 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cold-adapted arsenite oxidase from a psychrotolerant Polaromonas species†
Thomas H. Osbornea, Matthew D. Heatha, Andrew C. R. Martina, Jaroslaw A. Pankowskia, Karen A. Hudson-Edwardsb and Joanne M. Santini*a
aInstitute of Structural and Molecular Biology, University College London, Gower Street, London, WC1E 6BT, UK. E-mail: j.santini@ucl.ac.uk
bDepartment of Earth and Planetary Sciences, Birkbeck, University of London, Malet Street, London, WC1E 7HX, UK
First published on 31st October 2012
Abstract
Polaromonas sp. str. GM1 is an aerobic, psychrotolerant, heterotrophic member of the Betaproteobacteria and is the only isolate capable of oxidising arsenite at temperatures below 10 °C. Sequencing of the aio gene cluster in GM1 revealed the presence of the aioB and aioA genes, which encode the arsenite oxidase but the regulatory genes typically found upstream of aioB in other members of the Proteobacteria were absent. The GM1 Aio was purified to homogeneity and was found to be a heterodimer. The enzyme contained Mo and Fe as cofactors and had, using the artificial electron acceptor 2,6-dichlorophenolindophenol, a Km for arsenite of 111.70 ± 0.88 μM and a Vmax of 12.16 ± 0.30 U mg−1, which is the highest reported specific activity for any known Aio. The temperature-activity profiles of the arsenite oxidases from GM1 and the mesophilic betaproteobacterium Alcaligenes faecalis were compared and showed that the GM1 Aio was more active at low temperatures than that of A. faecalis. A homology model of the GM1 Aio was made using the X-ray crystal structure of the Aio from A. faecalis as the template. Structural changes that account for cold adaptation were identified and it was found that these resulted in increased enzyme flexibility and a reduction in the hydrophobicity of the core.
Introduction
Arsenic is a group 5 metalloid and is renowned for its toxicity.1 The soluble inorganic forms of arsenic, arsenite (+3) and arsenate (+5), are widespread in terrestrial and aquatic environments,2 where arsenite is the more toxic and mobile species.3 The consumption of naturally occurring inorganic arsenic in drinking water has caused poisoning of human populations across regions of West Bengal, Bangladesh, Chile, Taiwan and Vietnam, while anthropogenic activity (principally through the mining of valuable metals) has resulted in arsenic contamination of environments in Canada, USA, Argentina and Australia.4,5 Despite its toxicity, certain microbes are capable of gaining energy by oxidising arsenite to arsenate,6 and this process contributes to the global biogeochemical cycling of arsenic.7 Since the first discovery of bacterial arsenite oxidation by Green in 1918,8 over 50 strains of arsenite-oxidising bacteria have been isolated. The arsenite-oxidising bacteria are phylogenetically diverse and have been isolated from different environments (reviewed by Osborne and Santini).9Aerobic oxidation of arsenite is catalysed by arsenite oxidase (Aio) (see Lett et al. for recent changes to the nomenclature for the arsenite oxidase).10 Aio is a member of the DMSO reductase family of molybdoenzymes which is a family of redox enzymes that function in a diverse range of prokaryotic electron transport chains.11 The enzyme is a heterodimer that consists of a large α-subunit, AioA and a small β-subunit, AioB. The X-ray crystal structure of the Aio from the mesophilic, heterotrophic betaproteobacterium Alcaligenes faecalis has been resolved.12 The structure shows that AioA contains a 3Fe–4S cluster and a bis-molybdopterin guanine dinucleotide (bis-MGD) cofactor at the active site, while AioB contains a 2Fe–2S Rieske cluster. The Aio is exported to the periplasm by the Twin-arginine translocation (Tat) pathway conferred by an N-terminal signal sequence on AioB. The Aio that have been characterised to date have all been purified from mesophilic arsenite oxidisers (reviewed by Heath et al.).13
The aio operon structure always includes the aioA and aioB genes, with aioB located upstream of aioA. In some arsenite oxidisers of the Alpha- and Betaproteobacteria a cytochrome c-encoding gene (cytC) is located downstream of aioA9 and this cytochrome has been shown to serve as the electron acceptor to the Aio.14,15 Often found upstream of aioB are three genes (aioXSR) whose products are involved in the regulation of arsenite oxidase gene expression. AioS is a sensor histidine kinase and AioR a response regulator.16 Together with a third gene product, AioX, which has been shown to bind arsenite,17 they are thought to regulate transcription of the aioBA operon. Variation is seen in the other genes found in the aio cluster. Of the isolated arsenite oxidisers, only Polaromonas sp. str. GM1, an aerobic, heterotrophic, betaproteobacterium, has been shown to oxidise arsenite at temperatures below 10 °C.18 GM1 is also unusual in that aio gene expression is constitutive and it oxidises arsenite in the early log-phase of growth whereas most heterotrophic arsenite oxidisers oxidise arsenite in the late-log or stationary phase using arsenite as the secondary energy source. GM1's Aio is constitutively expressed which may be explained by the environment in which GM1 was isolated, an arsenic-contaminated biofilm that contained about 10 mM arsenic.18 In this paper we present a study of arsenite oxidation in GM1 and its cold-adapted arsenite oxidase. Understanding the basis of the enzyme's cold-adaptation could result after some improvement in its use as a broad-temperature range biosensor.
