Paraskevi, Athens, Greece
dKarl-Franzens-University of Graz, Institute of Physics, Universitätsplatz 5, 8010, Graz, Austria
eCurrently at Sabanci University, Nanotechnology Research and Application Center, Orhanli, Tuzla, 34956 Istanbul, Turkey
First published on 24th September 2013
Abstract
Magnetite nanoparticles embedded within the pores of a mesoporous silicon template have been characterized using electron tomography. Linear least squares optimization was used to fit an arbitrary ellipsoid to each segmented particle from the three dimensional reconstruction. It was then possible to calculate the demagnetizing factors and the direction of the shape anisotropy easy axis for every particle. The demagnetizing factors, along with the knowledge of spatial and volume distribution of the superparamagnetic nanoparticles, were used as a model for magnetic Monte Carlo simulations, yielding zero field cooling/field cooling and magnetic hysteresis curves, which were compared to the measured ones. Additionally, the local curvature of the magnetite particles' docking site within the mesoporous silicon's surface was obtained in two different ways and a comparison will be given. A new iterative semi-automatic image alignment program was written and the importance of image segmentation for a truly objective analysis is also addressed.
1 Introduction
Porous silicon (pSi), a versatile material achieved by nanostructuring of a silicon wafer, is employed in various fields such as in sensors, photovoltaics and energetic materials.1 Particularly, properties like photo-2,3 and electroluminescence4 in the visible range, its specific surface chemistry1 and the possibility to control the pore-size without pre-structuring make it highly attractive. Its biodegradability5 and bioactivity6 furthermore renders this material useful for biomedical applications.Due to the tunable size and the growth mechanism of the pores, as well as their quasi-regular arrangement,7 pSi is used as a template for the deposition of various molecules in controlled drug delivery.8 The embedding of iron oxide nanoparticles specifically holds a lot of promise in biomedicine due to their low toxicity. Thus the combination of both materials is of fundamental interest in basic research. In the case of magnetite nanoparticle infiltration within the pores, the magnetic properties can be tuned by the particle-size and their distance between each other.
Electron tomography9,10 (ET) is a method for directly accessing the three dimensional morphology of the material with nanometer resolution. It has proven to be an invaluable tool for the characterization of nanoparticles.11–13 In ET, first a series of projection images are acquired in a transmission electron microscope (TEM) by changing the tilt angle of the sample with respect to the electron beam. Secondly the acquired tilt series needs to be accurately aligned for obtaining a high quality reconstruction. This is usually done by tracking fiducial markers or by cross-correlation. In a third step, the aligned tilt series is then backprojected to form a digital three dimensional representation of the sample, usually either with a weighted backprojection (WBP) algorithm, or with a simultaneous iterative reconstruction technique (SIRT).14,15 Finally, for any kind of quantitative analysis, the reconstructed volume is subject to a segmentation process in order to partition the volume into specific objects of interest.
Here we propose a new iterative semi-automatic alignment method, where the image under alignment is compared with its reprojection from the initial reconstructed volume, thereby showing the translation needed for a correct alignment. Also to improve the highly important segmentation step, a public library of image manipulation algorithms Insight Toolkit (ITK)16 was employed and integrated as a dynamic library for Gatan Inc.'s Digital Micrograph™ (DM) software.17 To date it holds about 40 different algorithms written from the ITK library for image filtering, deconvolution, segmentation and optimization based image registration.16 These ITK algorithms were made publicly available and have been used to process data via scripting commands in DM.
High-angle annular dark-field scanning TEM (HAADF-STEM) imaging was used to acquire the tilt series, as it minimizes diffraction contrast, which violates the projection requirement.18 It states that the acquired intensity/signal has to be a monotonic function of some physical property of the sample (mass/thickness in conventional TEM or Z2 in HAADF-STEM).19
For optimized design of a catalytic material in terms of reactivity, the knowledge of the preferred adsorption sites of the particles in terms of surface curvature is advantageous. For this reason the analysis of the local curvature (LC) distribution of the magnetite particles' adsorption site on the surface of the pSi was studied.
The demagnetizing field generated within a particle as a response to the external magnetic field acts to reduce the magnetic moment of the particle. This gives rise to shape anisotropy and is determined by the demagnetizing factors (DFs). For a general ellipsoid, these factors can be calculated as defined in ref. 20. Linear least squares optimization was used to fit a general ellipsoid to the particles' surface mesh vertices. This allowed us to extract the area and volume of the particles, as well as the radii of the semi-axes along with their normalized directions. Therefore ET proves to be a unique tool to acquire the DF distribution of a nanoparticle assembly. The spatial and volume distribution were used as an input to magnetic Monte Carlo (MC) simulations with and without the knowledge of the DF distribution. A magnetic hysteresis curve and zero field cooling/field cooling (ZFC/FC) magnetization vs. T curves were simulated and compared to the measured ones from the same sample before the tomographic investigation.
The MC simulation technique with the implementation of the Metropolis21 algorithm was chosen here, since the influence of the finite temperature can be studied. The magnetostatic interactions of the nanoparticles in an assembly cannot be neglected due to their long range order; hence the dipolar interparticle interactions were included in the simulations. Performing simulations on magnetic nanoparticle assemblies usually requires assumptions to be made on the spatial and volume distribution of the sample. Interparticle interactions are characterized by three degrees of disorder: surface topology, volume distribution and the random distribution of the easy axes.22 In this study, we aim to prove that ET can readily solve the first two of these problems not forgetting the spatial distribution.
