Gobichelin A and B: mixed-ligand siderophores discovered using proteomics†
Yunqiu
Chen
,
Michelle
Unger
,
Ioanna
Ntai
,
Ryan A.
McClure
,
Jessica C.
Albright
,
Regan J.
Thomson
and
Neil L.
Kelleher
*
Department of Chemistry and Chemistry of Life Processes Institute, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA. E-mail: n-kelleher@northwestern.edu; Fax: +1-847-467-3276; Tel: +1-847-467-4362
First published on 24th September 2012
Abstract
“Omic” strategies have been increasingly applied to natural product discovery processes, with (meta-)genome sequencing and mining implemented in many laboratories to date. Using the proteomics-based discovery platform called PrISM (Proteomic Investigation of Secondary Metabolism), we discovered two new siderophores gobichelin A and B from Streptomyces sp. NRRL F-4415, a strain without a sequenced genome. Using the proteomics information as a guide, the 37 kb gene cluster responsible for production of gobichelins was sequenced and its 20 open reading frames interpreted into a biosynthetic scheme. This led to the targeted detection and structure elucidation of the new compounds produced by nonribosomal peptide (NRP) synthesis.
Introduction
Natural products and natural product derivatives have composed a major part of FDA-approved therapeutic drugs and continue to provide important sources for new drug discovery.1 Traditionally, natural product discovery is performed by a bio-assay guided approach, where the bioactive components are isolated by iterative fractionations coupled with bioactivity assays.2 The shortcoming of this approach is its tendency to rediscover already known natural products. Over the past decade, many systems biology strategies, including genomics, transcriptomics, metabolomics and proteomics, have been applied to natural product discovery. One of the challenges with genomics based approaches is to link the genomic and metabolomic data, as many biosynthetic genes remain “silent” in lab culture conditions.3,4 On the other hand, the proteomics based approach “PrISM” (Proteomic Investigation of Secondary Metabolism) permits the direct detection of the expressed genes for natural product biosynthesis, so that the resulting natural product can be discovered simultaneously and linked with the biosynthetic genes.5 Using the PrISM approach, new natural products have been discovered from strains both with known genomic sequences and without any genomic information.6,7As described previously,5 PrISM focuses primarily on the discovery of two types of natural products, nonribosomal peptides (NRPs) and polyketides (PKs). These two families contain many clinically important drugs, including the antibiotic daptomycin,8 the anticancer agent bleomycin9 and the immunosuppressant FK506.10 Both NRPs and PKs are synthesized by modularly organized enzymes, nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs), respectively. NRPSs and Type I PKSs are often much larger in size (>150 kDa) than most bacterial proteins, and consist of multiple domains acting as a molecular assembly line.11 The biosynthetic intermediates are covalently tethered to the carrier domains (thiolation domain) during the assembly.
In a PrISM screen, bacterial or fungal strains are grown under different culture conditions for different periods of time, and their proteomes are analyzed in discovery mode. The main approach used for fractionation of intact proteins is one-dimensional SDS-PAGE. The high molecular weight proteins (>150 kDa), often representing NRPSs or PKSs, are subjected to in-gel tryptic digestion and LC-MS analysis. The identified proteins/peptides are used for reconstructing the genetic information of the expressed biosynthetic gene clusters, which will guide the discovery of the natural product.
In the current investigation, we report the discovery of two new natural products from a Streptomyces strain with no genomic sequence available, using the PrISM platform. The compounds, gobichelin A and B, are related to amychelin that was discovered in 2011 (ref. 12) and proven to be iron-chelating agents (siderophores). The structures of gobichelins were solved by tandem mass spectrometry, stable isotopic labeling as well as 1D and 2D NMR, and have basic amino acids differing substantially from the neutral serine residues present in amychelin. The gob gene cluster is also sequenced and annotated into a biosynthetic mechanism proposed for assembly of the six building blocks comprising gobichelins.
