 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis and antiproliferative activity of some 3-(pyrid-2-yl)-pyrazolines†
Alexander
Ciupa
a,
Paul A.
De Bank
a,
Mary F.
Mahon
b,
Pauline J.
Wood
a and
Lorenzo
Caggiano
*a
aMedicinal Chemistry, Department of Pharmacy and Pharmacology, University of Bath, Claverton Down, Bath, BA2 7AY, UK. E-mail: l.caggiano@bath.ac.uk; Fax: +44 (0)1225 386114; Tel: +44 (0)1225 385709
bX-ray Crystallography Unit, Department of Chemistry, University of Bath, Claverton Down, Bath, BA2 7AY, UK
First published on 24th April 2013
Abstract
The synthesis and antiproliferative activity of eleven 3-(pyrid-2-yl)-pyrazolines in two cancer cell lines are reported. X-ray crystallography was obtained of the lead compound 8i which was screened in the NCI 60 human tumour cell line and displayed sub-micromolar activity. Cell cycle analysis, in vitro tubulin assay and confocal microscopy are also reported and suggest that the lead compound disrupts microtubule formation.
Introduction
Numerous pyrazoline derivatives possess potent biological activities,1,2 including antiproliferative,3–6 antimalarial7 and antibacterial activities.8 The inhibition of various processes such as xanthine oxidase,9 COX-2,10 monoamine oxidase11–17 and, of particular relevance to the current study, tubulin polymerisation18 have also been reported (selected examples are shown in Fig. 1).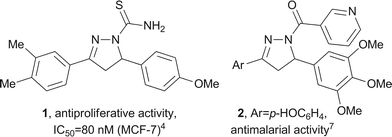 | ||
| Fig. 1 Examples of biologically active pyrazolines 1 and 2. | ||
We have previously reported the synthesis and physicochemical properties of the simple pyrazoline 8a (R1 = H, R2 = Me) and the corresponding oxidised pyrazole derivative, which are “turn on” fluorescent sensors for Cd2+ and Zn2+, respectively.19 Our interests in pyrazolines derived from 2-acetylpyridine 3 originated from possible chelation of ions in these systems by the pyridine nitrogen, the basic pyrazoline nitrogen (N2) and co-ordinating groups (R2, Scheme 1). Compounds of this structure have been reported as ligands for gold and display antiproliferative activities.20 In addition, the 3-pyridyl motif would afford compounds with improved pharmacological properties. N1-Substituted 3-(pyrid-2-yl)-pyrazolines display various biological activities, including antitubercular,21 antimycobacterial,22 antifungal,23 anti-inflammatory24,25 and antiproliferative20,24 activities. We therefore wished to investigate novel compounds of this class and examine their antiproliferative activity.
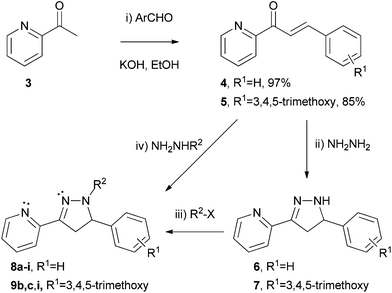 | ||
| Scheme 1 Synthesis of 3-(pyrid-2-yl)-pyrazolines 8 and 9 (see Table 1 for R2 groups and yields). | ||
Results and discussion
Continuing our research interests in pyrazolines derived from 2-acetylpyridine 3, we synthesised various derivatives following the sequence shown in Scheme 1. Claisen–Schmidt condensation of 2-acetylpyridine 3 with an aromatic aldehyde gave the corresponding chalcone 4 (97%) or 5 (85%) in excellent yields. Treatment with an excess of hydrazine hydrate gave the N–H pyrazoline derivative 6 or 7 which could be characterised by 1H NMR. Attempts to purify these compounds by silica gel column chromatography however led to decomposition, as was previously observed with similar substrates,22,23,26 possibly due to autoxidation.27 Instead, the N–H pyrazolines 6 and 7 were treated directly with the desired electrophile to afford the N-substituted pyrazolines 8 and 9 which could be purified, in a one-pot transformation in good yields.The N–Me and N–Ph substituted pyrazolines 8a,b and 9b were synthesised by reaction of the chalcone 4 or 5 with the commercially available substituted hydrazine NH2NHMe or NH2NHPh, affording the corresponding products 8a (72%),198b (49%) or 9b (34%) directly ((iv), Scheme 1). Low yields with arylhydrazines have been previously reported,26 and similar transformations suggest that the reaction proceeds via initial 1,2-addition followed by cyclisation and not 1,4-conjugate addition and subsequent cyclisation, which could generate different isomers.19,28
Compounds were examined for antiproliferative activity in vitro using the MTS assay29 with human colon carcinoma (HT29) and highly metastatic human breast carcinoma (MDA-MB-231) cell lines. The results are reported as the concentration required to inhibit 50% cell proliferation (IC50) and are shown in Table 1.
