Interaction of long telomeric DNAs with macrocyclic hexaoxazole as a G-quadruplex ligand†
Keisuke
Iida
a,
Gen
Tsubouchi
a,
Takahiro
Nakamura
a,
Satoki
Majima
a,
Hiroyuki
Seimiya
b and
Kazuo
Nagasawa
*a
aFaculty of Technology, Tokyo University of Agriculture and Technology (TUAT), 2-24-16 Naka-machi, Koganei-shi, Tokyo 185-0031, Japan. E-mail: knaga@cc.tuat.ac.jp; Fax: +81-42-388-7295; Tel: +81-42-388-7295
bCancer Chemotherapy Center, Japanese Foundation for Cancer Research (JFCR), 3-8-31 Ariake, Koto-ku, Tokyo 135-8550, Japan
First published on 7th November 2012
Abstract
The interactions of long telomeric DNAs, which mimic telomeres in living cells, with a macrocyclic hexaoxazole ligand L2H2-6OTD (2) were investigated by means of electrophoresis mobility shift assay, circular dichroism (CD) titration analysis, and DNA melting measurements. Our results indicate that G4 structure in long telomeric DNAs was mainly induced to take anti-parallel topology upon interaction with L2H2-6OTD (2). The overall higher-order G4 organizational structure of the long telomeric DNAs was suggested to be of beads-on-a-string type.
Introduction
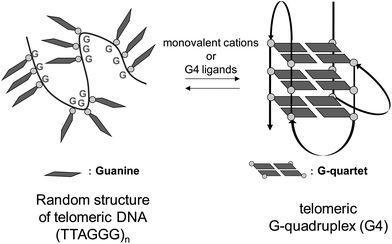
The ends of mammalian chromosomes carry specific chromatin structures called telomeres, which serve to protect chromosomal termini from inappropriate recombination, end-to-end fusion, and nuclease attack.1 Human telomeres have two general components, that is, the shelterin complex and telomeric DNA. The shelterin complex, which protects the telomeric DNA, consists of six proteins: TRF1, TRF2, POT1, TPP1, RAP1 and TIN2.2 The telomeric DNA contains repeating sequences of TTAGGG, with a single-stranded region of approximately 150 bases, the G-overhang, at its 3′ terminal. This guanine-rich single strand is believed to fold into characteristic three-dimensional structures called the G-quadruplex (G4) in living systems.3 G4 is formed by stacking of the G-quartet, a planar structure constructed from four guanines (Fig. 1). The G4 structure exists in equilibrium with a random structure, and it is stabilized by K+ under physiological conditions.Telomeric G4 structures are stabilized by monovalent cations and small molecules in vitro. Stabilization of telomeric G4 structures in cancer cells is reported to inhibit the activity of telomerase,4 an enzyme that is selectively expressed in most tumor cells.5 Since telomerase plays key roles in telomere maintenance and cellular immortalization, its inhibition leads to senescence and apoptosis of cancer cells.6 Therefore, telomeric G4 is regarded as a molecular target for cancer chemotherapy, and many small molecules that stabilize the G4 structure have been reported so far as candidate anti-cancer agents.7 In most studies to explore ligands that stabilize G4 structure, telo24 DNA, i.e., four sets of (TTAGGG) repeats comprising 24 bases in total, has generally been used as a telomere model. However, around 150 bases are involved in the case of telomeric DNA in vivo. Thus, it is important to examine the stabilization ability and binding mode of G4 ligands with long telomeric DNA sequences, which better mimic those in living cells.8 In the case of long telomeric DNAs in the presence of a G4 ligand, two points should be considered, i.e., (i) the topology of each G4 unit (Fig. 2) in the DNA and (ii) the overall higher-order G4 organizational structure (Fig. 3). Only limited studies have been reported on the interaction of long telomeric DNA with a G4 ligand.9 The topology of G4 units in long telomeric DNA was elucidated, but little is known about the overall higher-order organizational structures induced by G4 ligands.