Results and discussion
Using the PrISM platform, we screened 26 Actinomycetes for the expression of high molecular weight NRPS or PKS proteins.7 One of the strains, Streptomyces sp. NRRL F-4415, showed protein bands above 250 kDa by SDS-PAGE analysis when grown in ATCC 172 medium for 72 h (Fig. 1a). In-gel tryptic digestion of the high molecular weight proteins followed by LC-MS analysis identified 30 unique peptides matching NRPS proteins from disparate gene clusters in the database (Table S2†). Since these 30 peptides were identified as matching different proteins from different strains, we predicted that the actual full protein sequences are not present in the NCBI non-redundant protein database, i.e., a new NRPS gene cluster.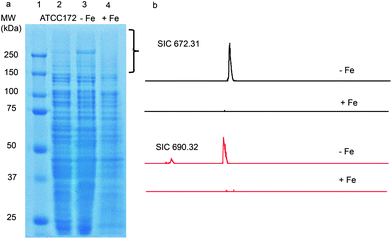 | ||
| Fig. 1 (a) SDS-PAGE analysis of the proteome from Streptomyces sp. NRRL F-4415 grown in ATCC 172 medium (lane 2), iron-deficient medium (lane 3) and in ATCC 172 medium supplemented with 1 mM FeSO4 (lane 4). The bracketed region was subjected to in-gel trypsin digestion and LC-MS analysis. (b) LC-MS analysis of supernatants from culture analogous to that from lanes 3 and 4. Shown are selected ion chromatograms (SICs) of m/z 672.3 and 690.3 in iron deficient medium and iron supplemented medium. | ||
The sequences of the identified peptides were reverse translated into nucleotide sequences for designing PCR primers to amplify the corresponding genomic regions (Tables S3 and S4†). The successful PCR amplicons showed relatively low sequence identities (51–74%) to the closest NRPS sequences in the NCBI non-redundant database (Table S5†). To retrieve the complete sequence of the detected gene cluster, a fosmid library was created on Streptomyces sp. NRRL F-4415, and screened by three pairs of PCR primers (5F, A7R; 18F, A7R; and 12F, A7R). A total of seven fosmids were identified to contain part of the gene cluster and subjected to 454 sequencing.
Assembly of the 454 sequencing results revealed a new NRPS biosynthetic gene cluster shown in Fig. 2a. It is 36.8 kb in size and contains 20 open reading frames. The gene cluster contains five genes homologous to siderophore transporter genes (gobMNOPQ shown in green), suggesting that it is likely to encode proteins that produce a siderophore. There are three modular NRPS genes (gobJRS shown in blue) that correspond to five peptide extension modules (Fig. 2b). A peptide extension module is usually composed of a condensation domain (C) catalyzing amide bond formation, an adenylation domain (A) for substrate recognition and activation, and a thiolation domain (T) as a transport unit.13GobJ encodes a 133 kDa protein comprising a thiolation domain (T) followed by an NRPS extension module with a cyclization domain (Cy) instead of a C domain. A Cy domain catalyzes not only condensation but also the heterocyclization between the side chain OH of threonine and serine or SH of cysteine and the carbonyl group of the peptide backbone to form a thiazoline or oxazoline ring. Similarly, both GobR (282 kDa) and GobS (251 kDa) consist of two NRPS extension modules, respectively. The second module of GobR contains an epimerization (E) domain, suggesting the stereochemistry of the corresponding amino acid in the final product has the D-configuration. GobS contains a thioesterase (TE) domain at the C-termini of the last peptide extension module that catalyzes the release of the peptide from the biosynthetic enzyme. GobH is homologous to the salicylate synthetase Irp9 from Yersinia enterocolitica (42% identity) and likely converts chorismate to salicylate.14 GobK contains an AMP-binding domain that is predicted to activate salicylate as the starter unit and load it to the first T domain on GobJ. GobG is homologous (87% similarity) to an amidinotransferase from Streptomyces griseus NBRC 13350 that converts L-arginine to L-ornithine. GobT encodes an NADPH dependent monooxygenase with 60% sequence identity to the ornithine monooxygenase in the coelichelin biosynthesis that forms N-hydroxyl-ornithine.