| Entry | Comp. | R1 | R2 | Yieldb (%) | IC50a (μM) | |
|---|---|---|---|---|---|---|
| HT29 | MDA-MB-231 | |||||
| a IC50 is the concentration that inhibits 50% cell proliferation. Values are the mean from three independent experiments, except * (one), with quadruplicate readings in each. b Yield over two steps from the chalcone (steps (ii) and (iii), Scheme 1). c Colchicine was used as a positive control. | ||||||
| 1 | 5 | 3,4,5-OMe | — | 85 | 9.6 ± 0.9 | 9.7 ± 4.3 |
| 2 | 8a | H | Me | 72 | 115.8 ± 11.6 | 135 ± 31.2 |
| 3 | 8b | H | Ph | 49 | 53.8 ± 3.94 | 60.2 ± 19.6 |
| 4 | 8c | H | Ac | 72 | 161.3 ± 21.9 | 294.1 ± 71.1 |
| 5 | 8d | H | CF3C=O | 65 | >500 | >500 |
| 6 | 8e | H | NH2C=S | 67 | 350.2 ± 42.0 | 140.5 ± 42.0 |
| 7 | 8f | H | NHMeC=S | 89 | 25.6 ± 4.0 | 20.7 ± 0.79 |
| 8 | 8h | H | Bz | 76 | >500 | >500* |
| 9 | 8i | H | 3,4,5-Trimethoxy-benzoyl | 75 | 2.53 ± 0.39 | 0.69 ± 0.10 |
| 10 | 9b | 3,4,5-OMe | Ph | 34 | 26.2 ± 3.4 | 4.36 ± 0.85 |
| 11 | 9c | 3,4,5-OMe | Ac | 80 | >500 | 52.5 ± 6.5 |
| 12 | 9i | 3,4,5-OMe | 3,4,5-Trimethoxy-benzoyl | 84 | 22.5 ± 2.50 | 11.4 ± 0.75 |
| 13 | Colchicinec | 0.008 ± 0.001 | 0.008 ± 0.001 | |||
The unsubstituted chalcone 4 has been reported to display modest antiproliferative activity in various cancer cell lines.30 Likewise, the 3,4,5-trimethoxy derivative 5 showed good activity in both the cell lines examined in this study (entry 1, Table 1). As can be seen in Table 1, the R2 substituent had a great effect on the antiproliferative activity of the pyrazolines. In the C5-phenyl pyrazoline series 8, the N–Me derivative 8a displayed weak activity, which improved two-fold with the N–Ph analogue 8b (entries 2 and 3). The introduction of an N-acetyl group (8c), which provides additional Lewis basic sites, resulted in partial loss of activity although it did exhibit some selectivity for the HT29 cell line (entry 4). The trifluoroacetyl analogue 8d, which has a similar steric requirement as the acetyl but is a much less effective Lewis base, resulted in complete loss of antiproliferative activity in both cell lines (entry 5).
