 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Shape recognition of microbial cells by colloidal cell imprints
Josef Borovičkaa, Simeon D. Stoyanovbc and Vesselin N. Paunov*a
aSurfactant and Colloid Group, Department of Chemistry, The University of Hull, Hull, HU6 7RX, UK. E-mail: V.N.Paunov@hull.ac.uk; Fax: +44 (0)1482 466410; Tel: +44 (0)1482 465660
bLaboratory of Physical Chemistry and Colloid Science, Wageningen University, 6703 HB Wageningen, The Netherlands
cDepartment of Mechanical Engineering, University College London, Torrington Place, London WC1E 7JE, UK
First published on 29th July 2013
Abstract
We have engineered a class of colloids which can recognize the shape and size of targeted microbial cells and selectively bind to their surfaces. These imprinted colloid particles, which we called “colloid antibodies”, were fabricated by partial fragmentation of silica shells obtained by templating the targeted microbial cells. We successfully demonstrated the shape and size recognition between such colloidal imprints and matching microbial cells. High percentage of binding events of colloidal imprints with the size matching target particles was achieved. We demonstrated selective binding of colloidal imprints to target microbial cells in a binary mixture of cells of different shapes and sizes, which also resulted in high binding selectivity. We explored the role of the electrostatic interactions between the target cells and their colloid imprints by pre-coating both of them with polyelectrolytes. Selective binding occurred predominantly in the case of opposite surface charges of the colloid cell imprint and the targeted cells. The mechanism of the recognition is based on the amplification of the surface adhesion in the case of shape and size match due to the increased contact area between the target cell and the colloidal imprint. We also tested the selective binding for colloid imprints of particles of fixed shape and varying sizes. The concept of cell recognition by colloid imprints could be used for development of colloid antibodies for shape-selective binding of microbes. Such colloid antibodies could be additionally functionalized with surface groups to enhance their binding efficiency to cells of specific shape and deliver a drug payload directly to their surface or allow them to be manipulated using external fields. They could benefit the pharmaceutical industry in developing selective antimicrobial therapies and formulations.
Introduction
Nanobiotechnology provides us with the opportunity to develop unconventional approaches for deactivating antibiotic resistant bacteria that do not rely on the existing pathways of antibiotic action.1,2 One possible route to address this involves nanoparticle formulations with engineered antibacterial action designed to target specific bacteria. This route has the potential to have high level of activity at very low concentrations and may also potentially circumvent the concern of resistance development. There is a significant amount of ongoing work on developing functional inorganic nanoparticles which exhibit strong and universal antibacterial action towards which bacteria have not been able to develop resistance.3–6 Recently, Dickert and Hayden7 used shape recognition of microbes to develop bioanalytical applications by producing patterned solid surfaces with the polyurethane and sol–gel process which imprints the surfaces of three different genera of yeast cells. The incubation of different genera of yeast cells with such a patterned surface led to their immobilisation and distinguishing between them.7 A similar approach of immobilising microbial organisms onto solid surfaces containing the sol–gel imprints was also employed by Cohen et al.8 Harvey et al. used the same principle for detection of bacterial spores on the surfaces of patterned microbeads.9 “Key–lock” interactions have also been recently demonstrated by Sacanna et al.10,11 in a colloid system capable of programmed binding into composite clusters.Recently, we pushed these ideas further and instead of binding free cells to their imprints on immobilised solid surfaces we created anisotropic colloid particles which are partial imprints of the original cells.12 Such particles are capable of specific recognition of microbial cells or other colloid objects, based “key–lock” colloidal interactions. Thus, we created the analogue of a whole cell “colloid antibodies”12 which can bind to cells based on their shape and size. These colloidal cell imprints were fabricated via several preparation steps which involve producing shells of an inert material onto the microbial cells that match closely in their shape and size, mimicking at a colloidal level the process of raising the protein antibodies in Nature. Further, these inorganic imprints were subjected to partial fragmentation which allowed the templated cells to be removed from the shells interior by a bleaching process while the shell fragments retained their ability to match and “recognise” the shape and size of the cells upon binding. We demonstrated that by pre-coating the target cells with gold nanoparticles, the latter can be integrated into the silica shell of the colloid imprint. When the target cell is recognised by such a photothermal colloid antibody the gold nanoparticles in the shell are delivered directly onto the target cell surface. Irradition of the cells dispersion with a laser led to selective killing of the target cells.12
The attraction between these colloidal cell imprints and the target cells is enhanced by several orders of magnitude by the increased contact area between their surfaces when such a cell “shape recognition” event takes place. The binding of these partially fragmented shells to the original target cells can be further fine-tuned by additional surface functionalisation with suitable bio-specific coating which promotes stronger adhesion between the shell fragments and the cell surface. Ideally, the inner surface of the colloidal cell imprint can be selectively functionalised with the protein antibody specific for the target cell membrane. Upon contact with the targeted microbes, the colloidal imprint can orientate and bind specifically to their surface only if there is a shape match. The colloidal imprints can be further functionalized to deliver a specific biocidal payload directly onto the target cell surface or deactivate them by impairing their biological activity and/or allowing the marked cell to be further manipulated using external fields or physical or chemical processes. This concept of a whole cell “colloid antibody” in-turn mimics the action of protein antibodies in Nature, where recognized (marked) antigens or microorganisms can be further targeted and processed by the immune system. We envisage that colloidal imprints can also be produced to match and capture target viruses. This concept is also expected to work for development of antiviral agents which can bind specifically to viruses matching their shape and size thus preventing the attachment of the viral capsid to the host cell and the subsequent release of the viral DNA. The specific shape and size recognition of bacteria by the colloidal imprints could potentially allow binding and delivery of additional antibacterial agents directly to the membrane of the targeted bacterial cell without affecting other cells with different sizes and morphologies. Note that in contrast to conventional antibiotics, only one colloidal imprint particle loaded with biocides would be sufficient to bind and deactivate the microbial cell. Our concept for preparation of colloidal cell imprints and their shape-selective action on targeted bacteria or viruses is illustrated in Scheme 1A and B.