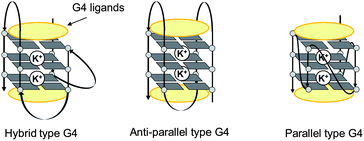 | ||
| Fig. 2 Schematic illustrations of representative G4 topologies. | ||
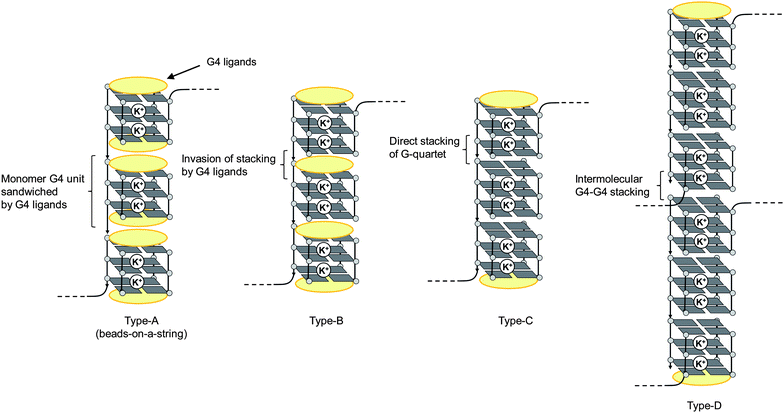 | ||
| Fig. 3 Schematic illustrations of some possible intramolecular (types A–C) and intermolecular (type D) higher-order G4 structures of long telomeric DNA. | ||
Recently, macrocyclic hexaoxazole compounds have been reported as G4 ligands, inspired by the natural G4 ligand telomestatin (1).10 A series of related compounds were reported by Rice and co-workers.11a–h We have also independently developed a similar series of macrocyclic hexaoxazole compounds almost at the same time, and we named the core skeleton 6OTD (Fig. 4a).11i–r,12 The 6OTDs interact with telo24 to form an anti-parallel-type G4 structure through an end-stacking mode with two molecules,11j,ni.e., interaction with the terminal G-quartet flat surfaces in telo24 via π–π stacking (Fig. 4b), which is similar to the mode of interaction of telomestatin (1) with telo24.10c–e However, the interaction modes of these G4 ligands with telomeric DNA in “cells” have not been addressed. In the present work, we examined the interaction of L2H2-6OTD (2) with long telomeric DNAs and evaluated the topologies of individual G4 units by means of electrophoresis mobility shift assay (EMSA) and circular dichroism (CD) titration analysis. The overall higher-order G4 organizational structures are also discussed on the basis of the results of CD titration and melting assay.
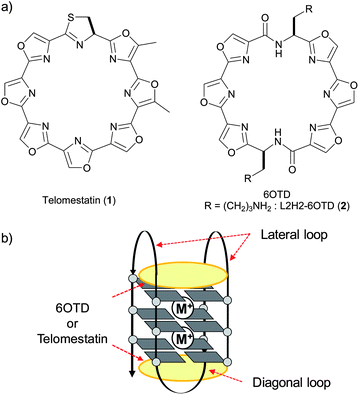 | ||
| Fig. 4 (a) Structures of telomestatin (1) and 6OTD; (b) their interaction modes with G4. | ||
Results and discussion
We prepared long telomeric DNAs consisting of 48, 72, and 96 bases, i.e., (TTAGGG)n (n = 8, 12, 16; telo48, telo72, and telo96, corresponding to two, three and four units of telo24, respectively).13 L2H2-6OTD (2),11c,j,12 which was one of the most potent telo24 G4-stabilizers among the 6OTD derivatives, was used for the present study (Fig. 4).First, EMSA was used to evaluate the complexation of L2H2-6OTD (2) with a variety of lengths of telomeric DNAs of telo24, telo48, telo72 and telo96. The concentration of G4 ligand 2 was systematically varied for each length of telomeric DNA, and electrophoresis was carried out (Fig. 5). Under these conditions, the original bands (without ligand) disappeared, and new bands with longer migration distances were observed in proportion to the concentration of ligand 2 in all cases. Moreover, these bands converged to one band in the presence of an excess of ligand 2. These observations indicate that L2H2-6OTD (2) binds to the DNAs to form “single” monomeric complexes in the presence of sufficient amounts of the ligand. We next examined the topologies of DNAs in the presence of L2H2-6OTD (2) by means of CD titration experiments.