The carbothioamide 1 shown in Fig. 1 displayed potent antiproliferative activity4 and similar compounds also show promise.5,6 The related derivative 8e was previously reported to exhibit activity as a gold complex with HeLa and A549 cancer cell lines.20 As the free ligand, however, the carbothioamide 8e gave poor inhibition of cell growth in the HT29 and MDA-MB-231 cell lines, while the monomethyl analogue 8f exhibited much improved activities for both cell lines (entries 6 and 7, Table 1).
X-ray crystallography of the carbothioamide 8e indicated a putative intramolecular H-bond to the basic pyrazoline nitrogen (shown in Fig. 2) and an intermolecular H-bond with the pyridine nitrogen (shown in the ESI†). Likewise, the monomethyl derivative 8f could also possess intramolecular co-ordination to the remaining N–H. Although these interactions are present in the solid state, in aqueous solution competing intermolecular H-bonding to the solvent will undoubtedly occur, affording different conformations such as that observed in co-ordination with gold.20 To investigate further we attempted to generate the dimethyl analogue 8g, as this would remove the interaction altogether. Attempts to obtain pyrazoline 8g either by methylation of the carbothioamide 8e or 8f, or by reaction of the N–H pyrazoline 6 with dimethylthiocarbamoyl chloride, were however unsuccessful as previously observed in similar substrates.26
![X-ray crystal structures of pyrazolines 8e and 8i with ellipsoids represented at 30% probability.Both pyrazolines 8e and 8i were racemic, but single compounds are shown to represent the structures obtained by X-ray crystallography. Crystal data for pyrazoline 8e, CCDC 926530. Formula: C15H14N4S1. M = 282.36, monoclinic. Unit cell parameters: a = 9.7950(2) Å, b = 14.7280(3) Å, c = 10.0360(2) Å. α = 90°, β = 107.768(1)°, γ = 90°, V = 1378.74(5) Å3, T = 150(2) K, space group P21/n, Z = 4, 24456 reflections collected, 3152 independent reflections [R(int) = 0.0669]. Final R indices [I > 2σ(I)] R1 = 0.0392 and wR2 = 0.0883. R indices (all data) R1 = 0.0608 and wR2 = 0.0979. Crystal data for pyrazoline 8i, CCDC 926531. Formula: C24H23N3O4. M = 417.45, orthorhombic. Unit cell parameters: a = 6.9770(1) Å, b = 22.0950(2) Å, c = 26.6010(3) Å. α = 90°, β = 90°, γ = 90°, V = 4100.73(8) Å3, T = 150(2) K, space group Pbca, Z = 8, 56599 reflections collected, 4679 independent reflections [R(int) = 0.0645]. Final R indices [I > 2σ(I)] R1 = 0.0422 and wR2 = 0.0882. R indices (all data) R1 = 0.0649 and wR2 = 0.0991.](/image/article/2013/MD/c3md00077j/c3md00077j-f2.gif) | ||
| Fig. 2 X-ray crystal structures of pyrazolines 8e and 8i with ellipsoids represented at 30% probability.‡ | ||
The effect of an N-benzoyl substituent (8h) was also investigated and exhibited no observable antiproliferative activity in the two cell lines examined (entry 8, Table 1). Following our previous studies using 3,4,5-trimethoxyphenyl substituents,31 which have been shown to play crucial roles in activity,32–36 we investigated this substitution pattern in the N-benzoyl group. Remarkably, the corresponding 3,4,5-trimethoxybenzoyl derivative 8i displayed potent activity and selectivity for the highly metastatic human breast carcinoma (MDA-MB-231, entry 9). The X-ray crystal structure of pyrazoline 8i was also obtained and is shown in Fig. 2. As with pyrazoline 8e, it shows that in the solid state the amide adopted the E-conformation.