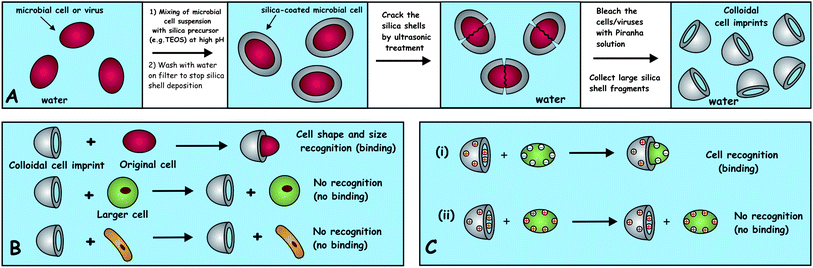 | ||
| Scheme 1 (A) The fabrication route and (B) the principle of action of colloidal cell imprints. The fragmented shells bind preferentially to target cells of matching shape and size of the shell inner surface due to the increased area of surface contact. This amplifies even weak attraction and results in much stronger and shape-specific binding. (C) The electrostatic (and other) interactions between the cells and their silica shell imprints can be augmented by the cell shape recognition: (i) the oppositely charged shell fragments and the cells attract much stronger by electrostatic forces upon contact when there is shape recognition. (ii) In the case of electrostatic repulsion between the shell inner surface and the cell surface the repulsion is amplified and there is no binding as a result of the shape recognition. | ||
In this paper we focus in more details on the interaction between the target cells and colloid imprints and we study the effect of their surface charge on the recognition and binding efficiency. We use yeast cells as model microbial cells in this work due to their robustness and easiness to culture and maintain. In this study we control the surface charge of both the yeast cells and their colloid imprint by pre-coating them with different polyelectrolytes. We also present experimental data for binding of colloid imprints of yeast cells to yeast in a mixture with rod-like bacterium, B. subtilis. Finally, we test the effect of the size of the recognised target particle by using latex particles of spherical shape and different sizes as templates. We also discuss the recognition effect from the point of increased surface area of contact between the target cells and the colloid imprints.
Experimental section
Materials
Methanol, isopropanol, toluene, aqueous ammonium hydroxide solution (35%) and the sulphuric acid (98%) were of analytical grade (purchased from Fisher Scientific). The hydrogen peroxide (BioReagent, 30%) was also purchased from Fisher Scientific. The poly(allylamine hydrochloride) (PAH) (average Mw ∼ 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and poly(sodium styrene sulfonate) (PSS) (average Mw ∼ 70
000) and poly(sodium styrene sulfonate) (PSS) (average Mw ∼ 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), the tetraethoxysiliane (TEOS) (ReagentPlus, ≥99%), and the 3-aminopropyltriethoxysilane (APTES) (≥99%) were purchased from Sigma-Aldrich. The sodium chloride (AnalaR) was purchased from VWR, UK. The 3, 6 and 10 μm carboxylate modified (CML) latex microparticles (4% w/v) were purchased from Invitrogen. Millipore water (specific resistivity 18 MΩ cm at 25 °C) was used in all experiments.
000), the tetraethoxysiliane (TEOS) (ReagentPlus, ≥99%), and the 3-aminopropyltriethoxysilane (APTES) (≥99%) were purchased from Sigma-Aldrich. The sodium chloride (AnalaR) was purchased from VWR, UK. The 3, 6 and 10 μm carboxylate modified (CML) latex microparticles (4% w/v) were purchased from Invitrogen. Millipore water (specific resistivity 18 MΩ cm at 25 °C) was used in all experiments.Fabrication of colloidal imprints using yeast cell templates
The experimental procedure for the fabrication of silica shells on yeast cells was as follows.12,13 3.0 g of Baker yeast cells (powder) were washed 3 times with Milli-Q water by centrifugation at 3000 rpm for 4 minutes and replacement of the supernatant water (using Auto Bench Centrifuge Mark IV, Baird & Tatlock, Oldham, UK), then suspended in 6 mL 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol–water mixture. This was followed by the addition of 0.5 mL of 25% ammonia as a catalyst and 5 mL of TEOS (Sigma). The suspension was then agitated at room temperature for 2 hours. The product was recovered by centrifugation (5 min at 300 rpm), washed 3 times with methanol and 2 times with Milli-Q water. Afterwards, the solid precipitate was dried overnight at 105 °C. Then, 0.5 g of these yeast core–silica shell particles were re-suspended in Milli-Q water whilst being agitated with a magnetic stirrer. This was followed by agitation using an ultrasonic bath (Grant MXB22, Grant Scientific, UK, RMS of 238 W and peak power of 475 W) for about 6–10 minutes which led to partial fragmentation of the shells. After centrifugation at 3000 rpm and removal of the supernatant, the sediment was dried up at 105 °C. The removal of the yeast cell from the core-shell particles was done only on a very small scale by treatment with Piranha solution (3