A list of abbreviations and symbols can be found in the ESI.†
2 Experimental section
2.1 Specimen preparation
The pSi templates were fabricated by anodization of a highly n-doped silicon wafer in aqueous hydrofluoric acid solution. By controlling the electrochemical parameters such as current density, electrolyte concentration and bath temperature, oriented pores were grown perpendicular to the surface of the (100) silicon wafer.23 For this work, a sample with an average pore diameter of 80 nm and a mean pore wall thickness of 40 nm was used. A thickness of about 35 μm of the porous layer has been obtained during the anodization procedure. In addition to the main pores, smaller dendritic side pores (<20 nm) were grown in the 〈111〉 direction, forming a complex three dimensional structure consisting of dendritic, oriented pores, which are separated from each other. Fig. 1(a) and (b) shows an overview of the cross-section and a high magnification scanning electron microscope (SEM) image of the investigated sample respectively before the infiltration of the magnetite nanoparticles. In Fig. 1(c) the quasi-regular arrangement of the pores is shown at the surface. Fe3O4 nanoparticles of ∼5 nm size have been infiltrated into the pores, which were prepared by high temperature decomposition in the presence of an organic precursor as explained in ref. 24. To avoid agglomeration and oxidation, the particles were coated with oleic acid, and dispersed in hexane solution.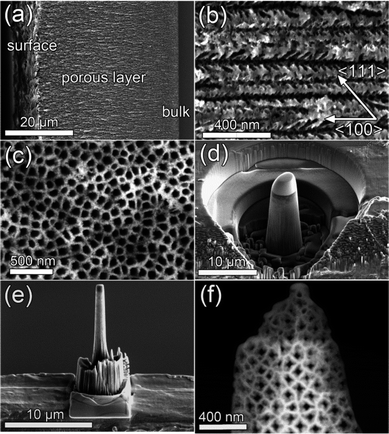 | ||
| Fig. 1 SEM image showing (a) an overview cross-section of the sample (b) the dendritic nature of the main pores at higher magnification and (c) the quasi-regular arrangement of the pores. (d) Pillar shaping of the sample via FIB milling using annular patterns. (e) The pre-cut was transferred on top of a Molybdenum semi-grid using a micromanipulator and thinned further on the edge applying low ion currents. (f) STEM image showing the resulting structure of the sample as used in the tomographic tilt series acquisition. | ||
The TEM preparation of the material was accomplished using a FEI Nova 200 Nanolab Dual-Beam (FIB-SEM) Instrument, where a pillar shaped sample was milled under cryo conditions (−160 °C measured at the stage) and transferred to a TEM grid as seen in Fig. 1(d) and (e). This geometry avoids self-shadowing, thickness changes during tilting and allows for higher maximum tilt angles, minimizing the missing wedge, which in turn deteriorates the resolution of reconstruction.10 Investigation of the reconstructions of samples milled at room temperature showed that some of the magnetite nanoparticles were fully embedded inside the silicon material. Since silicon is crystalline, this implies that the ion beam was redepositing silicon in the FIB preparation process. This effect was not found in cryo prepared samples. The final milling and polishing step was performed using low ion beam currents (1 pA). Before the TEM investigations, the sample was plasma cleaned using a Fischione Plasma Cleaner model 1020 for 2 minutes in order to minimize hydro-carbon build-up during acquisition. This procedure also effectively removed the oleic acid coating of the magnetite nanoparticles. The final structure of the sample used in the tomographic investigations is shown in Fig. 1(f).
2.2 Measurements and equipment
From previous studies, FTIR and EDX spectra show the presence of oleic acid and magnetite nanoparticles, respectively within the pSi template.25 Patterns of X-ray diffractograms of Fe3O4 nanoparticles correspond to an inverse spinel structure with lattice parameters of 8.38 Å, which is in good agreement with published magnetite patterns.26 Magnetization measurements have been carried out with a SQUID (Cryogenics) and a VSM (Quantum Design), respectively. To achieve field dependent magnetization curves a magnetic field between ±6 T was applied parallel and perpendicular to the pores. Temperature dependent magnetization was measured between 4 and 300 K.27A 200 kV Tecnai F20 equipped with a Schottky field-emission gun and a HAADF detector model Fischione 3000 was used to acquire a HAADF-STEM single axis tilt series from −79° to 78° with 1° tilt increments. The camera length was set to 200 mm and a spot size corresponding to 0.3 nm and a probe current of 20 pA was used. Dynamic focusing was not needed in this study because of the pillar shaped sample structure, staying in plane while tilted.
2.3 Projection-re-projection comparison alignment
The images were first roughly aligned using the common cross-correlation procedure followed by a method based on the center of mass as depicted in ref. 25. However, these methods were found not to be accurate enough to yield a high quality reconstruction. Hence, an iterative alignment method was implemented (realized in DM based on the ITK algorithms). Here the roughly aligned tilt series was first reconstructed using the WBP algorithm and then re-projected to form another tilt series, which was then cross-correlated with the original to find the correct translation. Alternatively one can calculate the maximum mutual information by exhaustive search to determine the misalignment.28 For this procedure the term projection-re-projection comparison (PRC) seems to be appropriate.The re-projections can be greatly enhanced by using a weighted sum of the maximum projection29 and the usual integral projection. In maximum projection, only the brightest pixel along the projected ray is displayed, therefore making the high intensity/high contrast objects clearly visible in HAADF-STEM images, which is crucial to obtain a correct alignment. In Fig. 2 one can see zero degree re-projections taken from an initial reconstruction using Fig. 2(a) maximum projection and Fig. 2(b) integral projection.