Continuing our interest in the 3,4,5-trimethoxyphenyl motif, we investigated the effect of this substitution pattern in three examples in the pyrazoline series 9. In all three examples, the compounds exhibited increased potency for the MDA-MB-231 cell line, as previously observed with derivative 8i which also possesses the 3,4,5-trimethoxyaryl motif. The N–Ph derivative 9b was more active in both cell lines than the corresponding C5-phenyl analogue 8b (entries 10 and 3, Table 1). Surprisingly, the N–Ac derivative 9c showed no activity in HT29 (>500 μM), yet exhibited improved activity and selectivity for MDA-MB-231 (52.5 μM) compared to pyrazoline 8c (entries 11 and 4). Incorporating the 3,4,5-trimethoxy-substitution in both the C5-aryl and N1-benzoyl positions of the pyrazoline ring (9i) did not further improve activity compared to compound 8i (entries 12 and 9). On the basis of these preliminary investigations, compound 8i was selected for further evaluation by the National Cancer Institute (NCI) and the results are shown in Table 2.
| Panel | Cell line | GI50a (μM) |
|---|---|---|
| a GI50 is concentration required to inhibit growth by 50% as defined by the National Cancer Institute (NCI). | ||
| Leukemia | CCRF-CEM | 2.08 |
| HL-60(TB) | 0.747 | |
| MOLT-4 | 4.46 | |
| RPMI-8226 | 66.3 | |
| SR | 0.415 | |
| Non-small cell | A549/ATCC | 0.952 |
| Lung | EKVX | >100 |
| HOP-62 | 1.82 | |
| HOP-92 | >100 | |
| NCI-H226 | 1.33 | |
| NCI-H23 | 3.45 | |
| NCI-H322M | >100 | |
| NCI-H460 | 0.505 | |
| NCI-H522 | 0.688 | |
| Colon | COLO 205 | 0.585 |
| HCC-2998 | 2.32 | |
| HCT-116 | 0.586 | |
| HCT-15 | 0.848 | |
| HT29 | 0.431 | |
| KM12 | 0.711 | |
| SW-620 | 0.567 | |
| CNS | SF-268 | 33.4 |
| SF-539 | 1.06 | |
| SNB-19 | 1.89 | |
| SNB-75 | 0.277 | |
| U251 | 0.868 | |
| Melanoma | LOX IMVI | 1.13 |
| MALME-3M | >100 | |
| M14 | 0.801 | |
| MDA-MB-435 | 0.273 | |
| SK-MEL-2 | 0.699 | |
| SK-MEL-28 | 1.51 | |
| SK-MEL-5 | 0.432 | |
| UACC-257 | >100 | |
| UACC-62 | 0.541 | |
| Ovarian | IGROV1 | 7.13 |
| OVCAR-3 | 0.719 | |
| OVCAR-4 | 5.95 | |
| OVCAR-5 | 4.01 | |
| OVCAR-8 | 3.28 | |
| NCI/ADR-RES | 0.519 | |
| SK-OV-3 | 0.762 | |
| Renal | 786-0 | 1.54 |
| A498 | 0.943 | |
| ACHN | 4.29 | |
| CAKI-1 | 0.493 | |
| RXF 393 | 1.03 | |
| SN12C | 3.53 | |
| TK-10 | >100 | |
| UO-31 | 5.01 | |
| Prostate | PC-3 | >100 |
| DU-145 | 4.26 | |
| Breast | MCF7 | 0.324 |
| MDA-MB-231/ATCC | 0.989 | |
| HS 578T | 9.18 | |
| BT-549 | 5.13 | |
| T-47D | >100 | |
| MDA-MB-468 | 0.417 | |
Pyrazoline 8i (NSC 761258) displayed promising GI50 values across the NCI 60 human tumour cell line panel and exhibited sub-micromolar activity in a range of cancer cell lines, including the multidrug resistant ovarian cell line NCI/ADR-RES (0.519 μM). Also of interest is that one of the most susceptible ovarian cancer cell lines, OVCAR-3, is particularly sensitive to tubulin disruptors,37 suggesting that pyrazoline 8i may be inhibiting cell proliferation through the disruption of tubulin. Although COMPARE analysis of the NCI 60 cancer cell line data did not afford strong correlations with compounds of known biological activity, drugs which inhibit the polymerization of tubulin did feature (see ESI†).