Results and discussion
GM1 can oxidise arsenite at 0 °C (data not shown) to 20 °C and does so in the early-log phase of growth,18 which is unusual for heterotrophic arsenite oxidisers. The Aio of GM1 is functional over a broad temperature range and its expression was found to be constitutively expressed and up-regulated in the late-log phase of growth.18 The data presented here contribute to a better understanding of arsenite oxidation and how the GM1 Aio is able to function at low temperatures.Identification of the aio gene cluster
The aioA gene of GM1 was mutated by targeted gene disruption generating GM1-A. This mutant was unable to oxidise arsenite and did not exhibit detectable Aio activity (using DCPIP as the electron acceptor) in total cell extracts (data not shown), confirming that aioA confers an arsenite-oxidising phenotype in GM1. Mutagenesis of aioA facilitated the cloning of a 5.3 kb fragment which was sequenced. The 5307 bp sequence contains four complete and one partial open reading frame: this includes a putative phage integrase (partial sequence), the arsenite oxidase small subunit (aioB), the arsenite oxidase large subunit (aioA), a putative c-type cytochrome (cytC) and a nitroreductase (Fig. 1). These genes are transcribed in the same direction. No consensus sequence for a promoter was identified. Genes involved in the regulation of arsenite oxidation (i.e. aioXSR) were not identified but this is not surprising given that arsenite oxidation in GM1 is constitutively expressed whereas in bacteria that contain aioXSR the expression of the aioBA operon is induced with arsenite.15,20,21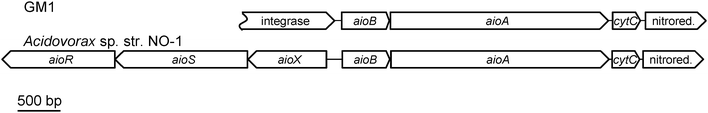 | ||
| Fig. 1 Arrangement of the aio gene clusters in GM1 and the putative arsenite oxidiser Acidovorax sp. str. NO-1.19aioR: encodes response regulator; aioS: encodes sensor histidine kinase; aioX: encodes arsenite-binding protein; aioA: encodes arsenite oxidase large catalytic subunit; aioB: encodes arsenite oxidase small subunit; cytC: encodes putative c-type cytochrome. Acidovorax sp. str. NO-1 accession number: HM357240.1. | ||
The partial open reading frame (1059 nt of a possible 1428 nt encoding 353/476 amino acids) upstream of aioB was found to share homology with putative phage integrases from various Betaproteobacteria such as Oxalobacteraceae sp. str. IMCC9480 (59% identity over 172 amino acids) and Cupriavidus (also known as Ralstonia) metallidurans (40% identity over 317 amino acids). The GM1 integrase sequence however, did not contain the conserved C-terminal catalytic domain present in the sequences of the closely related phage integrases, indicating that it is unlikely to be functional. The aioB gene is found downstream of the partial integrase sequence and encodes an apoprotein of 179 amino acids (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 561 Da). The protein shares closest sequence identity with the AioB homologues in the Betaproteobacteria such as Acidovorax sp. str. NO-1 (73% identity), A. faecalis (57% identity) and Herminiimonas arsenicoxydans (61% identity) compared with the alphaproteobacterial sequence from NT-26 (40% identity). The sequence contains a conserved motif involved in binding the Rieske 2Fe–2S cluster and in the case of A. faecalis (not in NT-26) conserved residues involved in disulphide bond formation, Cys-X-His-X-Gly-Cys*-X17-Cys-X-Cys*-His (cluster ligands are in italics; the asterisked residues form a disulphide bond which bridges over the cluster, connecting the two cluster-binding loops). In common with all other AioB sequences the GM1 sequence contains a Tat signal sequence (1–36 amino acids) with the conserved RRXFLK motif22 (RRFFLK in GM1 AioB) which directs the transport of the fully folded AioBA complex to the periplasm via the Tat pathway. A predicted signal cleavage site is present between amino acids 36 and 37 (i.e. ADA-AT) resulting in a processed protein of 14
561 Da). The protein shares closest sequence identity with the AioB homologues in the Betaproteobacteria such as Acidovorax sp. str. NO-1 (73% identity), A. faecalis (57% identity) and Herminiimonas arsenicoxydans (61% identity) compared with the alphaproteobacterial sequence from NT-26 (40% identity). The sequence contains a conserved motif involved in binding the Rieske 2Fe–2S cluster and in the case of A. faecalis (not in NT-26) conserved residues involved in disulphide bond formation, Cys-X-His-X-Gly-Cys*-X17-Cys-X-Cys*-His (cluster ligands are in italics; the asterisked residues form a disulphide bond which bridges over the cluster, connecting the two cluster-binding loops). In common with all other AioB sequences the GM1 sequence contains a Tat signal sequence (1–36 amino acids) with the conserved RRXFLK motif22 (RRFFLK in GM1 AioB) which directs the transport of the fully folded AioBA complex to the periplasm via the Tat pathway. A predicted signal cleavage site is present between amino acids 36 and 37 (i.e. ADA-AT) resulting in a processed protein of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 Da. Downstream of aioB is aioA and it encodes a protein of 827 amino acid (91
733 Da. Downstream of aioB is aioA and it encodes a protein of 827 amino acid (91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 690 Da unprocessed and 91
690 Da unprocessed and 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 559 Da without N-terminal methionine). The GM1 AioA shares the highest sequence identity with that from Acidovorax sp. str. NO-1 (80% identity), A. faecalis (70% identity) and Herminiimonas arsenicoxydans (69% identity) compared with that from NT-26 (47% identity). The conserved motif, Cys-X2-Cys-X3-Cys-X-Ser70 involved in binding the 3Fe–4S cluster, is conserved, as are the amino acids implicated in binding arsenite (i.e. His197, Glu205, Arg421 and His425). Downstream of aioB is cytC, which encodes a putative c-type cytochrome of 104 amino acids (10
559 Da without N-terminal methionine). The GM1 AioA shares the highest sequence identity with that from Acidovorax sp. str. NO-1 (80% identity), A. faecalis (70% identity) and Herminiimonas arsenicoxydans (69% identity) compared with that from NT-26 (47% identity). The conserved motif, Cys-X2-Cys-X3-Cys-X-Ser70 involved in binding the 3Fe–4S cluster, is conserved, as are the amino acids implicated in binding arsenite (i.e. His197, Glu205, Arg421 and His425). Downstream of aioB is cytC, which encodes a putative c-type cytochrome of 104 amino acids (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 884 Da unprocessed) and contains a single CX2CH heme-binding motif suggesting the presence of a single heme c. The protein also contains a general secretory pathway (Sec) signal sequence for its transport to the periplasm, with a predicted cleavage site between amino acids 22 and 23 (i.e. SFA-NL) resulting in a processed protein of 8503 Da. The putative cytochrome may act as a physiological electron acceptor to the GM1 Aio as was observed for the c552 cytochrome encoded by the cytC gene in the aio cluster of NT-26.15 The putative GM1 cytochrome and nitroreductase did not show significant similarity to cytochrome and nitroreductase encoding genes in other aio gene clusters.