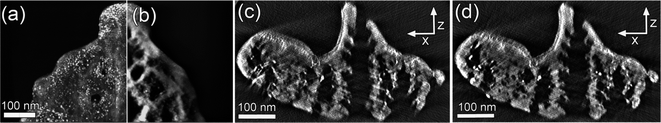 | ||
Fig. 2 (a) Re-projections from an initial reconstruction, using (a) maximum projection and (b) integral projection. Weighted sum (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) of these images was used in the cross-correlation with the acquired zero tilt image. (c) Reconstructed xz-plane (SIRT, 20 iterations) after the coarse alignment showing obvious misalignment errors as compared to (d), a reconstruction after three iterations of the PRC algorithm. 1) of these images was used in the cross-correlation with the acquired zero tilt image. (c) Reconstructed xz-plane (SIRT, 20 iterations) after the coarse alignment showing obvious misalignment errors as compared to (d), a reconstruction after three iterations of the PRC algorithm. | ||
The quality of the PRC alignment judged by a visual criterion is clearly better as seen in Fig. 2(c) and (d). Here the former image was reconstructed using SIRT with 20 iterations after aligning the tilt series with two iterations of the usual cross-correlation procedure and one refinement step based on the centre of mass. In cross-correlation, bandpass and hanning window filtering applied to a rectangular region of interest close to the tilt axis was used to optimize the process. The latter image was reconstructed approximately on the same plane, by using the same parameters. The reconstruction quality is clearly superior already after three iterations of the PRC alignment algorithm. The global tilt axis rotation and shift have been checked between each of the three iterations.
2.4 Segmentation
The aligned tilt series was reconstructed with a SIRT algorithm over 20 iterations. For any kind of quantitative analysis concerning ET data, it is paramount to obtain an objective segmentation and to minimize the user bias. The reconstructed volume was smoothed using a modified edge-preserving curvature anisotropic diffusion filter30 followed by a non-local means filter. To be short, the latter uses a search window to compare voxels' similarity to its neighbors, which is then used as a weighting function to be multiplied with that voxel. In the former, the strength of the smoothing parameter, i.e. the diffusion coefficient depends on the local gradient direction of the data. Diffusion takes place on voxels within a similar region rather than across boundaries, thus smoothing the data while preserving the edges.For the segmentation of the magnetite particles, the volume was first processed using a top hat filter that takes into account the area of the objects.31 This filter enhances the relative brightness of the nanoparticles thus allowing for a global threshold. The segmented particles were then processed by the common distance transform and watershed algorithm to label the particles and to separate the few agglomerated ones.32
For the segmentation of the template, a script was written within the DM environment to easily define seed points into the volume, which were used in a connected threshold algorithm.33 This segmentation routine uses a region growing method and includes those connected voxels to the segmentation, whose intensities fall in between the user determined minimum and maximum threshold levels. Fig. 3(a) shows the generated surface from the whole segmentation. For the actual analysis only the inner part of the pSi structure was studied since the outer surface was in direct contact with the ion beam in the FIB sample preparation process. Fig. 3(b) shows a region from the central slice (xy-plane) of the reconstruction and Fig. 3(c) is the segmentation of it. The dendritic growth of the pores in the 〈111〉 direction of the silicon wafer creates a highly complex network of the pores, making it difficult to separate out the main pores quantitatively. As shown in the ESI,† the main pores investigated here were found to be separated as claimed in ref. 23. In two pores, there were no magnetite nanoparticles found at all.
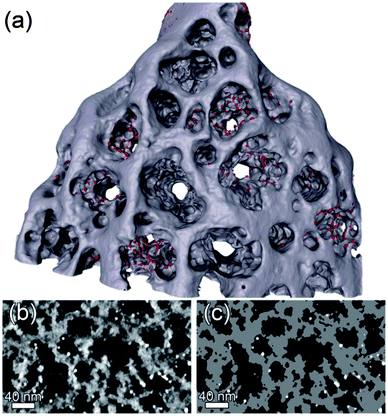 | ||
| Fig. 3 (a) Triangulated surface of the whole reconstruction (red: Fe3O4, grey: Si). (b) A detail of the central slice (xy-plane) of the reconstruction after filtering and (c) segmentation (white: Fe3O4, grey: Si). | ||
3 Analysis
3.1 Local curvature distribution
From a triangulated surface mesh (processed within the Amira34 programming environment), the principal curvatures (κ1, κ2) could be derived, opening the way for further calculations of the local curvature (LC) distribution. In particular, two approaches are proposed and discussed. In the first method, a set of rings were defined around the particles. The first ring goes through the branching points, which are the points shared between the particle and the template. The following rings were determined using the neighbourhood connectivity information of the triangles as illustrated in Fig. 4(d) for the first 3 rings. The local curvature for every particle was then calculated as the mean curvature value of these points, separately for every ring. The second method involved a fit of the points' three dimensional coordinates to a polynomial surface (carried out in Matlab35). This method approximates the materials surface at the particle location assuming it would have formed similar to its environment without particles.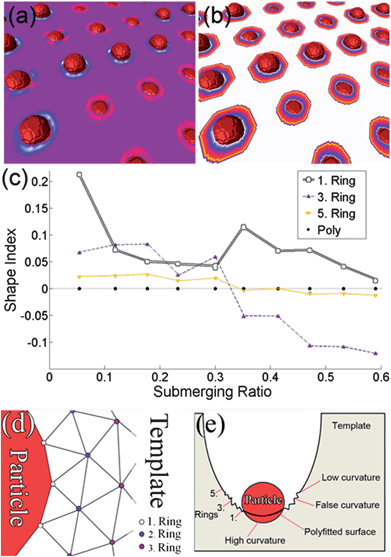 | ||
| Fig. 4 (a) Particles with different submerging ratios, showing negative (blue, white) or positive (red) local curvature instead of zero (purple) on a flat surface. (b) Visualization of the rings colored according to the ring value (white: 1. ring – > yellow: 5. ring). (c) Shape index of the test image with respect to the submerging ratio. (d) The triangulation scheme used to define the rings. (e) An overview image on both methods. | ||
The shape index36 can be defined as
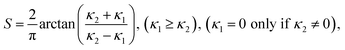 | (1) |
A similar approach to the ring method is reported in ref. 37, where the mean value of the shape index falling inside a sphere within a 1 nm radius was taken. This could lead to incorrect results if there is an adjacent surface near the particles. For both of these methods the quality of the triangulated mesh in the vicinity of sharp edges and at the transition of two different materials (the supporting template and the particle) is usually very poor. Depending on the submerging ratio rS of the surface area of the particle outside and inside the template, defined as rS = AOUT/(AOUT + AIN), the mesh of the template tends to be “upward” or “downward” in the vicinity of the particle, giving positive or negative curvature values respectively, regardless of the real curvature. As a remedy, an option was set up within the Amira module to exclude rings with obviously false points; however at the expense of locality.