To investigate potential interaction with tubulin dynamics, cell cycle analysis was performed with HT29 cells with pyrazoline 8i and colchicine, a known inhibitor of microtubule polymerisation. Cell cycle analysis was performed three times and the results shown in Fig. 3 are representative of the complete study (shown in the ESI†). The presence of 100 nM of colchicine led to cell cycle arrest in the G2/M phase, shown in Fig. 3B. In the presence of pyrazoline 8i at 1 μM (Fig. 3C) and 5 μM (Fig. 3D), 50% and 95% respectively of the cells also arrested in G2/M phase, suggesting that pyrazoline 8i affects microtubule polymerisation.
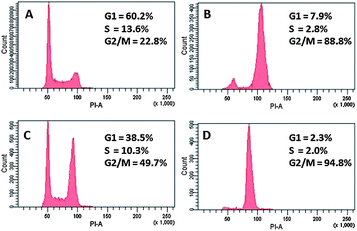 | ||
| Fig. 3 Cell cycle analysis. (A) HT29 cells only, (B) HT29 + colchicine (100 nM), (C) HT29 + 8i (1 μM), and (D) HT29 + 8i (5 μM). | ||
To examine this interaction an in vitro tubulin polymerisation assay was performed in the presence of pyrazoline 8i (20 μM) or paclitaxel (5 μM), which is known to polymerise microtubule formation. As can be seen in Fig. 4, the presence of paclitaxel led to an increased optical density (OD) compared to the control experiment, due to increased microtubule formation. The presence of pyrazoline 8i however gave a lower OD reading than the control, implying that pyrazoline 8i inhibits microtubule polymerisation and is consistent with the cell cycle analysis.
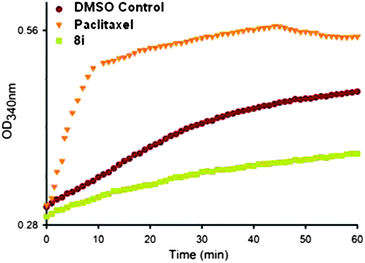 | ||
| Fig. 4 In vitro tubulin polymerisation assay. Paclitaxel (5 μM) and pyrazoline 8i (20 μM). | ||
Confocal microscopy was performed on HT29 cells to visualise the effects of pyrazoline 8i in microtubule formation, using Dm1a tubulin antibodies (Fig. 5). The effects of 100 nM colchicine, which inhibits microtubule polymerisation, can be seen in Fig. 5B, which contains much less tubulin compared to the control experiment (Fig. 5A). Likewise, compared to the control there is less tubulin present in cells treated with 5 μM pyrazoline 8i (Fig. 5C), which again supports the data obtained from the cell cycle analysis and in vitro tubulin polymerisation assay that pyrazoline 8i disrupts microtubule formation. The results are consistent with previous reports of similar compounds which were also shown to interfere with microtubule assembly.18
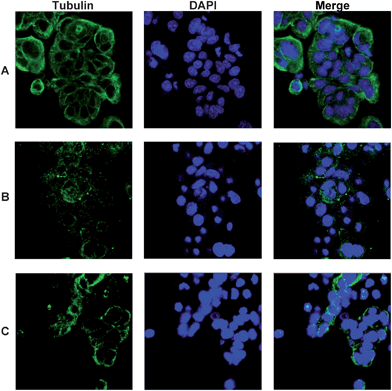 | ||
| Fig. 5 Confocal microscopy showing tubulin, Dm1a (green) and DNA, DAPI (blue). (A) HT29 cells only, (B) HT29 + colchicine (100 nM) and (C) HT29 + 8i (5 μM). | ||
Conclusions
In conclusion, we report the two-step synthesis of some 3-(pyrid-2-yl)-pyrazolines and their antiproliferative activities. The lead pyrazoline 8i was selected from this preliminary study and further investigated against the NCI 60 cancer cell line (NSC 761258) and exhibits sub-micromolar activity in a number of cell lines. The mode of action was explored by cell cycle analysis, in vitro microtubule assembly assay and confocal microscopy with α-tubulin antibody Dm1a which suggest that the novel compound 8i inhibits microtubule polymerisation.The promising antiproliferative activity of 3-(pyrid-2-yl)-pyrazoline 8i and the ease of its synthesis provide the potential for further development, which is currently under investigation and will be reported in due course.