1 methanol–water mixture. This was followed by the addition of 0.5 mL of 25% ammonia as a catalyst and 5 mL of TEOS (Sigma). The suspension was then agitated at room temperature for 2 hours. The product was recovered by centrifugation (5 min at 300 rpm), washed 3 times with methanol and 2 times with Milli-Q water. Afterwards, the solid precipitate was dried overnight at 105 °C. Then, 0.5 g of these yeast core–silica shell particles were re-suspended in Milli-Q water whilst being agitated with a magnetic stirrer. This was followed by agitation using an ultrasonic bath (Grant MXB22, Grant Scientific, UK, RMS of 238 W and peak power of 475 W) for about 6–10 minutes which led to partial fragmentation of the shells. After centrifugation at 3000 rpm and removal of the supernatant, the sediment was dried up at 105 °C. The removal of the yeast cell from the core-shell particles was done only on a very small scale by treatment with Piranha solution (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentrated sulphuric acid and hydrogen peroxide). About 500 mg sample of the dried yeast–silica core-shell particles was carefully added to 15 mL of Piranha solution placed in 200 mL glass beaker at room temperature in a fume hood behind a protective screen. The treatment with Piranha solution was done without any additional heating but the mixture gets pretty hot on its own. The duration was typically 10–20 minutes until the solution stopped bubbling, after which the mixture was left to naturally cool down overnight to room temperature for further decomposition of the unreacted hydrogen peroxide. The mixture was then carefully diluted about 10 times by pouring it into another glass beaker with 150 mL of milli-Q water. The produced silica shell fragments were separated by centrifugation using glass centrifugal tubes, further washed 3 times with milli-Q water and studied by optical and scanning electron microscopy. We found that this treatment cracked open most of the shells for silica-coated yeast. Caution: Piranha solution is a hazardous material and an extremely strong oxidant. It should be handled with great care! Mixing Piranha solution with a significant amount of organic material or organic solvents may cause bursts and even explosion! Please read the MSDS datasheets and the specialised literature on how to safely handle and dispose Piranha solution.
1 concentrated sulphuric acid and hydrogen peroxide). About 500 mg sample of the dried yeast–silica core-shell particles was carefully added to 15 mL of Piranha solution placed in 200 mL glass beaker at room temperature in a fume hood behind a protective screen. The treatment with Piranha solution was done without any additional heating but the mixture gets pretty hot on its own. The duration was typically 10–20 minutes until the solution stopped bubbling, after which the mixture was left to naturally cool down overnight to room temperature for further decomposition of the unreacted hydrogen peroxide. The mixture was then carefully diluted about 10 times by pouring it into another glass beaker with 150 mL of milli-Q water. The produced silica shell fragments were separated by centrifugation using glass centrifugal tubes, further washed 3 times with milli-Q water and studied by optical and scanning electron microscopy. We found that this treatment cracked open most of the shells for silica-coated yeast. Caution: Piranha solution is a hazardous material and an extremely strong oxidant. It should be handled with great care! Mixing Piranha solution with a significant amount of organic material or organic solvents may cause bursts and even explosion! Please read the MSDS datasheets and the specialised literature on how to safely handle and dispose Piranha solution.Fluorescent tagging of yeast cells and silica shell fragments
About 0.5 g of yeast cells were incubated in 10 mL of 10−4 M ethanolic solution of perylene (from Sigma) for 10 minutes before being washed and redispersed in Milli-Q water. The fluorescent tagging of the silica shell fragments was performed as follows. About 0.5 g of the silica shell fragments were washed once in methanol and once in toluene before being re-dispersed in 10 mL 10% aminopropylsilane (APTES, from Sigma) solution in toluene and incubated whilst being agitated by a magnetic stirrer for 1 hour. This was followed by a triple washing with methanol and 3 hour incubation in 1 mM solution of Rhodamine B isothiocyanate (RBITC, from Sigma) in methanol, further triple washing with methanol and Milli-Q water and re-dispersing in 20 mL of Milli-Q water. The silica surface functionalisation with APTES allows covalent grafting of the RBITC.Polyelectrolyte pre-treated cells and colloid imprints
The polyelectrolyte coating of the silica shell fragments and the yeast cells was done layer-by-layer.14,15 The cells and the silica fragments were incubated sequentially in the respective polyelectrolyte solutions (4 mg mL−1 PAH or PSS, in 0.15 M NaCl solution in Milli-Q water) for 10 minutes followed by a triple washing with Milli-Q water. The cell recognition experiments themselves were performed as follows. The dispersion of fragmented silica shells (0.025 g mL−1) was added drop-wise to the dispersions of the yeast cells (0.1 g mL−1) in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (typically 0.2
1 ratio (typically 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 mL) after being agitated with a IKA minishaker. In the experiments described in Fig. 3 and 4, native B. subtilis and yeast (0.02 g wet pellet) were incubated with 5 mL dispersion of silica shell fragments in Milli-Q water. The combined dispersions were then agitated for 1 hour on an orbital shaker (Vibrax VXR, IKA, Germany) at a frequency of 1500 min−1. The experimental results were analysed using optical and fluorescence microscopy.