To obtain a more quantitative assessment on the influence of this approach, a test image (1024 × 1024 × 50) was generated, where 6 pixel diameter spheres were brought into contact with a flat template (Fig. 4(a) and (b)) with different submerging ratios. The local curvature of the surface should therefore be zero everywhere. A flat surface was chosen here for simplicity even though the shape index is not explicitly defined. The exact number of points depends on the chosen mesh quality, on the size of the particles themselves and the submerging ratio, and differed consequently within the sample. The mesh quality was determined by a minimum edge length value, which was chosen to be non-limiting. In the test image (and in the reconstruction), the rings were separated approximately one pixel apart. The distance of the rings to the particle surface was hence directly correlated with the voxel size of the volume such that, within the pSi reconstruction, the first ring was touching the particle (branching points), the second ring was one pixel apart, etc., amounting to 0.76 nm each. The mean value for the number of points in rings 1, 3 and 5 can be exactly given and was 38.5, 50.5 and 55 respectively. Fig. 4(c) shows the value of the shape index as a function of the submerging ratio for each ring number. The shape index for the first ring (white squares) was always found to be positive. For the rings 2 to 4, the curvature's sign depends on the submerging ratio. Not until the 5th ring (yellow downward triangles), the shape index was zero within a 0.05 resolution as expected. Similar results were obtained using a test template possessing positive curvature with the exemption that the curvature was always lower than the real one regardless of the ring value, as shown later.
As a comparison, we also extracted the LC distribution by fitting a surface to the point coordinates with polynomial regression.38 The mean value of the point sets' normal directions was measured to ensure the correct sign for the curvature. For a reliable polynomial fit, it was crucial to include an arbitrary three dimensional rotation to the point sets, which was realized in the minimization process. Since the optimization routine did not support integer values, the optimization was performed for each polynomial degree independently in a loop, where the degree was limited to 4. Higher degrees tend to give better mean squared errors (MSE) for the distances of the original points to the fitted surface, but may produce unphysical zero-crossings in the area of the particle. In order to properly include or exclude fits, threshold values were set on the MSEs. To determine the lowest order polynomial degree, individual yet minimum values for the MSE were set as thresholds, rejecting inaccurate fits or higher order polynomials. The obtained polynomial surface was then used to calculate the shape index from analytical principal curvatures and the local curvature for each particle was taken as an averaged value of a small patch around the centre point of the first ring. In the case of the test sample in Fig. 4(c), this method yields zero curvature (black circles) for every submerging ratio. In the calculations, all the point coordinates from the second up to the fifth ring were included for the fitting process.
In Fig. 5(a) and (b) two examples of the fitting process are shown from the pSi surface. The blue line shows the mean value of the points' direction normal, that is, on which side the particle was and ensures the correct sign for the shape index after the rotation of the point set. The coloring depicts the shape index from the fit. In Fig. 5(c) one can see a triangulated mesh of one individual pore and in Fig. 5(d) a polyfitted surface representation of it.
 | ||
| Fig. 5 Fitted polynomial surface to predict the local curvature on the position of the particles. Examples of the process are shown in (a) and (b), where the original points are shown and the fitted surface with coloring indicating the shape index. The blue line is the mean direction normal of the points. (c) Triangulated mesh of the magnetite particles (red) in one pore. (pSi: grey) (d) Visualization of the results of the polynomial regression (fitted surface: blue). | ||
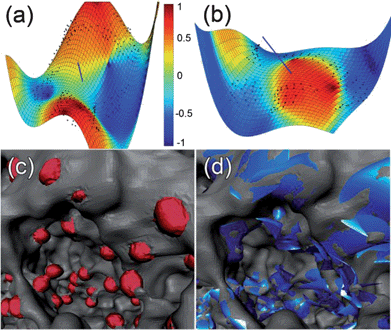
The LC distributions for different ring values are shown in Fig. 6(a)–(c) acquired using the ring method. As expected, the first ring's fitted Gaussian curve consists mainly of positive values, with a mean value of 0.27 and a standard deviation of 0.09. The peak tends to settle around zero within the next rings. The distribution of the shape index for the first method was always found to be a Gaussian curve centered at or close to zero irrespective of the number or locations of the rings used, therefore suggesting that the magnetite particles prefer near flat or saddle like surfaces for adsorption, which is in accordance with the results in ref. 37.