884 Da unprocessed) and contains a single CX2CH heme-binding motif suggesting the presence of a single heme c. The protein also contains a general secretory pathway (Sec) signal sequence for its transport to the periplasm, with a predicted cleavage site between amino acids 22 and 23 (i.e. SFA-NL) resulting in a processed protein of 8503 Da. The putative cytochrome may act as a physiological electron acceptor to the GM1 Aio as was observed for the c552 cytochrome encoded by the cytC gene in the aio cluster of NT-26.15 The putative GM1 cytochrome and nitroreductase did not show significant similarity to cytochrome and nitroreductase encoding genes in other aio gene clusters.
Purification and characterisation of the arsenite oxidase
The Aio from GM1 was purified from the periplasm by a two-step ammonium sulphate precipitation (50% and 75%) followed by cation-exchange and size-exclusion chromatography. The Aio was enriched 32.7-fold (Table 1). The purified Aio consisted of two heterologous subunits of 88 kDa (AioA) and 15 kDa (AioB) (Fig. 2), and had a native molecular mass of 105.5 kDa (determined by gel filtration chromatography) indicating that the enzyme is a heterodimer (α1β1). The oligomeric state of the GM1 Aio is like that of A. faecalis,23Arthrobacter sp. str. 15b24 and Ralstonia sp. str. 2214 but varies from that of NT-2625 and Hydrogenophaga sp. str. NT-1426 which form oligomers of α2β2 and α3β3, respectively.| Purification step | Total protein (mg) | Total activity (U) | Specific activity (U mg−1) | Purification (-fold) |
|---|---|---|---|---|
| The GM1 Aio was purified from the periplasm after the preparation of spheroplasts. The periplasm was fractionated by ammonium sulphate precipitation and the Aio-containing fraction desalted and purified by cation exchange and gel filtration chromatography. | ||||
| Total cell extract | 74.4 | 26.83 | 0.36 | 1 |
| Periplasm | 12.9 | 12.71 | 1.02 | 2.8 |
| Desalting | 6.74 | 12.94 | 1.92 | 5.3 |
| Cation exchange | 0.51 | 3.73 | 7.32 | 20.3 |
| Gel filtration | 0.14 | 1.65 | 11.79 | 32.7 |
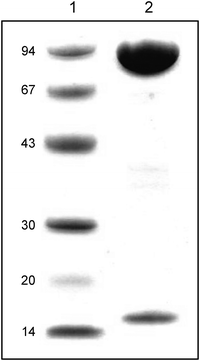 | ||
| Fig. 2 SDS-polyacrylamide gel of purified GM1 Aio stained with Coomassie Blue R350. 1: low molecular weight standards (size in kDa); 2: purified GM1 Aio. | ||
Purified GM1 Aio was analysed for metal cofactors using ICP-MS and was found to contain Mo (1.2 mol mol−1 enzyme) and Fe (6 mol mol−1 enzyme), indicating that like the mesophilic enzymes, the GM1 Aio contains Mo at the active site and two Fe–S clusters, which was also confirmed by sequence analyses (see above). The enzyme had a Km for arsenite of 111.70 ± 0.88 μM and a Vmax of 12.16 ± 0.30 μmol arsenite oxidised min mg−1, corresponding to a Kcat of 21.38 ± 0.52 s−1 (based on a native molecular mass of 105.5 kDa). All three of these values are the highest observed for any Aio purified to date, when DCPIP was used as the electron acceptor. The higher Km value shows that the GM1 Aio has a lower affinity for arsenite than the mesophilic enzymes, while the high Vmax value is at least double that of the most active mesophilic Aio (i.e. that of NT-14).14,25,26
A variety of c-type cytochromes and the copper-containing protein azurin can serve as electron acceptors to the Aio. NT-26 cytochrome c552 (electron acceptor to the NT-26 Aio),15 commercially purchased horse heart cytochrome c and Pseudomonas aeruginosa azurin, were tested for their ability to accept electrons from the GM1 Aio. Cytochrome c552 and horse-heart cytochrome c were not reduced by the GM1 Aio in the presence of arsenite (ESI,† Fig. S1 and S2). The oxidised P. aeruginosa azurin exhibited a characteristic absorption band at 625 nm which disappeared upon its reduction in the presence of the GM1 Aio and arsenite (Fig. 3). Azurin was not reduced by either the Aio or arsenite alone (ESI,† Fig. S3). The Aio of A. faecalis and Ralstonia sp. str. 22 can also use azurin as the electron acceptor but like the GM1 Aio are unable to use horse heart cytochrome c and the NT-26 c55214,23 whereas the NT-26 Aio which can use the c552 and horse heart cytochrome c cannot use azurin.15 The reason why the enzymes react differently with the various electron acceptors is presently unknown, but it appears to be unrelated to the redox potentials of the Rieske cluster.14,15,27
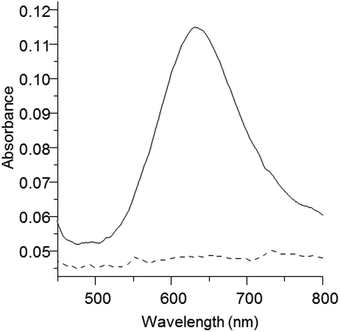 | ||
| Fig. 3 Oxidised and reduced spectra of P. aeruginosa azurin. Oxidised azurin (solid line) was reduced (dashed line) in the presence of the GM1 Aio and arsenite. | ||
Cold adaptation of GM1 Aio
GM1 is a psychrotolerant arsenite oxidiser with a growth range between 0–20 °C. We therefore tested the effect of temperature on Aio activity making comparisons with the mesophilic enzyme from the only organism with a solved X-ray crystal structure A. faecalis.12 The enzyme from A. faecalis was purified after successful heterologous expression in E. coli. GM1 Aio had a maximum observed activity at 45 °C (13.8 U mg−1), which was reduced to 8.0 U mg−1 at 60 °C and 9.2 U mg−1 at 10 °C corresponding to reductions of 42% and 33% of maximum observed activity, respectively (Fig. 4). In contrast, the Aio from A. faecalis had a maximum observed activity at 65 °C (8.1 U mg−1), which was reduced to 7.3 U mg−1 at 75 °C and 1.2 U mg−1 at 10 °C, corresponding to reductions of 10% and 75% of maximum observed activity, respectively. As can be seen in Fig. 4, the GM1 Aio was active at lower temperatures (Fig. 4). The fact that the GM1 Aio had a lower optimum temperature (45 °C) than the A. faecalis enzyme (65 °C) and was also more active at lower temperatures is a common trait of cold-adapted enzymes.28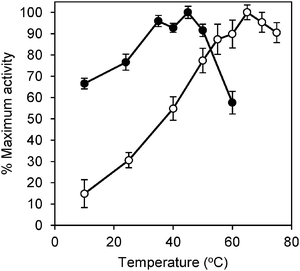 | ||