0.1 mL) after being agitated with a IKA minishaker. In the experiments described in Fig. 3 and 4, native B. subtilis and yeast (0.02 g wet pellet) were incubated with 5 mL dispersion of silica shell fragments in Milli-Q water. The combined dispersions were then agitated for 1 hour on an orbital shaker (Vibrax VXR, IKA, Germany) at a frequency of 1500 min−1. The experimental results were analysed using optical and fluorescence microscopy.Fabrication of fragmented silica shells using latex particle templates
The silica shells were prepared according to the method of Lu et al.17 using 6 μm CML latex particles as templates. Briefly, 0.5 mL of the latex dispersion was dispersed in a solution of 20 mL of isopropanol, 3.5 mL of Milli-Q water and 0.34 mL of TEOS to which 0.5 mL of 30% ammonia hydroxide catalyst was added. The mixture was agitated by stirring then left to react at room temperature for 3 hours. The dispersions were then centrifuged at 3000 rpm for five minutes which was followed by the removal of the milky supernatant containing homo-nucleated silica. The core/shell particles were then washed 4 times with isopropanol and two times with Milli-Q water. This was then followed by agitation with an ultrasonic tip for 2 minutes set to 50 W, washing with isopropanol and subsequent introduction into toluene in order to dissolve the cores and washing twice with toluene and transfer into water via isopropanol. The dissolution was checked using bright field microscopy as was sufficient fragmentation of the shells. The silica shell fragments were fluorescently tagged with RBITC in order to allow for their visualisation using fluorescent microscopy. A sample of these colloidal imprint particles was combined with CML latex microspheres firstly using a single size of particle, of 3, 6 and 10 μm diameters respectively, and then with a mixture of particles of all three sizes, in order to investigate the target particle size recognition specificity of the silica shell fragments. In the first series of experiments virtually no recognitions between the mismatching colloidal imprints and microsphere pairs was observed. Very similar results were also noted in the experiments involving the mixture of latex particles of different sizes. A high fraction of binding events was observed between the shell fragments and the target latex particles of the matching size (6 μm) and very few binding events were observed between either smaller (3 μm) or larger latex particles (10 μm).Polyelectrolyte-mediated surface charge induction
The coating of the yeast cells and their fluorescently tagged colloid imprints with polyelectrolytes was done as follows. The cells and the silica shell fragments were incubated sequentially in respective polyelectrolyte solutions (PAH or PSS, 4 mg mL−1 in 0.15 M NaCl solution) for 10 minutes in order to obtain the desired surface charge. The incubation steps were intermitted by a triple washing with deionised water. The cell recognition experiments between the yeast cells and their colloid imprints were performed in the following fashion. The shell dispersion (0.025 g mL−1) was added drop-wise to the dispersions of the cells (0.1 g mL−1) whilst the latter was being agitated on an IKA minishaker in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (typically 0.2
1 ratio (typically 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 mL), the combined dispersions were then left for 1 hour whilst being agitated on an orbital shaker (Vibrax VXR basic, IKA, Germany) at a frequency of 1500 min−1. In the case of latex microparticle–silica shell recognition experiments the procedure was as follows. The colloid imprints originating from 0.5 mL of the latex microsphere templates were combined with 50 μL of the 6 μm latex particles and were left on a bench top shaker at 1200 min−1 for 20 minutes. In the case of the recognition experiments involving the mixture of differently sized latex microspheres and the 6 μm shells, 1/3 of 50 μL of each latex suspension was used. The recognition was subsequently evaluated using bright field and fluorescent microscopy.
0.1 mL), the combined dispersions were then left for 1 hour whilst being agitated on an orbital shaker (Vibrax VXR basic, IKA, Germany) at a frequency of 1500 min−1. In the case of latex microparticle–silica shell recognition experiments the procedure was as follows. The colloid imprints originating from 0.5 mL of the latex microsphere templates were combined with 50 μL of the 6 μm latex particles and were left on a bench top shaker at 1200 min−1 for 20 minutes. In the case of the recognition experiments involving the mixture of differently sized latex microspheres and the 6 μm shells, 1/3 of 50 μL of each latex suspension was used. The recognition was subsequently evaluated using bright field and fluorescent microscopy.Characterisation of “cell–colloid imprint” binding events
Bright field and fluorescence microscopy images were obtained using a BX-51 fluorescence microscope (Olympus, Japan). In the case of fluorescence microscopy, excitation occurred with light from an Hg-arc lamp housed in a U-RFL-T power supply (Olympus, Japan). All filter sets used were manufactured by Olympus. For visualisation of the cells or shells treated with Rhodamine stains the MW1BA2 filter set (460–490 nm excitation λ; 510–550 nm emission λ) was used and the MNUA2 filters (360–370 nm excitation λ; 420–460 nm emission λ) were employed in the case of viewing the samples treated with perylene. Digital images were taken using a DP70 camera (Olympus, Japan) and analysed using Image Pro Plus or ImageJ software. SEM images were obtained using a Zeiss Evo 60 Scanning Electron Microscope. A drop of sample dispersion was placed onto a clean glass slide. The samples were left to dry at room temperature and stored in a sealed container. A 5 nm coating of electrically conducting gold/palladium was applied onto the samples using an Edwards High Vacuum E12E2 vacuum deposition coating unit.Results and discussion
Here we provide a proof of principle that such “tailor made” shell fragments can bind to the original cells based on their specific shape and size. We used a sol–gel process to produce silica shells onto the cell templates. Following ultrasonic treatment to crack the silica shells and the bleaching process with Piranha solution to remove the templated cells, we produced silica shell fragments which “remember” the shape and size of the templated microbes.This led to the formation of nearly hemi-spherical shells and smaller shell fragments which allow their effective binding when incubated with the original cells. In our proof of concept experiments, we used non-pathogenic cells to demonstrate the cell recognition. We chose baker's yeast (S. cerevisiae) as model target cells for templating with silica and produced hollow shells via the Stöber process13 which were subsequently fragmented by ultrasonic agitation and bleaching process.