 | ||
| Fig. 6 (a) LC distribution of the (a) first ring, (b) third ring and (c) fifth ring with Gaussian fits (red line). (d) LC distribution obtained with the polynomial regression. (e) The submerging ratio of the segmented particles and the Gaussian fit (red line). (f) Volume distribution obtained from the segmentation using 3D convex hull and (g) using the fitted ellipsoids with log-normal fits (red lines). (h) 3D Feret ratio of the particles and a Gaussian fit (red line). | ||
However, the second method paints a completely different picture. There two main Gaussian curves are centered at −0.43 and 0.51 accompanied by a smaller one at 0.07, indicating that the particles prefer convex or concave locations for adsorption. A possible explanation for the difference in the results can be found in the way the data are extracted. For the ring method, the values are closer to zero simply because of the extension of the particle, and even more, the distance of the rings from the high curvature adsorption site. This naturally explains why the two main peaks in the polyfitted distribution in Fig. 6(d) are both shifted towards zero; forming a similar Gaussian curve as in Fig. 6(c). This is also illustrated in Fig. 4(e), where the high curvature location is correctly estimated using the polyfit method. Near the particle the curvature is false because of the triangulation process and the 5th ring gives lower curvature values than the polyfit method.
In order to prove that the polyfit method can accurately fit high curvature locations, a test image was created (available in the ESI†). There, a particle was situated on a location with a shape index of 0.94 (Amira). With the ring method, strongly deviating results of 0.35, 0.4 and 0.5 were obtained on the first, third and fifth ring respectively. In contrast to that, the polyfit method yielded a value of 0.99 using the mean coordinates of the fitted points. Alternatively, when taking the mean shape index value of a small patch of points (as was done in the pSi sample), i.e. points that are within a Euclidian distance of 0.5 to the mean value of the fitted point coordinates, a value of 0.96 was achieved. Hence it is reasonable to conclude that the polyfit method is closer to the real situation than the ring method. In Fig. 6(e) the histogram of the submerging ratios for the particles is also given.
3.2 Demagnetizing factors
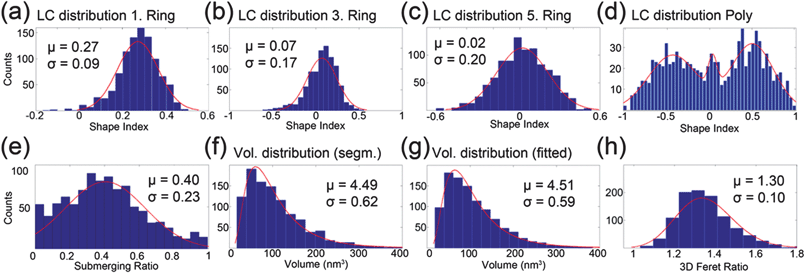
A module was written to export the coordinates of the particles' point sets from Amira to Matlab environment, where an arbitrary ellipsoid was fitted to the points using the linear least squares method.39 Thresholds were used such that the surface area and volume of the particles were similar to the fitted ones. The surface area of the particles was calculated in Amira as the sum of the triangle areas and the volume was calculated using the 3D convex hull of the point set. From the fit it was then possible to calculate the semi-radii of the particles along with the directions of these semi-axes. The longest semi-axis can be interpreted as the shape anisotropy easy axis since the demagnetizing energy is minimized in this direction. More information on the calculation of the DFs is given in the ESI.† Essentially, the equations for the DFs can be directly applied by inserting the semi-axes (a, b and c), which are obtained from the ellipsoidal fitting procedure. In Fig. 6(f) and (g) the volume distribution of the segmented particles and their ellipsoidal fits are presented respectively and the mean diameter of the particles from the former was found to be 5.9 nm. Additionally the 3D Feret ratio is shown in Fig. 6(h) as an overview of the eccentricity of the assembly. The Feret ratio was calculated by dividing the longest semi-axis with the smallest one from the ellipsoidal fit.In Fig. 7(a), the fitted particles of one pore are shown, with the blue lines showing the direction of the shape anisotropy easy axis and its magnitude is the inverse of the DF of that axis. Fig. 7(b) presents the DF distribution as a polar plot, where the angular axis is the angle between the shape anisotropy easy axis and the principal z-coordinate axis. The radial axis is the value of the DF. The z-coordinate is parallel to the pores and also the direction of the optical axis in the microscope. There were no perfectly spherical particles on the red line, which draws the DF value of 1/3. However care should be taken when interpreting this plot, since for example, the closest point to the red line has semi-axes of 2.6 nm, 2.6 nm and 2.5 nm. With a 0.76 nm voxel size in the reconstruction, the 0.1 nm difference in the semi-axes becomes problematic. Also there is a clear tendency of counts in the vicinity of the zero angle with respect to the z-axis. This could be an effect of the missing wedge, which causes elongation in the same z-axis direction, although the nature of the SIRT algorithm should minimize this effect. The maximum difference of the minor diameter from the major diameter is 4.4 nm, while the average is 1.6 nm and minimum of 0.1 nm. Both the average and the maximum values are well above the error coming from the voxel size.