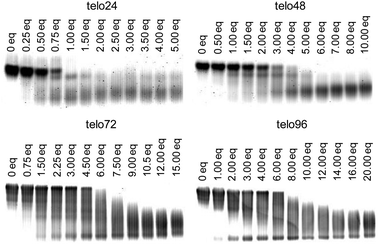 | ||
| Fig. 5 Evaluation of the interaction of telomeric DNAs (telo24, telo48, telo72, and telo96) with L2H2-6OTD (2). | ||
Telomeric DNAs are known to form a variety of topologies,14 including parallel, anti-parallel, and hybrid types, and these are usually present as a mixture or in equilibrium, depending upon the sequences and conditions. In the presence of high concentration of potassium cations, a hybrid-type structure is mainly observed, showing typical positive Cotton effects at 295 and 265 nm, and a negative Cotton effect at 240 nm in the CD spectrum. In the case of L2H2-6OTD (2) it induced a change of topology of telo24 from hybrid-type to anti-parallel form in the presence of KCl (Fig. 6a). Thus, we next titrated each length of telomeric DNA, i.e., telo48, telo72, and telo96, with G4 ligand 2 in the presence of KCl (100 mM), and the results (including telo24) are summarized in Fig. 6. In all cases, typical spectral changes from hybrid-type8a,g,h to anti-parallel form were observed during the titration, i.e., increments of positive Cotton effects at 230–240 nm and 280 nm and the negative effect at 255 nm were observed. At the end point of saturation, similar CD intensities (y-axis in Fig. 6) were obtained with each length of DNA. The concentrations of the DNAs used in these titration experiments were 10 μM for telo24, 5 μM for telo48 (half the concentration of telo24), 3.3 μM for telo72 (one-third of telo24), and 2.5 μM for telo96 (a quarter of telo24). Thus, the generation of two, three, and four times as many G4 structures, compared with the case of telo24, was suggested to occur with telo48, telo72 and telo96, respectively.8g,h
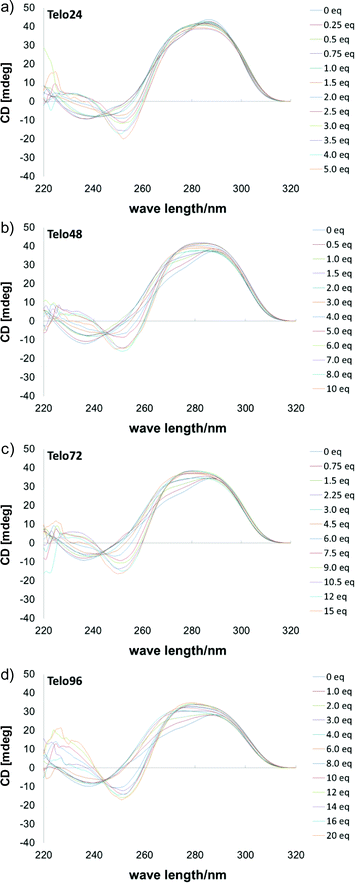 | ||
| Fig. 6 CD spectra of telomeric DNAs in the presence of KCl (100 mM) upon titration with 2, (a) telo24 (10 μM) + 2 (0–5 eq.), (b) telo48 (5 μM) + 2 (0–10 eq.), (c) telo72 (3.3 μM) + 2 (0–15 eq.) and (d) telo96 (2.5 μM) + 2 (0–20 eq.). | ||
That is, the number of units of anti-parallel G4 induced by 2 increased according to the length of telomeric DNA of telo24–telo96. In the case of anti-parallel type G4 topology, it has loops with top and bottom faces (Fig. 4b), and these loops prevent intramolecular G4–G4 interactions because of their steric hindrance. Thus, in the case of consecutive G4 units with anti-parallel topology, each G4 unit exists separately from others. Therefore, the higher-order G4 organizational structure of longer telomeric DNAs of telo48, 72 and 96 was suggested to be of the beads-on-a-string type (type-A in Fig. 3).8
Then, DNA melting experiments were carried out to evaluate the stabilization of long telomeric DNAs in the presence of L2H2-6OTD (2). Each size of telomeric DNA was treated with sufficient amounts of 2,15 and the Tm (melting temperature) values of the complexes were determined (Table 1). The Tm values for telo48, 72, and 96 were found to be 76, 73, and 70 °C, respectively, which are similar to the Tm value of 75 °C obtained for telo24, and this supports the consecutive formation of G4 units in long telomeric DNA in the presence of ligand 2. Since the G4 structure in telo24 interacts with two molecules of L2H2-6OTD (2),16 the longer sizes of telomeric DNAs may also interact with 2 in a similar manner to telo24, as shown in Fig. 7.