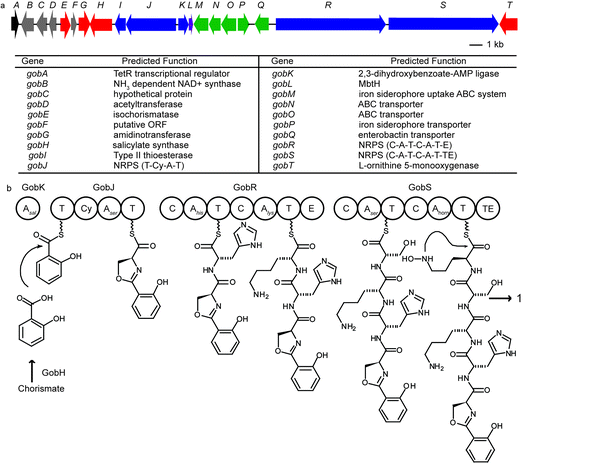 | ||
| Fig. 2 (a) Annotated gob gene cluster responsible for gobichelin biosynthesis. (b) Proposed biosynthetic mechanism for gobichelin A: A, adenylation domain; T, thiolation domain; Cy, cyclization domain; C, condensation domain; E, epimerization domain; TE, thioesterase. | ||
By comparing the active-site residues of known NRPS A domains, we used standard bioinformatics tools to predict the amino acid specificity of the individual modules.15–17 The amino acids incorporated were predicted as: salicylate for GobK, threonine/cysteine for GobJ, tryptophan/phenylalanine and lysine/ornithine for GobR, serine and N-hydroxyl-ornithine for GobS. Based on this analysis, the peptide product from this gene cluster can be predicted as a pentapeptide with a salicylate as the N-terminal cap, likely serving as an iron-chelating siderophore.
Subsequently, we found that supplementation of iron in the culture media strongly suppressed the expression of the high molecular weight NRPS proteins, while depletion of iron boosted protein expression (Fig. 1a). This agrees with expectations of regulatory behavior of siderophore biosynthetic gene clusters.18 To detect the natural product, Streptomyces sp. NRRL F-4415 was grown in iron deficient medium and the culture supernatant was analyzed by high resolution LC-MS. A peak at m/z 672.3, along with a related peak at m/z 690.3, was found to increase with culturing time and was absent when grown in iron supplemented media (Fig. 1b). High resolution tandem MS on m/z 672.3 revealed a sequence tag corresponding to 190 Da-His-Lys-Ser-(cyclic OHOrn), and tandem MS on m/z 690.3 corresponding to Sal-Ser-His-Lys-Ser-(cyclic OHOrn) (Fig. 3b). Given the Cyclization (Cy) domain predicted above, the side chain hydroxyl of serine was predicted to form an oxazoline ring with the carbonyl group of salicylate in the 672.3 species whereas the 690.3 species is consistent within 1 ppm of the ring open form. Based on these tandem MS results, we predicted the structures of the siderophore natural products, that we named gobichelin A (1) and gobichelin B (2), to be shown in Fig. 3a. The high resolution mass spectra and predictions based on the rules of co-linear NRPS biosynthesis also firmly support these structures (1: calc. 671.3027 Da, expt. 671.3022 Da; 2: calc. 689.3133 Da, expt. 689.3127 Da). The iron bound form of gobichelin A was also observed directly, with m/z 725.2224 (calc. 725.2221 for C30H39N9O9Fe [M − 2H + Fe(III)]+). The stereochemistry of gobichelins is predicted based on the NRPS domain organization, where lysine is epimerized to the D-conformation. The combination of three different iron chelating moieties, hydroxylbenzoate, oxazoline and hydroxamate in one siderophore compound has precedent with only oxazoline19 and amychelin (3, Fig. 3a).12
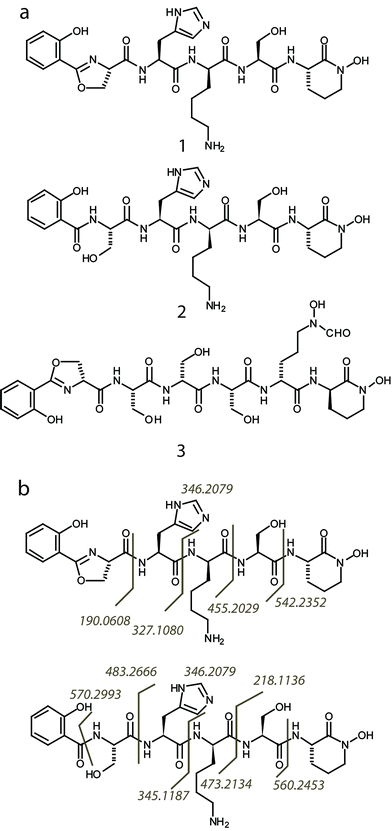 | ||
| Fig. 3 (a) Predicted structures of gobichelin A (1) and B (2), and amychelin (3). (b) The interpreted MS/MS fragmentation patterns for gobichelin A and B are indicated by matching fragment ions observed to their cleavage point in the molecule. | ||
Stable isotope labeled amino acids were added to the culture medium to confirm the amino acid composition of gobichelin A (Fig. 4). When supplemented with D7-ornithine, 13C6-lysine and D3-histidine, clear mass shifts of 7, 6 and 3 Da were observed, respectively, supporting the incorporation of one ornithine, one lysine and one histidine residue into the final product. When supplemented with D3-serine, 3 and 6 Da mass shifts were observed indicating two serine residues incorporated, although the 3 and 6 Da mass shift species have a skewed isotopic distribution where the monoisotopic peaks are not the most intense peaks. This skewed distribution is likely due to the conversion of labeled serine to other amino acids through primary metabolism.