| Fig. 4 Temperature-activity profiles of the GM1 and A. faecalis arsenite oxidases. Percentage of maximum activity is plotted as a function of temperature of GM1 (●) and A. faecalis (○) Aio. Data points and error bars represent mean and standard deviation of at least three assays, respectively. | ||
The properties that make enzymes cold-adapted are well known.28,29 The rate of an enzyme-catalysed reaction will change exponentially with a change in temperature hence temperature is in the exponent of the Arrhenius equation. Temperature is a function of activation energy and so a decrease in activation energy will make a reaction less dependent on temperature. In general, cold-adapted enzymes have lower activation energies than their mesophilic homologues increasing their activity at lower temperatures, which results from reduced enthalpy-requiring conformational changes needed to achieve the enzyme-substrate transition complex. The reduction in enthalpy-requiring changes often manifests itself as an increase in overall enzyme flexibility or localised flexibility around the active site, however, the increase in flexibility also decreases the protein's stability.28,29 The increase in flexibility to achieve increased reaction rates means that often cold-adapted enzymes also have decreased affinity for the substrate (i.e. higher Km)30,31 compared to their mesophilic homologues. An increased reaction rate and a decreased affinity for arsenite are observed for the GM1 Aio when compared to the mesophilic homologues suggesting that the former has increased flexibility.
Homology modelling of GM1 Aio
To investigate the molecular basis of cold-adaptation in the GM1 Aio, a model was built of the enzyme using the A. faecalis Aio crystal structure as a template. The model shows that the majority of amino acid differences between the two enzymes occur on the surface of the enzyme and that the entry channel to the active site is well conserved (Fig. 5). Destabilising effects on the structure, caused by amino acid differences between the GM1 and A. faecalis Aio were predicted using the SAAPdap pipeline of SAAPdb.32 44 amino acid differences (40 in AioA and 4 in AioB) were predicted to have destabilising structural effects on GM1 Aio, and are summarised in Table 2. It should be noted that as the analysis treats each amino acid difference as an individual mutation, many structural changes may be compensated by other changes. Increased flexibility/decreased stability of cold-adapted enzymes can be caused by several structural changes, including a decrease in stabilising bonds (H-bonds, van der Waals interactions and salt bridges), an increase in the surface hydrophobicity and a decrease in the core hydrophobicity. The SAAPdap analysis shows that a decrease in H-bonds and insertion of hydrophilic residues into the core are the major destabilising factors in the GM1 Aio. Of the 44 structure-affecting amino acid differences between the GM1 and the A. faecalis Aio, eight amino acids are unique to GM1 that appear to be conserved, or are synonymous, in sequences of A. faecalis, NT-26 (a mesophilic alphaproteobacterium with a similar Aio temperature-activity profile to A. faecalis;)33 and the thermophilic arsenite oxidiser Thermus thermophilus str. HB8.34 Of these eight amino acids, all in AioA, five introduce hydrophilic residues into the core of the structure: C32: Val-Thr; C231: Ile-Thr; C312: Gly-Thr; C469: Leu-His; C664: Gly-Asn; one disrupts a H-bond C668: Asp-Lys; one causes a clash: C779: Gly-Ala and one removes a buried charge: C713: Asp-Asn (Residue changes are written in the following format: ‘PDB number: A. faecalis residue-GM1 residue’). The main mechanism for cold-adaptation of the GM1 Aio seems to be the insertion of hydrophilic residues into the hydrophobic core, which leads to destabilization of the enzyme structure, resulting in its increased activity at lower temperatures.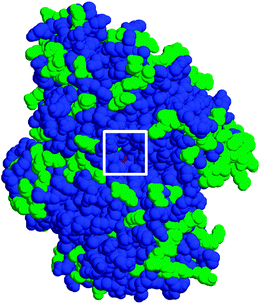 | ||
| Fig. 5 Space-filling homology model of GM1 Aio. The channel to the active site is boxed. Conserved residues between the GM1 and A. faecalis structure are blue; different residues are green. | ||
| Structural effect | No. of amino acid differences |
|---|---|
| a Disrupts a hydrogen bond.b Introduces a hydrophilic residue into the protein core.c Introduces a hydrophobic residue on the surface.d Introduces or removes a charged residue from the protein core.e Occurs at an interface according to difference in solvent accessibility.f Shows a specific binding interaction (hydrogen bond or van der Waals contact) at an interface.g Replacement sidechain clashes with its surroundings.h Introduces a void into the protein core.i Parent residue was a glycine with a backbone conformation not favoured for other residues. | |