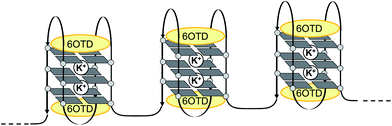 | ||
| Fig. 7 Schematic model of interactions of a L2H2-6OTD (2) with long telomeric DNA. | ||
Conclusions
In summary, complexation of long telomeric DNAs with G4 ligand of L2H2-6OTD (2) was investigated from the viewpoint of G4 topology and overall higher-order organizational structure. The G4 structure in long telomeric DNAs was mainly induced to take anti-parallel topology by interaction with L2H2-6OTD (2). The overall higher-order G4 organizational structure of the long telomeric DNAs was also suggested to be of the beads-on-a-string type (type A in Fig. 3). Thus, the longer DNAs were stabilized in a similar manner to telo24 by G4 ligand 2.Experimental
CD spectrometry
CD spectra were recorded on a J-720 spectropolarimeter (JACSO, Tokyo, Japan) using a quartz cell of 1 mm optical path length and an instrument scanning speed of 500 nm min−1 with a response time of 1 s, over a wavelength range of 220–320 nm. The purified nucleotides (Sigma Genosys) were dissolved as 1.0 mM stock solutions in sterilized water to be used without further purification. These nucleotides were diluted to 10 μM (telo24), 5.0 μM (telo48), 3.3 μM (telo72) and 2.5 μM (telo96) with 50 mM Tris–HCl, pH 7.5, and 100 mM KCl buffer, respectively. Subsequently, these solutions were annealed by heating at 96 °C for 2 min, then slowly cooled to room temperature, and then corresponding concentrations of 2 were titrated with these DNA solutions. Finally the CD spectra are representative of ten scans taken at 25 °C.DNA melting assay
A solution of all DNA (10 μM: telo24, 5.0 μM: telo48, 3.3 μM: telo72, 2.5 μM: telo96) was prepared in 50 mM Tris–HCl, pH 7.5, 100 mM KCl buffer. Subsequently, corresponding concentrations of 2 were added. Melting and annealing curves were obtained by monitoring the CD intensity at 295 nm on a J-720 spectropolarimeter (JASCO, Tokyo, Japan) by using a quartz cell of 1 mm optical path length; the temperature was changed as follows: 4 to 99 °C then 99 to 4 °C at 1.0 °C min−1.Electrophoresis mobility shift assay (EMSA)
EMSA was performed as follows; 2.0 μL of oligonucleotides (50 μM of telo24, 25 μM of telo48, 16.7 μM of telo72 and 12.5 μM of telo96) in TE buffer was added to 5.0 μL of Tris–HCl buffer (50 mM, pH 7.1) with 250 mM of KCl. This solution was heated at 94 °C for 2 min and then slowly cooled to 25 °C. The corresponding concentration of 2 in DMSO was added to the DNA solution. After a 14 h incubation period, the samples were mixed with 2.0 μL of Ficoll 400 solution (100 mg mL−1). Next, 0.5 μL of each mixture was run on 12% nondenatured polyacrylamide gels in 1 × TBE buffer (100 V for 10 min, then 200 V for 15 min for telo24, 30 min for telo48, 45 min for telo72 and 60 min for telo96, respectively), and stained with GelStar®. The gels were scanned with a phosphorimager (Typhoon 8600, GE Healthcare) using a 580–640 band pass filter.Acknowledgements
This study was supported in part by a Grant-in-Aid for Scientific Research (B).Notes and references
- T. de Lange, Genes Dev., 2005, 19, 2100 CrossRef CAS.