![Incorporation of stable isotope labeled amino acids into gobichelin A. All amino acids were added to iron-deficient medium at 1 mM. (1) Control – no supplement. (2) l-[2,3,3,4,4,5,5-d7]ornithine. (3) l-[13C6]lysine. (4) l-[2,3,3-d3]serine. (5) l-[α-d1; imidazole-2,5-d2]histidine. The labeled amino acid subunits are colored in the gobichelin structure, where the heavy isotope labeled atoms are denoted with ‘*’.](/image/article/2013/MD/c2md20232h/c2md20232h-f4.gif) | ||
| Fig. 4 Incorporation of stable isotope labeled amino acids into gobichelin A. All amino acids were added to iron-deficient medium at 1 mM. (1) Control – no supplement. (2) L-[2,3,3,4,4,5,5-D7]ornithine. (3) L-[13C6]lysine. (4) L-[2,3,3-D3]serine. (5) L-[α-D1; imidazole-2,5-D2]histidine. The labeled amino acid subunits are colored in the gobichelin structure, where the heavy isotope labeled atoms are denoted with ‘*’. | ||
Our first attempt to isolate pure gobichelin A and B under weakly acidic HPLC conditions yielded only gobichelin B, leading us to suspect that gobichelin A spontaneously hydrolyzed into gobichelin B during the purification process. Different from the hydrolysis of amychelin, where the cyclic hydroxyl-ornithine is hydrolyzed into a linear form,12 gobichelin A converts through hydrolysis of the oxazoline ring into a free serine. The opening of oxazoline rings in weakly acidic conditions has been observed in previous cases, such as photobactin,20 anachelin,21 and agrobactin.22 We subsequently performed the purification again under non-acidic conditions, and both gobichelin A and B were isolated to more than 85% purity. Detailed NMR analysis of gobichelin A using 1H, 13C, COSY, TOCSY, HSQC and HMBC experiments was able to confirm the amino acid contents of gobichelin A (Table 1, Fig. S1–S6†).