Acknowledgements
We wish to thank Dr Timothy J. Woodman, Dr Anneke Lubben and Dr Adrian Rogers (University of Bath) for their assistance with the NMR, mass spectra and confocal spectroscopy, respectively. We also wish to thank Dr Andrew Chalmers for the antibodies used in the confocal experiments and to Cancer Research at Bath (CR@B) for funding the confocal imaging. We are extremely grateful to the National Cancer Institute (NCI) for conducting the in vitro testing and the University of Bath for providing a studentship for AC. We also wish to acknowledge RCUK and the University of Bath for the fellowship to LC.Notes and references
- M. Abdel-Aziz and A. M. Gamal-Eldeen, Pharm. Biol., 2009, 47, 854–863 CrossRef CAS.
- M. R. Shaaban, A. S. Mayhoub and A. M. Farag, Expert Opin. Ther. Pat., 2012, 22, 253–291 CrossRef CAS.
- S. A. F. Rostom, M. H. Badr, H. A. Abd El Razik, H. M. A. Ashour and A. E. Abdel Wahab, Arch. Pharm. Chem. Life Sci., 2011, 344, 572–587 CrossRef CAS.
- P.-C. Lv, H.-Q. Li, J. Sun, Y. Zhou and H.-L. Zhu, Bioorg. Med. Chem., 2010, 18, 4606–4614 CrossRef CAS.
- B. Insuasty, L. Chamizo, J. Muñoz, A. Tigreros, J. Quiroga, R. Abonía, M. Nogueras and J. Cobo, Arch. Pharm. Chem. Life Sci., 2012, 345, 275–286 CrossRef CAS.
- A.-S. S. Hamad Elgazwy, D. H. S. Soliman, S. R. Atta-Allah and D. A. Ibrahim, Chem. Cent. J., 2012, 6, 50 CrossRef CAS.
- B. N. Acharya, D. Saraswat, M. Tiwari, A. K. Shrivastava, R. Ghorpade, S. Bapna and M. P. Kaushik, Eur. J. Med. Chem., 2010, 45, 430–438 CrossRef CAS.
- P. M. Sivakumar, S. Ganesan, P. Veluchamy and M. Doble, Chem. Biol. Drug Des., 2010, 76, 407–411 CAS.
- K. Nepali, G. Singh, A. Turan, A. Agarwal, S. Sapra, R. Kumar, U. C. Banerjee, P. K. Verma, N. K. Satti, M. K. Gupta, O. P. Suri and K. L. Dhar, Bioorg. Med. Chem., 2011, 19, 1950–1958 CrossRef CAS.
- R. Fioravanti, A. Bolasco, F. Manna, F. Rossi, F. Orallo, F. Ortuso, S. Alcaro and R. Cirilli, Eur. J. Med. Chem., 2010, 45, 6135–6138 CrossRef CAS.
- R. Cirilli, R. Ferretti, B. Gallinella, L. Turchetto, A. Bolasco, D. Secci, P. Chimenti, M. Pierini, V. Fares, O. Befani and F. La Torre, Chirality, 2004, 16, 625–636 CrossRef CAS.