| HBonda | 22 |
| CorePhilicb | 10 |
| SurfacePhobicc | 6 |
| Buriedcharged | 6 |
| Interfacee | 5 |
| Bindingf | 4 |
| Clashg | 6 |
| Voidsh | 4 |
| Glycinei | 1 |
Our results are in agreement with the study of another member of the DMSO reductase family of molybdoenzymes, the cold-adapted periplasmic nitrate reductase (Nap) from Shewanella gelidimarina.35 Cold-adaptation in the S. gelidimarina Nap also resulted from destabilising changes to the structure and not changes to the active site or the presence of different cofactors.
Experimental
GM1 growth conditions
GM1 was grown in a Minimal Salts Medium (MSM) containing 0.04% yeast extract in the absence or presence of 5 mM arsenite as described previously.18 All incubations were performed at 10 °C unless stated otherwise. Arsenic analyses were determined as described previously.18 The GM1-A strain, in which aioA was mutated, was grown in the presence of 5 μg ml−1 kanamycin. For the preparation of cell extracts and the purification of the Aio, GM1 was grown in the absence of arsenite in 2 L batch cultures (in 5 L flasks) until late-log phase (72 h; OD600: 0.15–0.17).Mutagenesis of aioA
The GM1 aioA gene was disrupted by targeted gene disruption as has been done previously.15,25 Briefly, a ∼0.5 kb portion of the aioA gene18 was cloned into the suicide plasmid pJP5603 (KmR).36 The recombinant plasmid was transformed into Escherichia coli str. S17-1 λpir37 and transferred into a spontaneous rifampicin-resistant mutant of GM1 (GM1-R) by filter mating at 20 °C for 24 h on a Luria Bertani (LB) agar plate. The conjugation mixture was plated on MSM containing 1.5% purified agar (Oxoid), 0.04% yeast extract (Oxoid), 100 μg ml−1 rifampicin and 5 μg ml−1 kanamycin. The transconjugants were purified and selected for their ability to oxidise 5 mM arsenite in MSM with 0.04% yeast extract as described previously.18Cloning and sequencing of the aio gene cluster
The aio gene cluster was cloned by a ‘plasmid rescue’ technique. Genomic DNA from GM1-A was isolated using the Wizard® genomic DNA purification kit (Promega) and digested with the restriction enzyme ClaI. The linear fragments were circularised with T4 DNA ligase (Promega), transformed into E. coli str. JM109 λpir by electroporation and transformants containing pJP5603 and flanking GM1 genomic DNA were selected on LB containing 20 μg ml−1 Km. Both strands of the aio gene cluster were sequenced by primer-walking. Sequencing was performed at the Wolfson Institute for Biomedical Research (WIBR) by capillary electrophoresis using a 3730XL Genetic Analyser with BigDye 3.1 (Applied Biosystems). Databases were searched using the BLAST suite (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and open reading frames (ORFs) allocated using ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). Signal peptide motifs were identified using SignalP38 and TatP.39 The GenBank accession number for the sequence of the GM1 aio gene cluster is EU106602.Preparation of GM1 cell fractions
Total cell extracts were prepared as described previously6 using the previously determined optimum buffer of the Aio with the artificial electron acceptor 2,6-dichlorophenolindophenol (DCPIP), 50 mM MES (pH 5.5).18 For the preparation of spheroplasts, cells were harvested and washed and spheroplasts prepared as described previously40 with the following modifications: (1) the harvested cells were suspended in ice-cold 750 mM sucrose, 30 mM Tris/HCl (pH 8) at a ratio of 0.5 g wet cells per ml, (2) lysozyme was added at a concentration of 10 mg g−1 wet cells, (3) cells were incubated with lysozyme at room temperature for 1 h, (4) after the addition of 15 mM EDTA (pH 8, 0 °C), the suspension was incubated at 0 °C for 30 min.Purification of the GM1 Aio
The GM1 Aio was purified from the periplasm after spheroplast preparation. The periplasm was subjected to two ammonium sulphate precipitation steps, 50% and 75% saturation (0 °C). The protein pellet was suspended in 50 mM MES (pH 5.5) and desalted using a PD-10 desalting column (GE Healthcare) according to the manufacturer's instructions, with 50 mM MES (pH 5.5) as the elution buffer. The sample (3.5 ml) was passed through a 0.22 μm filter (Millipore) and concentrated using a Vivaspin 20 device (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO) (Sartorius Stedium Biotech) to <500 μl. The sample was applied to a SP sepharose fast flow HiPrep 16/10 cation exchange column (GE Healthcare) which had been pre-equilibrated with 50 mM MES (pH 5.5). Chromatography was carried out at room temperature (20–25 °C) at a flow rate of 5 ml min−1. The Aio was eluted using a 1 M NaCl gradient in 50 mM MES (pH 5.5). The NaCl gradient was as follows: (1) 0 M for 2.25 column volumes and (2) a linear increase to 0.6 M over 12 column volumes. Fractions containing Aio activity were pooled and concentrated. The sample was then applied to a Superdex 200 10/300 gel filtration column (GE Healthcare) pre-equilibrated with 50 mM MES, 150 mM NaCl (pH 5.5). Chromatography was carried out at a flow rate of 0.5 ml min−1 at room temperature. Fractions containing Aio activity were pooled and de-salted using a PD-10 desalting column.