These silica shell fragments can be further functionalized to increase the recognition efficiency. To illustrate this in the simplest model system, here we explored the cell recognition depending on the surface charge of the cells and the shell fragments, respectively. We coated the targeted cells and the corresponding fragmented silica shells with polyelectrolytes, polyallylamine hydrochloride (PAH) and polystyrene sulfonate (PSS) to enhance their surface charge and explore the effect of the surface adhesion on the binding efficiency depending on the cell shape recognition (Scheme 1C). Note that unlike the cell recognition by patterned surfaces,7,8 in our study the target microbial cells are interacting with separate individual fragments of silica shells which geometrically match their shape and size. Due to the mobility of the individual silica shell fragments, this approach gives significantly higher probability of cell capture than using patterned flat surfaces with the same cell imprints.7,8
The fabrication of the silica shells on yeast cell templates was performed by adapting the method of Weinzierl et al.13 Subsequent treatment with Piranha solution which removed the cell templates led to formation of shell fragments matching the yeast cells which were then used in the shell–cell recognition experiments (Fig. 1).
 | ||
| Fig. 1 (a) Low and (b) high magnification scanning electron microscopy images of a dried sample of fragmented silica shells produced by templating yeast cells followed by ultrasonic treatment and bleaching of the cells with Piranha solution. | ||
To aid the visualisation of the cell recognition experiments, the silica shells were functionalised with APTES and grafted with Rhodamine B isothiocyanide (RBITC) while the cells were stained with perylene. In all experiments, the cells and the silica shell fragments were dispersed in Milli-Q water. Optical micrographs of mixed dispersions of yeast cells incubated with APTES/RBITC-tagged silica shell fragments showed that the latter “recognise” and bind to their matching cell counterparts. We performed a set of 15 experiments with all combinations of PAH and PSS polyelectrolyte coatings of the cells and the shell fragments: (i) hydrated yeast untreated with polyelectrolytes, (ii) yeast coated with a monolayer of PAH, and (iii) yeast coated with PAH and PSS. The following types of silica shell fragments were used: (i) untreated, (ii) treated with either PAH or PSS, or (iii) treated consequently with each of those polyelectrolytes in both possible orders. As expected, there was no effect of the order of coating (e.g. PAH or PSS–PAH) of the fragmented silica shells with polyelectrolyte layers on the type of electrostatic interaction with the cells of opposite surface charge. The only governing factor of the recognition was the affinity of the terminal polyelectrolyte coating of the respective species.
Fig. 2 and Table 1 show the results of the cell recognition experiments after incubation of the yeast cells with the fragmented silica shells of different surface charge augmented by polyelectrolytes. The samples were assessed for recognition events by fluorescence microscopy with TRITC (green/red) and perylene (UV/blue) filter sets and bright-field microscopy. Both images were superimposed in one composite image to facilitate the analysis of the positions of the shell fragments and the cells. We counted the number of cells with attached shell fragments and calculated the fraction of the shell–cell recognition events relative to the total number of cells on the counting plate. The percentage of the recognition events presented in Fig. 2 gives a quantitative estimate for the effectiveness of the shell fragments in recognising and binding to the cell target with an appropriate orientation.
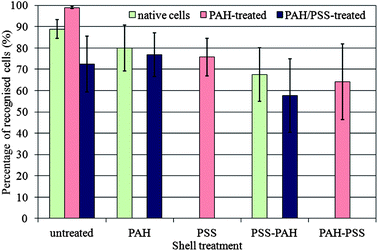 | ||
| Fig. 2 Percentage of recognised yeast cells with silica shell fragments of various polyelectrolyte coats. The corresponding error bars represent the standard deviations. | ||
| Cell treatment | Shell fragment treatment | Percentage of recognition of cells ± standard deviation/% |
|---|---|---|
| — | — | 89 ± 4 |
| PAH | — | 99 ± 1 |
| PAH–PSS | — | 72 ± 13 |
| — | PAH | 80 ± 11 |
| PAH | PAH | No recognition |
| PAH–PSS | PAH | 77 ± 10 |
| — | PAH–PSS | No recognition |
| PAH | PAH–PSS | 76 ± 9 |
| PAH–PSS | PAH–PSS | No recognition |
| — | PSS | No recognition |
| PAH | PSS | 64 ± 18 |
| PAH–PSS | PSS | No recognition |
| — | PSS–PAH | 68 ± 13 |
| PAH | PSS–PAH | No recognition |
| PAH–PSS | PSS–PAH | 58 ± 17 |
Note that the thickness of the polyelectrolyte coating may also play a minor role in the recognition events. Single coats with PAH or PSS are approximately 1.5 nm thick,16 but a double layer on both PAH and PSS on the shell and the cell has a combined thickness of 6 nm which may cause small shape distortion between the shell fragments and the cells (only 6 μm in diameter), thus resulting in less favourable interaction and a lower recognition rate. In addition, multiple coating of silica shell fragments with two polyelectrolyte layers led to partial aggregation of the shell fragments which were difficult to redisperse by ultrasonic treatment without their further disintegration to much smaller fragments. This also resulted into a slightly lower cell recognition rate with shell fragments coated with multiple polyelectrolyte layers. Upon recognition, the shell fragments are “docked” to their cell counterparts via the concave side which corresponds to the largest achievable area of surface contact with the cell and maximal shell–cell adhesion. Our observations revealed that when cell recognition occurred most of the cells did have attached silica shell counterparts – see Fig. 3(I). A large number of images were analysed but only a very few events of fragmented shells bound solely via their outer convex surface were encountered, except in the case of additional cell aggregation. Fig. 3(II) shows the result of the incubation of a mixture of two different types of microbial cells, yeast and Bacillus subtilis (both native) with a PAH-coated colloid imprint for yeast. The results indicate that the silica shell fragments bind preferentially to yeast (round shaped) but not to the rod-shaped B. subtilis.