|
|
|||||
|---|---|---|---|---|---|
| Position | δH (multiplicity, J, Hz) | δC | Position | δH (multiplicity, J, Hz) | δC |
| Salicylate | Lys | ||||
| 1 | 7.48 (dt, 7.8, 15.9) | 134.5 | 20 | ||
| 2 | 7.03 (t, 9.8) | 116.3 | 21 | 4.28 (dd, 8.8, 5.7) | 53.8 |
| 3 | 7.68 (d, 7.7) | 128.6 | 22 | 1.75 (m), 1.66 (m) | 29.7 |
| 4 | 109.9 | 23 | 1.24 (m) | 21.8 | |
| 5 | 158.0 | 24 | 1.61 (dt, 7, 14) | 26.1 | |
| 6 | 7.03 (t, 9.8) | 116.3 | 25 | 2.92 (dd, 8.7, 6.5) | 39.0 |
| 7 | 26 | ||||
| 8 | 167.5 | 27 | 173.9 | ||
| Ser | Ser | ||||
| 9 | 4.99 (dd, 10.3, 6.7) | 67.2 | 28 | ||
| 10 | 4.57 (t, 9.5), 4.44 (t, 7.8) | 69.4 | 29 | 4.40 (m) | 55.7 |
| 11 | 173.3 | 30 | 3.83 (dd, 5, 2.7) | 60.9 | |
| His | 31 | ||||
| 12 | 32 | 170.9 | |||
| 13 | 4.65 (dd, 8.6, 6.5) | 53.2 | OH-Orn | ||
| 14 | 3.03 (dd, 14.7, 8.6), 3.11 (dt, 16.1, 7.8) | 27.6 | 33 | ||
| 15 | 131.1 | 34 | 4.39 (m) | 50.3 | |
| 16 | 6.98 (s) | 119.8 | 35 | 1.74 (m)/1.95 (m) | 26.5 |
| 17 | 7.91 (s) | 134.5 | 36 | 1.92 (m)/1.87 (m) | 19.9 |
| 18 | 37 | 3.60 (t, 5.4) | 60.8 | ||
| 19 | 172.6 | 38 | |||
| 39 | 166.3 | ||||
Based on the structures of gobichelins and the annotations of the gene cluster, we proposed the biosynthetic mechanism for gobichelin A shown in Fig. 2b. The mechanism shows some similarity with the biosynthesis of amychelin, a siderophore produced by Amycolatopsis sp. AA4.12 Both gene clusters contain a dihydroxybenzoate-AMP ligase for the incorporation of the N-terminal salicylate moiety, a cyclization domain to form an oxazoline ring, and both load a hydroxyl-ornithine at the C-termini. However, while gobichelin A is directly released from the NRPS assembly line by the thioesterase on the last domain of GobS, the NRPS for amychelin biosynthesis lacks a TE domain, and the release of the product is predicted to be performed by an external α/β hydrolase. In addition, the amino acid composition differs substantially between amychelin and gobichelin. Amychelin contains three consecutive serine residues that are predicted to help decrease solvation energy and form a favorable structure for iron binding, whereas gobichelins contain the basic amino acids histidine and lysine. These basic amino acids might help deprotonate the OH groups on hydroxyl-ornithine and the salicylate moieties through intramolecular hydrogen bonding to facilitate their iron chelation, although this hypothesis needs to be confirmed by more detailed studies, like X-ray crystallography, on the Fe–gobichelin complex structure.
Conclusion
Using proteomics, we discovered two new peptide natural products from Streptomyces sp. NRRL F-4415, termed gobichelin A and B. The biosynthetic gene cluster for the gobichelins was also found and associated with the compound, and a biosynthetic mechanism proposed accordingly. Due to the presence of hydroxamate and salicylate functional groups, as well as the unusual basic amino acid residues, the gobichelins serve as unique mixed ligand iron-chelating agents. This example, together with our previous work, has shown that the “protein-first” natural product discovery strategy is capable of linking biosynthetic genes with the resulting natural product, even when no genomic information is available. Future effort will be focused on advancing the overall efficiency of this “structure-based” discovery pipeline by varying a larger number of culture conditions, including co-culturing, improving the ability to detect expression of gene clusters at lower copy number, and by applying more sophisticated bioinformatics to allow for early dereplication of known natural products.Experimental
Materials
Streptomyces sp. NRRL F-4415 was from US Department of Agriculture, Agricultural Research Service. All chemicals were from Sigma unless otherwise noted. Sequencing grade trypsin was from Promega. Stable-isotope labeled amino acids were from Cambridge Isotopes or CDN isotopes. 454 sequencing was performed at the University of Illinois Biotechnology Center. Traditional sequencing services were performed at the Northwestern University Genomics Core Facility.PrISM detection