000 MWCO) (Sartorius Stedium Biotech) to <500 μl. The sample was applied to a SP sepharose fast flow HiPrep 16/10 cation exchange column (GE Healthcare) which had been pre-equilibrated with 50 mM MES (pH 5.5). Chromatography was carried out at room temperature (20–25 °C) at a flow rate of 5 ml min−1. The Aio was eluted using a 1 M NaCl gradient in 50 mM MES (pH 5.5). The NaCl gradient was as follows: (1) 0 M for 2.25 column volumes and (2) a linear increase to 0.6 M over 12 column volumes. Fractions containing Aio activity were pooled and concentrated. The sample was then applied to a Superdex 200 10/300 gel filtration column (GE Healthcare) pre-equilibrated with 50 mM MES, 150 mM NaCl (pH 5.5). Chromatography was carried out at a flow rate of 0.5 ml min−1 at room temperature. Fractions containing Aio activity were pooled and de-salted using a PD-10 desalting column.Heterologous expression and purification of the Alcaligenes faecalis Aio
The aioB and aioA (aioBA) genes from A. faecalis were amplified without the native twin-arginine translocation (Tat) leader sequence using the following primers: Forward; 5′-GC![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) CGTACAACATTGCAGTACCCT-3′ and Reverse: 5′-CG
CGTACAACATTGCAGTACCCT-3′ and Reverse: 5′-CG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCAGGCGAAGCGGCGGCTTTT-3′. The restriction enzymes XbaI and XhoI (underlined above) were used for cloning into the expression vector, pPROEX-HTb in frame with the N-terminal histidine tag. The PCR reaction was performed for 35 cycles using the KOD proofreading DNA polymerase (Applied Biosystems) with a 55 °C annealing temperature as per the manufacturer's instructions.
TCAGGCGAAGCGGCGGCTTTT-3′. The restriction enzymes XbaI and XhoI (underlined above) were used for cloning into the expression vector, pPROEX-HTb in frame with the N-terminal histidine tag. The PCR reaction was performed for 35 cycles using the KOD proofreading DNA polymerase (Applied Biosystems) with a 55 °C annealing temperature as per the manufacturer's instructions.The E. coli JM109 λpir strain was used for Aio expression. Expression of the Aio involved growing E. coli aerobically at 21 °C for 24 h in LB containing 100 μg ml−1 Ap, 40 μM IPTG (isopropyl β-D-1-thiogalactopyranoside) and 1 mM Na2MoO4.
Cells were harvested by centrifugation at 5464 × g for 10 min (4 °C) and the pellets washed by suspending them in binding buffer (20 mM KH2PO4, 500 mM NaCl, 20 mM imidazole, pH 7.3). The cells were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180 × g for 10 min (4 °C) and the pellet resuspended in binding buffer (10 ml g−1 wet weight cells). Cells were disrupted by two passages through a French press (13
180 × g for 10 min (4 °C) and the pellet resuspended in binding buffer (10 ml g−1 wet weight cells). Cells were disrupted by two passages through a French press (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi) and the cell debris removed by centrifugation at 30
000 psi) and the cell debris removed by centrifugation at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g (4 °C). The supernatant was filtered through a 0.22 μM filter (Millipore) and loaded onto a HisTrap-Ni Sepharose High Performance column (GE Healthcare) that had been pre-equilibrated with binding buffer. Chromatography was carried out at 1 ml min−1 at room temperature and unbound proteins removed with 30 column volumes of binding buffer followed by a gradient of 30 column volumes with elution buffer (20 mM KH2PO4 500 mM NaCl, 500 mM imidazole, pH 7.3). Fractions that contained Aio activity (Aio was eluted at 295 mM imidazole) were pooled and the sample was concentrated using a Vivaspin 20 (MWCO 50
000 × g (4 °C). The supernatant was filtered through a 0.22 μM filter (Millipore) and loaded onto a HisTrap-Ni Sepharose High Performance column (GE Healthcare) that had been pre-equilibrated with binding buffer. Chromatography was carried out at 1 ml min−1 at room temperature and unbound proteins removed with 30 column volumes of binding buffer followed by a gradient of 30 column volumes with elution buffer (20 mM KH2PO4 500 mM NaCl, 500 mM imidazole, pH 7.3). Fractions that contained Aio activity (Aio was eluted at 295 mM imidazole) were pooled and the sample was concentrated using a Vivaspin 20 (MWCO 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) (Satorius Stedium Biotech) centrifugal concentrator and loaded on to a Superdex 200 10/300 gel filtration column (GE Healthcare) that had been pre-equilibrated with 50 mM MES, 150 mM NaCl (pH 6.0). Gel filtration chromatography was carried out at a flow rate of 0.5 ml min−1 at room temperature. Fractions containing Aio activity were pooled and the buffer exchanged using a PD-10 desalting column (GE Healthcare), according to the manufacturer's instructions, with 50 mM MES (pH 6.0) serving as the elution buffer. The sample was concentrated using a Vivaspin 20 (MWCO 50
000) (Satorius Stedium Biotech) centrifugal concentrator and loaded on to a Superdex 200 10/300 gel filtration column (GE Healthcare) that had been pre-equilibrated with 50 mM MES, 150 mM NaCl (pH 6.0). Gel filtration chromatography was carried out at a flow rate of 0.5 ml min−1 at room temperature. Fractions containing Aio activity were pooled and the buffer exchanged using a PD-10 desalting column (GE Healthcare), according to the manufacturer's instructions, with 50 mM MES (pH 6.0) serving as the elution buffer. The sample was concentrated using a Vivaspin 20 (MWCO 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) (Satorius Stedium Biotech) and Micron YM-30 (Millipore) centrifugal concentrators and enzyme assays performed immediately.