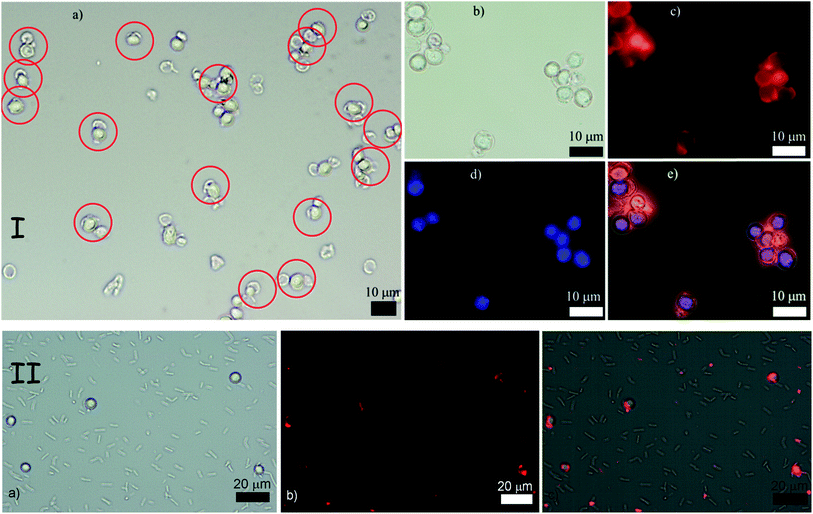 | ||
| Fig. 3 (I) Top row of images: “Colloid imprint–cell” recognition experiment between PAH–PSS treated cells and PSS–PAH treated silica shell fragments. (a) Typical low resolution image of the mixed shell–cell sample with bright field optical microscopy. (b) Bright field optical image. (c) Fluorescence microscopy image highlighting the RBITC tagged shell fragments, (d) fluorescence microscopy image of the yeast cells stained with perylene. (e) Composite image of (b), (c) and (d). (II) Bottom row of images: (a) bright field optical microscopy image of a mixture of yeast (round) and B. subtilis cells (rod-like) incubated with colloid imprints for yeast. (b) Fluorescence microscopy image of this sample with the TRITC filter set. The colloidal imprints are fluorescently tagged with RBITC. (c) Overlay of the images (a) and (b) showing the location of the silica shell fragments (colloidal imprint for yeast). | ||
Another possible approach for imaging the binding events between cells and colloid imprints could be to dry the sample and use SEM at high resolution. However, we have reservations that this would be representative for the frequency of the binding events between cells and shell fragments which we are aiming to characterise in this study. The reason is that upon drying both cells and shell fragments are brought together by lateral capillary forces20 of the liquid meniscus which cause them to aggregate and cluster indiscriminately. Also, the clustering of cells and shells additionally obscures the counting. In addition, the cells also significantly change their shape upon drying. Although we looked at dried samples, we do not think that the results of the dried cell–imprint mixture are representative for what occurs in solution where different interaction forces are in operation. Therefore, for the purposes of counting cell–imprint binding we restrict only to imaging with a combination of optical and fluorescence microscopy with suitable fluorescence markers.
Following the successful demonstration of the recognition of the yeast cells by matching silica shell fragments we probed the importance of the target particle size on the recognition rate. To simplify the experiments and to exclude the effect of microbial cell shape variation, we used monodisperse carboxylate modified latex (CML) microspheres of three different diameters. We fabricated silica shell fragments using 6 μm monodisperse latex microsphere templates which were then incubated with an aqueous suspension of a mixture of latex microspheres that were smaller, larger and equal to the diameter of the original sacrificial latex templates, all with identical surface chemistry. This allowed us to investigate exclusively the role of the inner curvature of the colloid imprint in proportion to the surface curvature the target particles themselves (see Fig. 4).
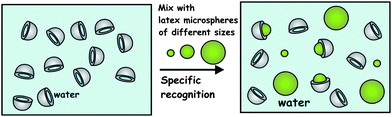 | ||
| Fig. 4 Schematics of our experiments probing the role of the target particle size in the colloid imprint–target recognition. The silica shell fragments, fabricated using 6 μm sacrificial CML latex particles, were incubated with 3, 6 and 10 μm latex microspheres, respectively. | ||
Analysing the microscope images obtained from these experiments it is possible to put values to the frequency of the recognition events. As previously, the numbers only give a guideline to the rate of recognition seen; however, the results are conclusive in showing the preferential recognition of the intermediate size particles by the matching “colloid imprints”, despite the slightly elevated rate of recognitions observed in the other two sizes of latex particles. We analysed a large number of images of the incubated silica shell fragments produced by templating 6 μm latex particles with a separate sample of single size latexes of three different sizes (3, 6 and 10 μm). A recognition rate of 93% was found for the scenario where the silica shell fragments matched the microspheres, whilst the number of mismatches was very low and primarily between “shapeless”, very small fragments (not specific in shape). Fig. 5 shows a typical micrograph of the mixture of the differently sized latex particles incubated with the silica shell fragments templated for 6 μm microspheres. As in the study of the interaction of yeast cells with matching colloidal imprints, the recognition rate as a percentage of recognition per average image was calculated.