 | ||
| Fig. 7 (a) Fitted ellipsoids (red). Blue points are from the mesh of the generated surface of the particles and the lines are the directions of the shape anisotropy easy axis. The length of the line is the inverse of DF. (b) The angular axis is the angle of the shape anisotropy easy axis with respect to the z-axis vs. the DF on the radial axis. | ||
3.3 Monte Carlo simulations and magnetic measurements
The measured magnetic hysteresis curve and the ZFC/FC magnetization vs. T curve can be seen in Fig. 8(a) and (b) respectively. In the former, a coercive field of 60 Oe was measured and a slight difference on the saturation magnetization between parallel and perpendicular field to the surface can be distinguished. This is due to the difference in the spatial distribution of the particles in the parallel and perpendicular directions to the sample surface. The dipolar coupling between the particles is less in the parallel direction because the pore walls are separating the pores and the particles therein. This behavior becomes stronger as the particle radius grows. The blocking temperature TB in the latter figure was found to be 12 K, which is higher than in non-interacting assemblies of magnetite nanoparticles. The blocking temperature of 4.05 K can be calculated for the non-interacting case using KμV = 25kBTB, where Kμ is the anisotropy energy density, V is the volume and kB is the Boltzmann constant. A mean diameter of 5.9 nm was used from Fig. 6(f) assuming spherical particles.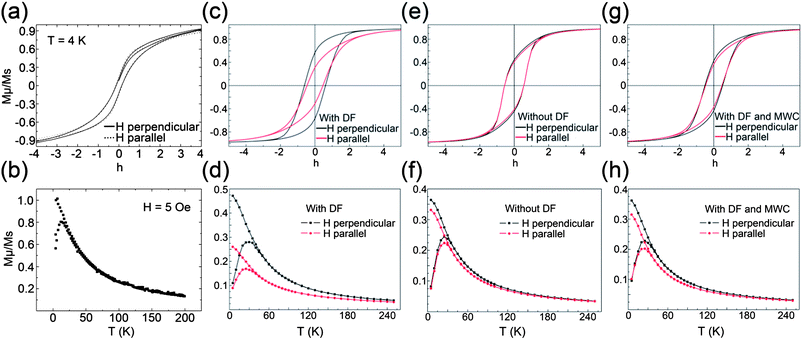 | ||
| Fig. 8 (a) Measured hysteresis loops with magnetic field applied perpendicular (z-axis) and parallel (xy-plane) to the surface. (b) Measured ZFC/FC magnetization vs. T curves. (c) Simulated hysteresis loops including the shape anisotropy (DFs). (d) Simulated ZFC/FC magnetization vs. T curves including the shape anisotropy. (e) Simulated hysteresis loops omitting the shape anisotropy. (f) Simulated ZFC/FC magnetization vs. T curves omitting the shape anisotropy. (g) Simulated hysteresis loops with missing wedge correction and including the shape anisotropy. (h) Simulated ZFC/FC magnetization vs. T curves with missing wedge correction and including the shape anisotropy. The magnetic hysteresis simulations were performed at T = 4.2 K and the ZFC/FC vs. T simulations were performed at a cooling field of Hcool = 50 Oe. | ||
To obtain the magnetic configuration under an applied field H and finite temperature T, the MC simulation, using the standard Metropolis algorithm, was employed.40,41 More detailed information about the simulation can be found in the ESI.† At a given temperature and applied field, the system was allowed to relax towards equilibrium for the first 103 MC steps per spin, and thermal averages were calculated over the subsequent 104 steps. The measurements were averaged over 10 different initial conditions (random configurations of the anisotropy easy axis and initial spin orientations). The error bars were too small to be included in the figures. The mean particle volume was 〈V〉 = 108.01 nm3 according to the log-normal fit in Fig. 6(f). The material values of the bulk Fe3O4, namely, the saturation magnetization MS = 4.8 × 105 A m−1 and the anisotropy energy density Kμ = 1.3 × 104 J m−3 were used in the simulations.
The simulations were performed in two directions for the applied field, namely along the z-axis (perpendicular to the surface) and on the xy-plane (parallel to the surface) and by including or omitting the DFs. The xy-plane simulations were performed with the field rotated in steps of 30° and the final curve was taken by averaging the values over all directions. As can be seen in Fig. 8(c), where the DFs were included, there is a clear difference between the hysteresis loops taken for the two different directions of the applied field. With the field along the z-axis, the hysteresis loop presents a square-like shape, whereas when in plane, its shape is thinner and more inclined. Consequently, the coercive field is much larger when the field is perpendicular to the surface Hc,perp = 0.611 compared to the case of the field parallel to the surface Hc,par = 0.462. In Fig. 8(d) the simulated ZFC/FC magnetization vs. T curves, performed at cooling field Hcool = 50 Oe are shown. One can see that the blocking temperature is higher in the perpendicular applied field (along the z-axis) case (TB,perp = 29 K) than that of the field applied in the xy-plane (TB,par = 24.5 K). Both values were bigger than the measured one. The difference between the two directions can be explained by looking at Fig. 7(b), where a clear accumulation of counts was found to be close to the zero angle to the z-axis. This means that the demagnetizing energy in the Hamiltonian equation (see ESI†) is stronger in the z-direction.
Next, by omitting the demagnetizing (shape) energy term in the Hamiltonian, the hysteresis (Fig. 8(e)) and ZFC/FC vs. T (Fig. 8(f)) curves were calculated. The particles were assumed to be spherical and the radius was calculated from the volume distribution. The difference between the curves calculated with the two directions of the applied field becomes smaller. The coercive field of Hc,no_demagn = 0.557 and a blocking temperature of TB,no_demagn = 25.5 K were found for both directions of the applied field. Therefore the difference between the two hysteresis loops in Fig. 8(c), in this assembly model, can be attributed to the shape anisotropy. The demagnetizing energy is equivalent to a bi-directional anisotropy (shape anisotropy) with anisotropy constants Kshape1 = 0.5μ0(Na − Nc)M2S, Kshape2 = 0.5μ0(Na − Nc)M2S and Kshape1 ≥ Kshape2,42–44 where μ0 is the permeability constant, Na is the DF of the easy axis, Nc of the hard axis. Each particle in the assembly (apart from the uniaxial crystallographic anisotropy, which is randomly oriented in the assembly) has also shape anisotropy. The shape anisotropy easy axes have a narrow, non-uniform distribution around the z-axis. So adding the demagnetizing energy term in the Hamiltonian actually includes another anisotropy term (shape anisotropy), approximately directed along the z-axis. The average shape anisotropy density to the crystallographic anisotropy is Kshape/Kμ = 0.5μ0〈(Na − Nc)V〉M2S/Kμ〈V〉 = 1.26, meaning that the shape anisotropy plays an important role. More particles have shape anisotropy easy axis along the z-axis and this accounts for the bigger value of the coercive field in this direction and the difference in the shape of the hysteresis loop. It explains also the increase of the blocking temperature when the cooling field is perpendicular to the surface.