000) (Satorius Stedium Biotech) and Micron YM-30 (Millipore) centrifugal concentrators and enzyme assays performed immediately.
Enzyme assays and protein determination
Aio and malate dehydrogenase enzyme assays were done as described previously.6 The results of the kinetics are from two independent experiments with three replicates for each arsenite concentration tested, so there are a total of 6 values for each concentration. All assays were performed at 24 °C unless otherwise stated. The temperature of the enzyme assays was controlled with a Varian Cary dual cell Peltier. Reaction mixtures were incubated at the cell temperature for 5 minutes prior to the addition of arsenite.Protein concentrations were determined using the Bradford method41 and BioRad reagent according to the manufacturer's instructions and bovine serum albumin served as the standard.
The oxidised and reduced spectra of the Rhizobium sp, str. NT-26 cytochrome c552,15 horse heart cytochrome c (Sigma), and P. aeruginosa azurin (Sigma) were recorded as described previously15,26 with 20 μg of electron acceptor and 3 μg of Aio.
Subunit composition and cofactor analysis
SDS-PAGE of denatured protein was performed as described previously40 with low molecular weight standards (GE Healthcare). For cofactor analysis purified Aio was prepared as described previously25 and analysed for Fe, Co, Ni, Cu, Zn, Mo and W, by inductively coupled plasma-mass spectrometer (ICP-MS) (Varian) at the Mineralogy Department, Natural History Museum, London, UK.Homology modelling
Modelling of the GM1 Aio was performed with Modeller42 using the A. faecalis Aio crystal structure as a template (PDB code 1g8j, chains C and D).12 The alignment was hand-modified to accommodate the insertion at C505–C506 and deletion at C651 to minimise disruption of the parent structure. Structural effects of amino acid differences were analysed using the SAAPdap pipeline of the SAAPdb database32 treating each difference as a single mutation in the A. faecalis structure.Conclusions
GM1 is a psychrotolerant arsenite oxidiser that possesses an Aio that is adapted to function at cold temperatures. Compared to its mesophilic counterparts the GM1 Aio has the same subunit composition and contains the same cofactors but it has a much higher reaction rate and a lower affinity for arsenite. Cold-adaptation of GM1 Aio is probably caused by an increase in the enzyme's flexibility which appears to result from changes in the overall protein structure and not by alterations specific to the active site or cofactors. The insertion of hydrophilic residues into the core is a major destabilising factor in the GM1 Aio. The aerobic arsenite oxidase is an ancient bioenergetic enzyme27,43,44 and understanding the reaction mechanism from psychrophilic, mesophilic and thermophilic homologues will not only improve our understanding of its evolution but should also aid our development of a broad-temperature range arsenite biosensor.Acknowledgements
THO was supported by a Natural Environment Research Council PhD studentship (14404A). MDH is supported by a Medical Research Council PhD studentship (G0800112). We would like to thank Barbara Schoepp-Cothenet and Gretchen Anderson for supplying the A. faecalis strain and helpful discussions. We would also like to thank Stanislav Strekopytov for performing the metal analyses of the arsenite oxidase.References
- J. O. Nriagu, in Environmental chemistry of arsenic, ed. W. T. Frankenberger, Marcel Dekker, New York, 2002, ch. 1, pp. 1–26 Search PubMed.
- P. L. Smedley and D. G. Kinniburgh, Appl. Geochem., 2002, 17, 517 CrossRef CAS.
- M. F. Hughes, Toxicol. Lett., 2002, 133, 1 CrossRef CAS.
- P. Ravenscroft, H. Brammer and K. Richards, Arsenic pollution a global synthesis, Wiley-Blackwell, UK, 2009 Search PubMed.
- D. K. Nordstrom, Science, 2002, 296, 2143 CrossRef CAS.
- J. M. Santini, L. I. Sly, R. D. Schnagl and J. M. Macy, Appl. Environ. Microbiol., 2000, 66, 92 CrossRef CAS.
- R. S. Oremland and J. F. Stolz, Science, 2003, 300, 939 CrossRef CAS.
- H. H. Green, S. Afr. J. Sci., 1918, 14, 465 CAS.
- T. H. Osborne and J. M. Santini, in The Metabolism of Arsenite, ed. J. M. Santini and S. A. Ward, CRC Press, Boca Raton, Florida, 2012, ch. 5, pp. 61–69 Search PubMed.
- M. C. Lett, D. Muller, D. Lièvremont, S. Silver and J. M. Santini, J. Bacteriol., 2012, 194, 207 CrossRef CAS.
- A. G. McEwan, J. P. Ridge, C. A. McDevitt and P. Hugenholtz, Geomicrobiol. J., 2002, 9, 3 CrossRef.
- P. J. Ellis, T. Conrads, R. Hille and P. Kuhn, Structure, 2001, 9, 125 CrossRef CAS.