- N. Gökhan-Kelekçi, S. Yabanoğlu, E. Küpeli, U. Salgin, Ö. Özgen, G. Uçar, E. Yeşilada, E. Kendi, A. Yeşilada and A. A. Bilgin, Bioorg. Med. Chem., 2007, 15, 5775–5786 CrossRef.
- A. Sahoo, S. Yabanoglu, B. N. Sinha, G. Ucar, A. Basu and V. Jayaprakash, Bioorg. Med. Chem. Lett., 2010, 20, 132–136 CrossRef CAS.
- F. Chimenti, A. Bolasco, F. Manna, D. Secci, P. Chimenti, O. Befani, P. Turini, V. Giovannini, B. Mondovì, R. Cirilli and F. La Torre, J. Med. Chem., 2004, 47, 2071–2074 CrossRef CAS.
- F. Chimenti, E. Maccioni, D. Secci, A. Bolasco, P. Chimenti, A. Granese, O. Befani, P. Turini, S. Alcaro, F. Ortuso, R. Cirilli, F. La Torre, M. C. Cardia and S. Distinto, J. Med. Chem., 2005, 48, 7113–7122 CrossRef CAS.
- F. Chimenti, A. Bolasco, F. Manna, D. Secci, P. Chimenti, A. Granese, O. Befani, P. Turini, S. Alcaro and F. Ortuso, Chem. Biol. Drug Des., 2006, 67, 206–214 CAS.
- R. Fioravanti, A. Bolasco, F. Manna, F. Rossi, F. Orallo, M. Yáñez, A. Vitali, F. Ortuso and S. Alcaro, Bioorg. Med. Chem. Lett., 2010, 20, 6479–6482 CrossRef CAS.
- M. Abdel-Aziz, O. M. Aly, S. S. Khan, K. Mukherjee and S. Bane, Arch. Pharm. Chem. Life Sci., 2012, 345, 535–548 CrossRef CAS.
- A. Ciupa, M. F. Mahon, P. A. De Bank and L. Caggiano, Org. Biomol. Chem., 2012, 10, 8753–8757 CAS.
- S. Wang, W. Shao, H. Li, C. Liu, K. Wang and J. Zhang, Eur. J. Med. Chem., 2011, 46, 1914–1918 CrossRef CAS.
- S. G. Kini, A. R. Bhat, B. Bryant, J. S. Williamson and F. E. Dayan, Eur. J. Med. Chem., 2009, 44, 492–500 CrossRef CAS.
- M. G. Mamolo, D. Zampieri, V. Falagiani, L. Vio and E. Banfi, Il Farmaco, 2001, 56, 593–599 CrossRef CAS.
- M. G. Mamolo, D. Zampieri, V. Falagiani, L. Vio and E. Banfi, Il Farmaco, 2003, 58, 315–322 CrossRef CAS.
- S. M. Sondhi, S. Kumar, N. Kumar and P. Roy, Med. Chem. Res., 2012, 21, 3043–3052 CrossRef CAS.
- B. P. Bandgar, L. K. Adsul, H. V. Chavan, S. S. Jalde, S. N. Shringare, R. Shaikh, R. J. Meshram, R. N. Gacche and V. Masand, Bioorg. Med. Chem. Lett., 2012, 22, 5839–5844 CrossRef CAS.
- C. D. Cox, M. J. Breslin and B. J. Mariano, Tetrahedron Lett., 2004, 45, 1489–1493 CrossRef CAS.
- A. L. Baumstark, M. Dotrong and P. C. Vasquez, Tetrahedron Lett., 1987, 28, 1963–1966 CrossRef CAS.
- Z.-L. Gong, L.-W. Zheng, B.-X. Zhao, D.-Z. Yang, H.-S. Lv, W.-Y. Liu and S. Lian, J. Photochem. Photobiol., A, 2010, 209, 49–55 CrossRef CAS.