When the demagnetizing energy is omitted, the shapes of the hysteresis loops are typical of a random anisotropy assembly. The small differences between the loops along the two directions are due to the dipolar interactions between the particles in the non-symmetric shaped assembly. Considering the importance of the demagnetizing energy term in the Hamiltonian as explained above, and the better qualitative agreement of the simulated data compared to the measured ones, it is suggested that the particles are more spheroid-like. One obvious explanation for this discrepancy can be found in the missing wedge problem, which causes elongation of the particles in the z-direction making them ellipsoidal. The elongation factor in the z-direction spawning from the missing wedge is 1.15 for an averaged maximum tilt angle of 78.5° degrees.10 If the z-coordinates of the particles are divided by the elongation factor employing a missing wedge correction (MWC) and proceed with the simulations again, the hysteresis loops as seen in Fig. 8(g) and ZFC/FC vs. T curves in Fig. 8(h) are found. Here the qualitative agreement is better than when omitting the DF's. The directions of the shape anisotropy easy axis of the particles are more randomly oriented and therefore the ZFC/FC vs. T curves taken from different directions to the surface are also more similar.
The concentration of particles is different in the vicinity of the surface and the bulk silicon and here we studied just a small fraction of the pores approximately in the middle of the sample (Fig. 1(a)). Higher concentrations will bring exchange interactions and fanning effects. The shape of the measured FC curve at low temperatures is descending linearly, which implies weak dipolar interactions and the measured hysteresis curve does not saturate, which could be explained by a non-random assembly of ellipsoidal particles with the hard axis perpendicular to the field.
4 Conclusions
Magnetic nanoparticle simulations have not been performed hitherto using ET data according to the authors' knowledge. In this work we have shown the usefulness and power of such a technique to acquire three dimensional quantitative data using fewer assumptions on the nanoparticle assemblies.The two different methods for the distribution of the local curvature shown here are applicable to any catalytic material. The ring method is sufficient when the radius of the particle is much smaller than the radius of curvature of the surface, otherwise polynomial regression will give more reliable results provided that they are visually certified. The implementation of the ITK library commands to DM might provide an important bridge between the electron microscopy community and the biological and medical field, the primal users of the ITK software. Finally the introduction of the new alignment method PRC can be useful when the deposition of fiducial markers like colloidal gold is not possible in practice.
The physical understanding of nature lies in the equations that are continuously tested and scrutinized yet are still not falsified. These equations are submerged in the simulations that are performed in various fields to predict and understand the response and behavior of materials and nature itself. For the field of materials science, ET can be extremely useful as it provides nanoscale three dimensional information of real materials, which can be further used for any kind of simulations, not just limited to magnetic ones, for example the optical response of porous materials or electron transport in solar cells. The models used in these kinds of simulations for the material's morphology are often extremely basic compared to the real acquired three dimensional data produced by ET. Therefore as a general outlook, it is the author's view that it would be highly interesting and rewarding to use ET in combination with physical simulations within various fields to produce more accurate predictions and results with fewer assumptions on the materials.
Acknowledgements
The authors would like to thank Prof. Ferdinand Hofer, Dr Mihaela Albu, Dr Peter Pölt, Dr Matthew Weyland, Dr Prof. Heinz Krenn and Margit Wallner for their help in this publication. Thanks to Dr M. P. Morales (ICMM, CSIC Madrid) for the fabrication of the iron oxide nanoparticles. Also the help from Bernhard Schaffer, Paul Thomas and Robin Harmon from Gatan, Inc. is appreciated. This work was supported by FP7 under Copper project, Austrian Cooperative Research (ACR) and the Austrian Science Fund FWF under project P21155.Notes and references
- M. J. Sailor, in Porous Silicon in Practice: Preparation, Characterization and Applications, Wiley-VCH, Weinheim, 2011, p. 218 Search PubMed.
- L. T. Canham, Appl. Phys. Lett., 1990, 57, 1046–1048 CrossRef CAS.
- V. Lehmann and U. Gösele, Appl. Phys. Lett., 1991, 58, 856 CrossRef CAS.
- N. Koshida and H. Koyama, Appl. Phys. Lett., 1992, 60, 347 CrossRef CAS.
- L. T. Canham, Adv. Mater., 1995, 7, 1033 CrossRef CAS.
- E. Pastor, E. Matveeva, V. Parkhutik, J. Curiel-Esparza and M. C. Millan, Phys. Status Solidi C, 2007, 4, 2136–2140 CrossRef CAS.
- P. Granitzer, K. Rumpf, M. Albu and P. Poelt, ECS Trans., 2010, 25, 139–145 CrossRef CAS PubMed.
- Z. Li, J. C. Barnes, A. Bosoy, J. F. Stoddart and J. I. Zink, Chem. Soc. Rev., 2012, 41, 2590–2605 RSC.
- J. Banhart, in Advanced Tomographic Methods in Materials Research and Engineering, Oxford University Press, USA, 2008 Search PubMed.
- P. A. Midgley, E. P. W. Ward, A. B. Hungría and J. M. Thomas, Chem. Soc. Rev., 2007, 36, 1477–1494 RSC.