- M. D. Heath, B. Schoepp-Cothenet, T. H. Osborne and J. M. Santini, in The Metabolism of Arsenite, ed. J. M. Santini and S. A. Ward, CRC Press, Boca Raton, Florida, 2012, ch. 7, pp. 81–97 Search PubMed.
- A. Lieutaud, R. van Lis, S. Duval, L. Capowiez, D. Muller, R. Lebrun, S. Lignon, M. L. Fardue, M. C. Lett, W. Nitschke and B. Schoepp-Cothenet, J. Biol. Chem., 2010, 285, 20442 CrossRef.
- J. M. Santini, U. Kappler, S. A. Ward, M. J. Honeychurch, R. N. vanden Hoven and P. V. Bernhardt, Biochim. Biophys. Acta, 2007, 1767, 189 CrossRef CAS.
- S. Sardiwal, J. M. Santini, T. H. Osborne and S. Djordjevic, FEMS Microbiol. Lett., 2010, 313, 20 CrossRef CAS.
- G. Liu, M. Liu, E. H. Kim, W. S. Maaty, B. Bothner, B. Lei, C. Rensing, G. Wang and T. R. McDermott, Environ. Microbiol., 2012, 14, 1624 CrossRef CAS.
- T. H. Osborne, H. E. Jamieson, K. A. Hudson-Edwards, D. K. Nordstrom, S. R. Walker, S. A. Ward and J. M. Santini, BMC Microbiol., 2010, 10, 205 CrossRef.
- Y. Huang, H. Li, C. Rensing, K. Zhao, L. Johnstone and G. Wang, J. Bacteriol., 2012, 194, 1635 CrossRef CAS.
- S. Koechler, J. Cleiss-Arnold, C. Proux, O. Sismeiro, M. A. Dillies, F. Goulhen-Chollet, F. Hommais, D. Lièvremont, F. Arsène-Ploetze, J. Y. Coppée and P. N. Bertin, BMC Microbiol., 2010, 10, 53 CrossRef.
- D. R. Kashyap, L. M. Botero, W. L. Franck, D. J. Hassett and T. R. McDermott, J. Bacteriol., 2006, 188, 1081 CrossRef CAS.
- N. R. Stanley, T. Palmer and B. C. Berks, J. Biol. Chem., 2000, 275, 11591 CrossRef CAS.
- G. L. Anderson, J. Williams and R. Hille, J. Biol. Chem., 1992, 267, 23674 CAS.
- K. S. Prasad, V. Subramanian and J. Paul, Biometals, 2009, 22, 711 CrossRef CAS.
- J. M. Santini and R. N. vanden Hoven, J. Bacteriol., 2004, 186, 1614 CrossRef CAS.
- R. N. vanden Hoven and J. M. Santini, Biochim. Biophys. Acta, 2004, 1656, 148 CrossRef CAS.
- S. Duval, A. L. Ducluzeau, W. Nitschke and B. Schoepp-Cothenet, BMC Evol. Biol., 2008, 8, 206 CrossRef.
- G. Feller and C. Gerday, Nat. Rev. Microbiol., 2003, 1, 200 CrossRef CAS.
- K. S. Siddiqui and R. Cavicchioli, Annu. Rev. Biochem., 2006, 75, 403 CrossRef CAS.
- D. Georlette, B. Damien, V. Blaise, E. Depiereux, V. N. Uversky, C. Gerday and G. Feller, J. Biol. Chem., 2003, 278, 37015 CrossRef CAS.
- S. D'amico, J. C. Marx, C. Gerday and G. Feller, J. Biol. Chem., 2003, 278, 7891 CrossRef CAS.
- J. M. Hurst, L. E. M. McMillan, C. T. Porter, J. Allen, A. Fakorede and A. C. R. Martin, Hum. Mutat., 2009, 30, 616 CrossRef CAS.
- T. P. Warelow and J. M. Santini, Unpublished work.
- T. M. Gihring, G. K. Druschel, R. B. McClesky, R. J. Hamers and J. F. Banfield, Environ. Sci. Technol., 2001, 19, 3857 CrossRef.
- P. J. Simpson and R. Codd, Biochem. Biophys. Res. Commun., 2011, 414, 783 CrossRef CAS.
- R. J. Penfold and J. M. Pemberton, Gene, 1992, 118, 145 CrossRef CAS.
- R. Simon, U. Priefer and A. Pühler, Biotechnology, 1983, 1, 784 CrossRef CAS.
- T. N. Petersen, S. Brunak, G. von Heijne and H. Nielsen, Nat. Methods, 2011, 8, 785 CrossRef CAS.
- D. Bendtsen, H. Nielsen, D. Widdick, T. Palmer and S. Brunak, BMC Bioinf., 2005, 6, 167 CrossRef.
- T. Krafft and J. M. Macy, Eur. J. Biochem., 1998, 255, 647 CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248 CrossRef CAS.
- N. Eswar, M. A. Marti-Renom, B. Webb, M. S. Madhusudhan, D. Eramian, M. Shen, U. Pieper and A. Sali, Curr. Protoc. Bioinformatics, 2006, Suppl. 15, 5.6.1 Search PubMed.
- R. van Lis, W. Nitschke, S. Duval and B. Schoepp-Cothenet, in The Metabolism of Arsenite, ed. J. M. Santini and S. A. Ward, CRC Press, Boca Raton, Florida, 2012, ch. 10, pp. 125–144 Search PubMed.
- E. Lebrun, M. Brugna, F. Baymann, D. Muller, D. Lièvremont, M.-C. Lett and W. Nitschke, Mol. Biol. Evol., 2003, 20, 686 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2mt20180a |
| This journal is © The Royal Society of Chemistry 2013 |