- M. A. Blasco, Nat. Chem. Biol., 2007, 3, 640 CrossRef CAS.
- A. T. Phan, FEBS J., 2010, 277, 1107 CrossRef CAS.
- A. M. Zahler, J. R. Williamson, T. R. Cech and D. M. Prescott, Nature, 1991, 350, 718 CrossRef CAS.
- (a) N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich and J. W. Shay, Science, 1994, 266, 2011 CAS; (b) J. W. Shay and W. E. Wright, Nat. Rev. Drug Discovery, 2006, 5, 577 CrossRef CAS.
- S. A. Stewart and R. A. Weinberg, Annu. Rev. Cell Dev. Biol., 2006, 22, 531 CrossRef CAS.
- S. Neidle, FEBS J., 2010, 277, 1118 CrossRef CAS.
- For structural analysis of long telomeric DNAs see: (a) Y. Xu, T. Ishizuka, K. Kurabayashi and M. Komiyama, Angew. Chem., Int. Ed., 2009, 48, 7833 CrossRef CAS; (b) L. Petraccone, J. O. Trent and J. B. Chaires, J. Am. Chem. Soc., 2008, 130, 16530 CrossRef CAS; (c) D. Renciuk, I. Kejnovska, P. Skolakova, K. Bednarova, J. Motlova and M. Vorlıckova, Nucleic Acids Res., 2009, 37, 6625 CrossRef CAS; (d) M. Vorlıckova, J. Chladkova, I. Kejnovska, M. Fialova and J. Kypr, Nucleic Acids Res., 2005, 33, 5851 CrossRef; (e) V. Singh, M. Azarkh, M. Drescher and J. S. Hartig, Chem. Commun., 2012, 48, 8258 RSC; (f) L. Petraccone, C. Spink, J. O. Trent, N. C. Garbett, C. S. Mekmaysy, C. Giancola and J. B. Chaires, J. Am. Chem. Soc., 2011, 133, 20951 CrossRef CAS; (g) H.-Q. Yu, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 15461 CrossRef CAS; (h) H. Yu, X. Gu, S. Nakano, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2012 DOI:10.1021/ja305384c.
- C.-C. Chang, C.-W. Chien, Y.-H. Lin, C.-C. Kang and T.-C. Chang, Nucleic Acids Res., 2007, 35, 2846 CrossRef CAS.
- For isolation of telomestatin, see: (a) K. Shin-ya, K. Wierzba, K. Matsuo, T. Ohtani, Y. Yamada, K. Furihata, Y. Hayakawa and H. Seto, J. Am. Chem. Soc., 2001, 123, 1262 CrossRef CAS; Selected reports of telomestatin as G4 ligands, see: (b) P. Shirude, E. Gillies, S. Ladame, F. Godde, K. Shin-Ya, I. Huc and S. Balasubramanian, J. Am. Chem. Soc., 2007, 129, 11890 CrossRef CAS; (c) M.-Y. Kim, H. Vankayalapati, K. Shin-ya, K. Wierzba and L. H. Hurley, J. Am. Chem. Soc., 2002, 124, 2098 CrossRef CAS; (d) F. Rosu, V. Gabelica, K. Shin-ya and E. D. Pauw, Chem. Commun., 2003, 2702 RSC; (e) E. M. Rezler, J. Seenisamy, S. Bashyam, M.-Y. Kim, E. White, W. D. Wilson and L. H. Hurley, J. Am. Chem. Soc., 2005, 127, 9439 CrossRef CAS.