- J. C. Hindson, Z. Saghi, J.-C. Hernandez-Garrido, P. A. Midgley and N. C. Greenham, Nano Lett., 2011, 11, 904–909 CrossRef CAS PubMed.
- S. Sueda, K. Yoshida and N. Tanaka, Ultramicroscopy, 2010, 110(9), 1120–1127 CrossRef CAS PubMed.
- R. Leary, Z. Saghi, M. Armbrüster, G. Wowsnick, R. Schlögl, J. M. T. Thomas and P. A. Midgley, J. Phys. Chem. C, 2012, 116(24), 13343–13352 CAS.
- J. Fessler, in Analytical Tomographic Image Reconstruction Methods, 2009, http://web.eecs.umich.edu/%7Efessler/course/516/l/c-tomo.pdf, retrieved May 22, 2012 Search PubMed.
- A. Alpers, R. J. Gardner, S. König, R. S. Pennington, C. Boothroyd, L. Houben, R. E. Dunin-Borkowski and K. J. Batenburg, arXiv:1205.5738v1 [math-ph], 2012.
- The Insight Segmentation and Registration Toolkit, http://www.itk.org.
- Digital Micrograph™, Gatan Inc., http://www.gatan.com/imaging/dig_micrograph.html.
- M. Weyland, P. A. Midgley and J. M. Thomas, J. Phys. Chem. B, 2001, 105, 7882–7886 CrossRef CAS.
- P. A. Midgley, M. Weyland, J. M. Thomas and B. F. G. Johnson, Chem. Commun., 2001, 907–908 RSC.
- J. A. Osborn, Phys. Rev., 1945, 67, 351–357 CrossRef.
- N. Metropolis, A. Rosenbluth, M. A. Teller and E. Teller, J. Chem. Phys., 1953, 21, 1087–1092 CrossRef CAS.
- J. L. Dormann, D. Fiorani and E. J. Tronc, J. Magn. Magn. Mater., 1999, 202, 251–267 CrossRef CAS.
- P. Granitzer, K. Rumpf, M. Venkatesan, A. G. Roca, L. Cabrera, M. P. Morales, P. Poelt and M. Albu, J. Electrochem. Soc., 2010, 157(7), K145–K151 CrossRef CAS PubMed.
- A. G. Roca, M. P. Morales and C. J. Serna, IEEE Trans. Magn., 2006, 42(10), 3025–3029 CrossRef.
- M. C. Scott, C.-C. Chen, M. Mecklenburg, C. Zhu, R. Xu, P. Ercius, U. Dahmen, B. C. Regan and J. Miao, Nature, 2012, 483, 444–447 CrossRef CAS PubMed.
- M. E. Fleet, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1984, 40, 1491–1493 CrossRef.
- P. Granitzer and K. Rumpf, Materials, 2011, 4, 908–928 CrossRef.
- L. Ibanez, W. Schroeder, L. Ng and J. Cates, in The ITK Software Guide, 2nd edn, Kitware Inc., 2005 Search PubMed.
- G. Lehmann, The Insight Journal, 2006, January–June, http://hdl.handle.net/1926/164 Search PubMed.
- http://www.itk.org/Doxygen/html/classitk_1_1CurvatureNDAnisotropicDiffusionFunction.html, retrieved June 2012.
- R. Beare, The Insight Journal, 2007, July–December, http://hdl.handle.net/1926/1316 Search PubMed.
- R. Leary, Z. Saghi, M. Armbrüster, R. Schlögl, J. M. Thomas and P. Midgley, J. Phys.: Conf. Ser., 2012, 371, 012024 CrossRef.
- S. Dugas, The Insight Journal, RPI, OpenSource, 2007, http://hdl.handle.net/1926/1307 Search PubMed.
- Amira, Version 5.3, Visage Imaging, San Diego, CA, USA Search PubMed.
- Matlab, Version R2010b, The MathWorks Inc., Natick, MA, USA Search PubMed.
- J. J. Koenderink and A. van Doorn, Image. Vis. Comput., 1992, 10, 557–565 CrossRef.
- E. P. W. Ward, T. J. V. Yates, J.-J. Fernández, D. E. W. Vaughan and P. A. Midgley, J. Phys. Chem. C, 2007, 111, 31 Search PubMed.
- J. D'Errico, Polyfitn, http://www.mathworks.com/matlabcentral/fileexchange/34765-%20polyfitn, MATLAB Central File Exchange, retrieved Jan 25, 2012 Search PubMed.
- Y. Petrov, Ellipsoid fit, http://www.mathworks.com/matlabcentral/fileexchange/24693-%20ellipsoid-fit, MATLAB Central File Exchange, retrieved Jan 25, 2012 Search PubMed.
- E. Stoner and E. Wohlfarth Philos, Philos. Trans. R. Soc. London, Ser. A, 1948, 240, 599 CrossRef.
- K. Binder and D. W. Heermann, in Monte Carlo Simulation in Statistical Physics, Springer-Verlag, Berlin, 1992 Search PubMed.
- A. Aharony, in Introduction to the Theory of Ferromagnetism, Clarendon Press, Oxford, 1996 Search PubMed.
- B. D. Cullity and C. D. Graham, in Introduction to Magnetic Materials, 2nd edn, IEEE Press & Wiley, New Jersey, 2009 Search PubMed.
- J. M. D. Coey, in Magnetism and Magnetic Materials, Cambridge University Press, New York, 2010 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: List of abbreviations and symbols are provided. Information about the calculation of the demagnetizing factors, pore connectivity and details of the Hamiltonian model used in the simulations are presented. See DOI: 10.1039/c3nr02922k |
| This journal is © The Royal Society of Chemistry 2013 |