Streptomyces sp. NRRL F-4415 was cultured in ATCC 172 medium (10 g L−1 glucose, 20 g L−1 soluble starch, 5 g L−1 yeast extract, 5 g L−1 N-Z amine type A, 1 g L−1 CaCO3) at 30 °C. Cells from 24 h, 48 h and 72 h were collected and lysed by sonication. The proteome lysate was separated on a one-dimensional SDS-PAGE, and the proteome region above 150 kDa was excised for in-gel trypsin digestion. The extracted peptides were separated on a self-packed nano-capillary column (5 μm Jupiter C18, 100 mm × 75 μm) using a Dionex UltiMate® 3000 Nano LC system (Thermo Fisher Scientific). LC solvents included 5% acetonitrile in water with 0.1% formic acid (mobile phase A) and 95% acetonitrile with 0.1% formic acid (mobile phase B). The LC gradient was set as follows: 0 min, 0% B; 55 min, 45% B; 63 min, 80% B; 67 min, 0% B with re-equilibration of 0% B until 90 min. Peptides were eluted into a nanoelectrospray ionization (nESI) source on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Intact peptide data were collected in the 400–2000 m/z range, and MS/MS spectra were acquired using data dependent mode where the top ten most abundant peaks from the full scan were selected for Higher Energy Collision Dissociation (HCD) fragmentation. The LC-MS data was searched against the non-redundant protein database NCBInr using Open Mass Spectrometry Search Algorithm (OMSSA).PCR, fosmid library creation and sequencing
Peptides identified as deriving from NRPS or PKS genes were reverse translated to nucleotide sequences, using the codon usage table for Streptomyces coelicolor A3(2). Primers were designed based on both the most likely codons and degenerate codons. Primers designed from reverse translation were paired with degenerate primers for the A3 and A7 conserved regions of NRPSs.23 Genomic DNA of NRRL F-4415 was isolated from 5 mL cultures using a DNeasy Blood and Tissue DNA kit (Qiagen). PCR reactions were performed using GoTaq® Green Master Mix (Promega). Successful PCR products were sequenced using the same primer pairs. A fosmid library of NRRL F-4415 was created using CopyControl™ fosmid library production kit (Epicentre Biotechnologies, WI). A total of 1440 colonies were screened by three PCR reactions to yield seven positive fosmids containing the target gene cluster. The seven fosmids were pooled for 454 pyrosequencing on a GS FLX+ Sequencer (Roche). A total of 145.3 Mbp was used for sequence assembly using SeqMan Pro (DNAStar), resulting in a continuous contig of 84, 139 bp. BLASTX and FramePlot were used for ORF prediction.Natural product detection
Streptomyces sp. NRRL F-4415 was grown in a defined iron-deficient medium (2 g L−1 K2SO4, 3 g L−1 K2HPO4, 1 g L−1 NaCl, 5 g L−1 NH4Cl, 2 mg L−1 thiamine, 0.005 mg L−1 CuSO4, 0.035 mg L−1 MnSO4·H2O, 2 mg L−1 ZnSO4·7H2O, 80 mg L−1 MgSO4·7H2O, 100 mg L−1 CaCl2·2H2O, 2.5% glycerol and 2 mM 2,2′-bipyridine). 1 mM of stable isotope labeled amino acids were added to the medium when necessary. Culture supernatants were separated by a Phenomenex Luna C18 column (250 × 2 mm, 5 μm) eluting with acetonitrile/water with 0.1% formic acid from 0 to 100%. A Q-Exactive Orbitrap (Thermo Fisher Scientific) was used for full mass detection from m/z 300 to 1500 followed by data dependent fragmentation of the top five most intense ions.Isolation of gobichelin
A single colony of Streptomyces sp. NRRL F-4415 was used for inoculating 4 × 5 mL ATCC 172 medium. After 48 h growth, they were transferred to 4 × 500 mL of the iron-deficient medium mentioned above without 2,2′-bipyridine in 2 L baffled flasks. After 6 days of growth at 30 °C, the culture supernatant was separated from cells by filtration. The aqueous layer was concentrated in vacuo to 160 mL, and each 40 mL was passed through a Strata C8 (55 μm 70 Å) 10 g/60 mL SPE column (Phenomenex) and washed by 60 mL of 5% MeOH, 120 mL of 50% MeOH and 120 mL of 100% methanol. The 50% MeOH fraction, which contained gobichelins, was concentrated in vacuo to 5 mL, and separated on a preparative Phenomenex Luna C18 (2) column (5 μm, 10 × 250 mm) using 15 to 35% acetonitrile/water over 15 min, monitored by UV absorbance at 300 nm. The purified gobichelin A (∼20 mg) was dissolved in D2O. 1H, 13C and 2D NMR spectra were recorded in a Bruker AVANCE III NMR (500 MHz for 1H) with direct cryoprobe. Chemical shifts were referenced to the residual solvent peaks in D2O.Acknowledgements
We thank the Agricultural Research Service, United States Department of Agriculture for providing us the bacterial strain. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM085232. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2012, 75, 311–335 CrossRef CAS.