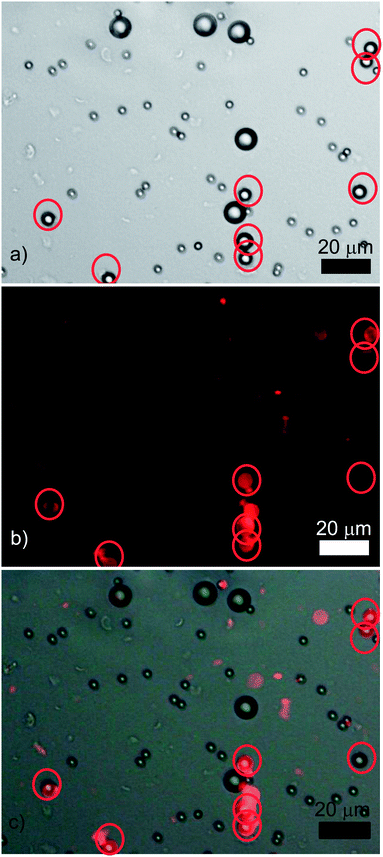 | ||
| Fig. 5 A typical result of an experiment where three sets of differently sized CML latex microspheres (3, 6, and 10 μm) were incubated together with colloid imprints (tagged with RBITC) matching the surface curvature of the intermediately sized particles: (a) an optical micrograph, (b) the corresponding fluorescence microscopy image, and (c) is the overlay of the images (a) and (b). The red circles indicate the location of the intermediately sized (6 μm) latex particles. | ||
The percentage recognition between the colloid imprints and the 6 μm target microparticles was found to be 92%, whilst that of the mismatches was about 1% – see Fig. 6 for the summary of the experimental results. The results demonstrate that colloid imprints can also differentiate between particles of similar shape but different sizes and selectively bind to them.
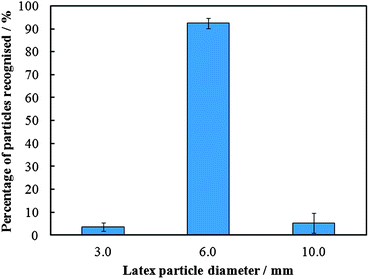 | ||
| Fig. 6 A graphical representation of the results from the experiments which investigated the role of the target particle (cell) size in the recognition with a colloidal imprint targeting only one specific particle size. Error bars represent standard deviations. | ||
From practical point of application of colloid imprints to non-spherical bacteria or viruses, it is interesting to consider if small shell fragments matching the shape of non-spherical bacteria would be able to bind to spherical bacteria. The results presented above show that the curvature match between the shell fragments for intermediate size particles leads to preferential binding to the particles of the same surface curvature. In this respect the size of the shell fragment is not so important if there is no surface curvature match. Therefore smaller fragments from shells matching the shape of non-spherical cells are not likely to bind to spherical cells as the probability for full surface curvature match between them is low. Since we have over 92% cell recognition rate upon cell shape match (see Fig. 2 and 3) we envisage that the probability of such events would be less than those of non-specific shape binding.
The shape recognition between the colloidal imprint and the target cell does not require moulding of the shape of the target cell at the molecular scale, but at the colloidal scale. While the molecular recognition can implemented via specific key–lock type of interactions, in this study we have modelled the cell–colloid imprint recognition with attractive and repulsive electrostatic interactions between their surfaces. This is true hierarchical length scale binding, which allows fine tuning the interactions between the colloid imprint and target cells. The integral effect comes from the overall increase of the contact area of the interacting surfaces which is achieved upon shape and size match. The colloid imprints of different sizes would differentiate between big yeast cells and very small bacterial spores. This is supported by our results in Fig. 4 in conjunction with the experimental results for the recognition of latex particles of different sizes by matching colloid imprints (Fig. 5 and 6).
One can also show theoretically that the specific cell shape recognition comes from the curvature match between the cell and the inner surface of the colloidal cell imprint. Let us denote the surface interaction energy between the two plane parallel surfaces at a distance h between each other by f(h) and δ0 is the minimal value of h, i.e. hmin = δ0. The value of δ0 is determined by the surface roughness, and for the silica surface we can approximate δ0 with the size of the primary silica domains,18i.e. ∼3–5 nm. One can approximately estimate that the interaction energy of the inner surface of a hemispherical silica shell fragment interacting with a spherical particle (cell) of radius R, upon perfect shape and size match between them is
| Uin(δ0) ≈ 2πR2f(δ0), | (1) |
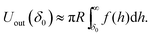 | (2) |
For example, at very small surface-to-surface separations, h, where the van der Waals interaction is dominating, the surface interaction energy is f(h) ≈ fvw(h) = −AH/(12πh2), where AH is the Hamaker constant.19 Then, the ratio of eqn (1) and (2) is
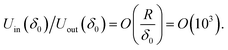 | (3) |
This is a rough estimate of the ratio of the interaction energies between cell and colloid imprint in favourable (inner) and unfavourable (outer) orientations. Hence for typical values of the cell/imprint parameters R = 3 μm and δ0 = 3 nm, the interaction energy of the cell and the colloidal imprint upon recognition, Uin is up to 3 orders of magnitude larger than that of non-shape specific binding, Uout. The cell recognition effect is coming from the surface geometry match and does not require additional molecular recognition unless the colloidal imprint surfaces are treated with bioligands capable of binding to specific groups on the matching cell surface.