- (a) G. S. Minhas, D. S. Pilch, J. E. Kerrigan, E. J. LaVoie and J. E. Rice, Bioorg. Med. Chem. Lett., 2006, 16, 3891 CrossRef CAS; (b) C. M. Barbieri, A. R. Srinivasan, S. G. Rzuczek, J. E. Rice, E. J. LaVoie and D. S. Pilch, Nucleic Acids Res., 2007, 35, 3272 CrossRef CAS; (c) S. G. Rzuczek, D. S. Pilch, E. J. LaVoie and J. E. Rice, Bioorg. Med. Chem. Lett., 2008, 18, 913 CrossRef CAS; (d) M. Satyanarayana, S. G. Rzuczek, E. J. LaVoie, D. S. Pilch, A. Liu, L. F. Liu and J. E. Rice, Bioorg. Med. Chem. Lett., 2008, 18, 3802 CrossRef CAS; (e) D. S. Pilch, C. M. Barbieri, S. G. Rzuczek, E. J. LaVoie and J. E. Rice, Biochimie, 2008, 90, 1233 CrossRef CAS; (f) Y.-C. Tsai, H. Qi, C.-P. Lin, R.-K. Lin, J. E. Kerrigan, S. G. Rzuczek, E. J. LaVoie, J. E. Rice, D. S. Pilch, Y. L. Lyu and L. F. Liu, J. Biol. Chem., 2009, 284, 22535 CrossRef CAS; (g) S. G. Rzuczek, D. S. Pilch, A. A. Liu, L. F. Liu, J. E. Rice and E. J. LaVoie, J. Med. Chem., 2010, 53, 3632 CrossRef CAS; (h) M. Satyanarayana, Y.-A. Kim, S. G. Rzuczek, D. S. Pilch, A. A. Liu, L. F. Liu, J. E. Rice and E. J. LaVoie, Bioorg. Med. Chem. Lett., 2010, 20, 3150 CrossRef CAS; (i) M. Tera, Y. Sohtome, H. Ishizuka, T. Doi, M. Takagi, K. Shin-ya and K. Nagasawa, Heterocycles, 2006, 69, 505 CrossRef CAS; (j) M. Tera, H. Ishizuka, M. Takagi, M. Suganuma, K. Shin-ya and K. Nagasawa, Angew. Chem., Int. Ed., 2008, 47, 5557 CrossRef CAS; (k) S. Majima, M. Tera, K. Iida, K. Shin-ya and K. Nagasawa, Heterocycles, 2011, 82, 1345 CAS; (l) K. Iida, S. Majima, T. Ohtake, M. Tera, K. Shin-ya and K. Nagasawa, Heterocycles, 2012, 84, 401 CrossRef CAS; (m) T. Nakamura, K. Iida, M. Tera, K. Shin-ya, H. Seimiya and K. Nagasawa, ChemBioChem, 2012, 13, 774 CrossRef CAS; (n) K. Iida, M. Tera, T. Hirokawa, K. Shin-ya and K. Nagasawa, Chem. Commun., 2009, 6481 RSC; (o) K. Iida, M. Tera, T. Hirokawa, K. Shin-ya and K. Nagasawa, J. Nucleic Acids, 2010, 217627 Search PubMed; (p) Y. Amemiya, Y. Furunaga, K. Iida, M. Tera, K. Nagasawa, K. Ikebukuro and C. Nakamura, Chem. Commun., 2011, 47, 7485 RSC; (q) M. Tera, K. Iida, H. Ishizuka, M. Takagi, M. Suganuma, T. Doi, K. Shin-ya and K. Nagasawa, ChemBioChem, 2009, 10, 431 CrossRef CAS; (r) M. Tera, K. Iida, K. Ikebukuro, H. Seimiya, K. Shin-ya and K. Nagasawa, Org. Biomol. Chem., 2010, 8, 2749 RSC.
- The core structure of macrocyclic hexaoxazole was named “HX” by Rice and co-workers. At nearly the same time, we also developed the same structure as a G4 ligand, and named it “6OTD”. L2H2–6OTD (2)11j is identical to HXDL.11c.
- The DNAs were purchased from Sigma Genosys. See table below for sequences of telomeric DNAs used in this work.
. - P. Balagurumoorthy and S. K. Brahmachari, J. Biol. Chem., 1994, 269, 21858 CAS.
- The amounts for melting analysis were determined by the results obtained from EMSA in Fig. 5.
- L2H2-6OTD (2) was found to form complexes with telo24 in 2
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 based on the electrospray ionization mass spectra (see ESI†).
1 based on the electrospray ionization mass spectra (see ESI†).
Footnote |
| † Electronic supplementary information (ESI) available: Normalized thermal melting and annealing profiles, electrospray ionization mass spectra. See DOI: 10.1039/c2md20234d |
| This journal is © The Royal Society of Chemistry 2013 |