- A. H. Cory, T. C. Owen, J. A. Barltrop and J. G. Cory, Cancer Commun., 1991, 3, 207–212 CAS . MTS is 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
- L. Mishra, R. Sinha, H. Itokawa, K. F. Bastow, Y. Tachibana, Y. Nakanishi, N. Kilgore and K.-H. Lee, Bioorg. Med. Chem., 2001, 9, 1667–1671 CrossRef CAS.
- A. Ciupa, N. J. Griffiths, S. K. Light, P. J. Wood and L. Caggiano, Med. Chem. Commun., 2011, 2, 1011–1015 RSC.
- K. Gaukroger, J. A. Hadfield, N. J. Lawrence, S. Nolan and A. T. McGown, Org. Biomol. Chem., 2003, 1, 3033–3037 CAS.
- T. M. Beale, R. M. Myers, J. W. Shearman, D. S. Charnock-Jones, J. D. Brenton, F. V. Gergely and S. V. Ley, Med. Chem. Commun., 2010, 1, 202–208 RSC.
- A. Kamal, Y. V. V. Srikanth, T. B. Shaik, M. N. A. Khan, M. Ashraf, M. K. Reddy, K. A. Kumar and S. V. Kalivendi, Med. Chem. Commun., 2011, 2, 819–823 RSC.
- A. Kamal, J. S. Reddy, M. J. Ramaiah, D. Dastagiri, E. V. Bharathi, M. V. P. Sagar, S. N. C. V. L. Pushpavalli, P. Ray and M. Pal-Bhadra, Med. Chem. Commun., 2010, 1, 355–360 RSC.
- A. Kamal, A. Viswanath, M. J. Ramaiah, J. N. S. R. C. Murty, F. Sultana, G. Ramakrishna, J. R. Tamboli, S. N. C. V. L. Pushpavalli, D. Pal, C. Kishor, A. Addlagatta and M. P. Bhadra, Med. Chem. Commun., 2012, 3, 1386–1392 RSC.
- R. H. Shoemaker, Nat. Rev. Cancer, 2006, 6, 813–823 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) is available: Experimental procedures, characterisation data, 1H and 13C NMR spectra, HPLC traces and MTS assays for all compounds. In addition, NCI data and cell cycle analysis of compound 8i together with X-ray crystallography data for pyrazolines 8e and 8i are provided. CCDC 926530 and 926531. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c3md00077j |
| ‡ Both pyrazolines 8e and 8i were racemic, but single compounds are shown to represent the structures obtained by X-ray crystallography. Crystal data for pyrazoline 8e, CCDC 926530. Formula: C15H14N4S1. M = 282.36, monoclinic. Unit cell parameters: a = 9.7950(2) Å, b = 14.7280(3) Å, c = 10.0360(2) Å. α = 90°, β = 107.768(1)°, γ = 90°, V = 1378.74(5) Å3, T = 150(2) K, space group P21/n, Z = 4, 24456 reflections collected, 3152 independent reflections [R(int) = 0.0669]. Final R indices [I > 2σ(I)] R1 = 0.0392 and wR2 = 0.0883. R indices (all data) R1 = 0.0608 and wR2 = 0.0979. Crystal data for pyrazoline 8i, CCDC 926531. Formula: C24H23N3O4. M = 417.45, orthorhombic. Unit cell parameters: a = 6.9770(1) Å, b = 22.0950(2) Å, c = 26.6010(3) Å. α = 90°, β = 90°, γ = 90°, V = 4100.73(8) Å3, T = 150(2) K, space group Pbca, Z = 8, 56599 reflections collected, 4679 independent reflections [R(int) = 0.0645]. Final R indices [I > 2σ(I)] R1 = 0.0422 and wR2 = 0.0882. R indices (all data) R1 = 0.0649 and wR2 = 0.0991. |
| This journal is © The Royal Society of Chemistry 2013 |