Conclusions
In summary, we have developed colloidal cell imprints which can be used for the shape-specific binding to cells and can be applied for shape-selective targeting and further deactivation of microorganisms. These colloid imprints represent large fragments of silica shells templated over the sacrificial target microorganisms. We demonstrated the shape recognition between colloid imprints and matching yeast cells which resulted in successful cell binding. We tested the effect of the surface charge of the colloid imprint and the target cell on the recognition efficiency. The effect of the target particle size was also studied. This concept is the first step in the further development of “colloid antibodies” for targeted immobilisation and deactivation of pathogens. We present experimental data on shape-specific cell binding and analyse the effect of the cell and its imprint surface coating on the binding efficiency.Future outlook
Although here we focus only on the role of the surface charge in the attraction/repulsion between cells and their colloidal imprints, the colloidal imprint surface could be functionalised with biospecific ligands (e.g. antibodies, carbohydrates) which may increase the specificity of surface binding and further exclude indiscriminate attachment of smaller biological objects/fragments to its surface. In addition, grafting polyethylene glycol chains on both the inner and the outer surfaces of the colloidal imprint particles could be used to suppress such non-specific binding. We envisage that colloid imprints of microbial cells could be further “equipped” with antimicrobial agents to bind and potentially deactivate specific strains of antibiotic resistant microbes where most conventional antibiotics are not working. Biocide-loaded colloidal cell imprints may also find applications as selective antimicrobial agents, as well as in the pharmaceutical industry for novel antibacterial therapies. In addition, cells recognised and docked by matching colloidal antibodies could be further manipulated, collected for diagnostic purposes or killed using external fields and processes, which mimic the action of protein-based antibodies in the immune system.Acknowledgements
This work was supported by a CASE PhD studentship funded by BBSRC (grant BB/F01807X/1).Notes and references
- C. A. Arias and B. E. Murray, N. Engl. J. Med., 2009, 360, 439 CrossRef CAS.
- D. Abbanat, M. Macielag and K. Bush, Expert Opin. Invest. Drugs, 2003, 12, 379 CrossRef CAS.
- P. Li, J. Li, C. Wu, Q. Wu and J. Li, Nanotechnology, 2005, 16, 1912 CrossRef CAS.
- H. J. Choi, S. W. Han, S. J. Lee and K. Kim, J. Colloid Interface Sci., 2003, 264, 458 CrossRef CAS.
- S. Shrivastava, T. Bera, A. Roy, G. Singh, P. Ramachandrarao and D. Dash, Nanotechnology, 2007, 18, 225103.1 CrossRef.
- F. Martinez-Gutierrez, P. L. Olive, A. Banuelos, E. Orrantia, N. Nino, E. M. Sanchez, F. Ruiz, H. Bach and Y. Av-Gay, Nanomedicine, 2010, 6, 681 CrossRef CAS.
- F. L. Dickert and O. Hayden, Anal. Chem., 2002, 74, 1302 CrossRef CAS.
- T. Cohen, J. Starosvetsky, U. Cheruti and R. Armon, Int. J. Mol. Sci., 2010, 11, 1236 CrossRef CAS.
- S. Harvey, G. Mong, R. Ozanich, J. McLean, S. Goodwin, N. Valentine and J. Fredrickson, Anal. Bioanal. Chem., 2006, 386, 211 CrossRef CAS.
- S. Sacanna, W. T. M. Irvine, P. M. Chaikin and D. J. Pine, Nature, 2010, 464, 575 CrossRef CAS.
- S. Sacanna, W. T. M. Irvine, L. Rossi and D. J. Pine, Soft Matter, 2011, 7, 1631 RSC.
- J. Borovička, M. J. Metheringham, L. A. Madden, C. D. Walton, S. D. Stoyanov and V. N. Paunov, J. Am. Chem. Soc., 2013, 135, 5282 CrossRef.
- D. Weinzierl, A. Lind and W. Kunz, Cryst. Growth Des., 2009, 9, 2318 CAS.
- G. Decher, Science, 1997, 277, 1232 CrossRef CAS.
- R. F. Fakhrullin, A. I. Zamaleeva, M. V. Morozov, D. I. Tazetdinova, F. K. Alimova, A. K. Hilmutdinov, R. I. Zhdanov, M. Kahraman and M. Culha, Langmuir, 2009, 25, 4628 CrossRef CAS.
- F. Caruso, in Colloids and Colloid Assembly, ed. F. Caruso, Wiley-VCH Verlag GmbH, 2004, ISBN 3527306609, p. 252 Search PubMed.
- Y. Lu, J. McLellan and Y. Xia, Langmuir, 2004, 20, 3464 CrossRef CAS.
- J. L. Roussett, A. Boukentert, B. Champagnont, J. Dumast, E. Duvalt, J. F. Quinsont and J. Serughetti, J. Phys.: Condens. Matter, 1990, 2, 8445 CrossRef.
- B. V. Derjaguin, Surface forces, B. V. Derjaguin, N. V. Churaev and V. M. Muller, translated from the Russian by V. I. Kisin, translation edited by J. A. Kitchener, Consultants Bureau, New York, 1987 Search PubMed.
- P. A. Kralchevsky, V. N. Paunov, I. B. Ivanov and K. Nagayama, J. Colloid Interface Sci., 1992, 151, 79–94 CrossRef CAS; P. A. Kralchevsky, N. D. Denkov, V. N. Paunov, O. D. Velev, I. B. Ivanov, H. Yoshimura and K. Nagayama, J. Phys.: Condens. Matter, 1994, 6, A395–A402 CrossRef.
| This journal is © The Royal Society of Chemistry 2013 |