- M. J. Weinstein and G. H. Wagman, Antibiotics, Isolation, Separation, and Purification, Elsevier Scientific Pub. Co., Amsterdam, 1978 Search PubMed.
- A. A. Brakhage, J. Schuemann, S. Bergmann, K. Scherlach, V. Schroeckh and C. Hertweck, Prog. Drug Res., 2008, 66(1), 3–12 Search PubMed.
- C. Johnston, A. Ibrahim and N. Magarvey, Med. Chem. Commun., 2012, 3, 932–937 RSC.
- S. B. Bumpus, B. S. Evans, P. M. Thomas, I. Ntai and N. L. Kelleher, Nat. Biotechnol., 2009, 27, 951–956 CrossRef CAS.
- B. S. Evans, I. Ntai, Y. Chen, S. J. Robinson and N. L. Kelleher, J. Am. Chem. Soc., 2011, 133, 7316–7319 CrossRef CAS.
- Y. Chen, I. Ntai, K. S. Ju, M. Unger, L. Zamdborg, S. J. Robinson, J. R. Doroghazi, D. P. Labeda, W. W. Metcalf and N. L. Kelleher, J. Proteome Res., 2012, 11, 85–94 Search PubMed.
- F. Ehlert and H. C. Neu, Eur. J. Clin. Microbiol., 1987, 6, 84–90 Search PubMed.
- H. Umezawa, K. Maeda, T. Takeuchi and Y. Okami, J. Antibiot., 1966, 19, 200–209 CAS.
- H. Motamedi, S. J. Cai, A. Shafiee and K. O. Elliston, Eur. J. Biochem., 1997, 244, 74–80 CrossRef CAS.
- M. A. Fischbach and C. T. Walsh, Chem. Rev., 2006, 106, 3468–3496 CrossRef CAS.
- M. R. Seyedsayamdost, M. F. Traxler, S. L. Zheng, R. Kolter and J. Clardy, J. Am. Chem. Soc., 2011, 133, 11434–11437 CrossRef CAS.
- D. Schwarzer, R. Finking and M. A. Marahiel, Nat. Prod. Rep., 2003, 20, 275–287 RSC.
- O. Kerbarh, A. Ciulli, N. I. Howard and C. Abell, J. Bacteriol., 2005, 187, 5061–5066 CrossRef CAS.
- B. O. Bachmann and J. Ravel, Methods Enzymol., 2009, 458, 181–217 Search PubMed.
- C. Prieto, C. Garcia-Estrada, D. Lorenzana and J. F. Martin, Bioinformatics, 2012, 28, 426–427 Search PubMed.
- M. Rottig, M. H. Medema, K. Blin, T. Weber, C. Rausch and O. Kohlbacher, Nucleic Acids Res., 2011, 39, W362–W367 CrossRef.
- S. Lautru, R. J. Deeth, L. M. Bailey and G. L. Challis, Nat. Chem. Biol., 2005, 1, 265–269 CrossRef CAS.
- B. Sontag, M. Gerlitz, T. Paululat, H. F. Rasser, I. Grun-Wollny and F. G. Hansske, J. Antibiot., 2006, 59, 659–663 CrossRef CAS.
- T. A. Ciche, M. Blackburn, J. R. Carney and J. C. Ensign, Appl. Environ. Microbiol., 2003, 69, 4706–4713 CrossRef CAS.
- H. Beiderbeck, K. Taraz, H. Budzikiewicz and A. E. Walsby, Z. Naturforsch., C: J. Biosci., 2000, 55, 681–687 Search PubMed.
- S. A. Ong, T. Peterson and J. B. Neilands, J. Biol. Chem., 1979, 254, 1860–1865 Search PubMed.
- A. Ayuso-Sacido and O. Genilloud, Microb. Ecol., 2005, 49, 10–24 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2md20232h |
| This journal is © The Royal Society of Chemistry 2013 |

