Two-in-one: a pH-sensitive, acridine-based, fluorescent probe binds G-quadruplexes in oncogene promoters†
Claudia
Percivalle
ab,
Tariq
Mahmood‡
a and
Sylvain
Ladame
*a
aDepartment of Bioengineering, Imperial College London, South Kensington campus, London SW7 2AZ, UK. E-mail: sladame@imperial.ac.uk; Fax: +44 (0)02075949817; Tel: +44 (0)02075945308
bDepartment of Chemistry, University of Pavia, 27100 Pavia, Italy
First published on 11th September 2012
Abstract
We report the synthesis of an acridine-containing cyanine dye and demonstrate its potential as a pH-responsive colorimetric indicator and fluorescent probe. Similarly to its parent molecule BRACO-19, this tri-substituted acridine also binds strongly to DNA quadruplexes found in the promoter region of oncogenes.
Fluorescent small molecules have found valuable applications as probes for physiological monitoring, imaging and/or sensing.1 Among them, cyanine dyes have received particular attention due to their relative chemical stability, high molar absorptivity and high fluorescence quantum yield.2 They generically consist of a conjugated system based on a polymethine chain linking two nitrogen-containing heterocycles. The length of the polymethine chain and the nature of the heterocycles will determine the spectroscopic properties of the dyes. While symmetrical cyanine dyes are commonly used as fluorescent labels, unsymmetrical cyanine dyes can also be engineered that are responsive to the environment they are put into.3 We recently reported the synthesis and fluorogenic properties of a series of acridine-containing cyanine dyes.4 Acridines are well-known pharmacophores developed for their antibacterial, trypanocidal, antimalarial and antitumor activities.5 The anticancer activity of acridines relies essentially on their capacity to interact with nucleic acids, either (i) via intercalation between double-stranded DNA base-pairs and inhibition of a DNA topoisomerase II enzyme,6 or (ii) via stabilization of alternative four-stranded DNA structures called G-quadruplexes.7 The best known example in the latter category is the so-called BRACO-19,8 a 3,6,9-trisubstituted acridine capable of inhibiting telomerase activity and also showing significant in vivo anticancer activity in tumour xenografts.9 Herein, we report the synthesis of a water-soluble and acridine-containing monomethine cyanine dye directly inspired from BRACO-19. We also demonstrate its dual applications (1) as a pH-sensitive, colorimetric and fluorescent sensor and (2) as a G-quadruplex binding ligand.
A pH-sensitive probe
A cyanine dye 1§ was obtained from a 3,6-bis-[3-pyrrolidino-propionamide] acridone precursor (BRACO-14)10 by adapting a protocol previously reported by us (Fig. 1).4 Absorbing in the green area of the spectrum, this water-soluble monomethine cyanine dye proved to be sensitive to proton concentration over a pH range of 5–9. Intracellular pH is an essential physiological factor that modulates the function of many organelles. It plays a key role in cell proliferation mechanisms and intracellular acidification has been associated with apoptosis, notably in cancer cells.11 Whilst colorimetric pH indicators are highly valuable for in vitro applications due to their ease of use, pH fluorosensors are more suited for in vivo sensing/imaging and offer the advantage of much greater sensitivity and accuracy.12 Ideally, the dye will be water soluble and able to sense pH values within the critical intracellular pH window of 5–8. If applied to the specific detection of intracellular acidification, it will also be characterised by an increased fluorescence in acidic media (for a more reliable readout). In recent years, a significant number of fluorescent probes have been reported that are responsive to pH and cover a broad spectral range, and some of them are suitable for in vivo applications.12,13 | ||
| Fig. 1 Structures of the G-quadruplex binding ligand BRACO-19 and the pH-sensitive monomethine cyanine dye 1. | ||
Herein, the absorption and emission spectra of dye 1 were recorded in potassium phosphate buffers covering a broad spectrum of pH values ranging from 4 to 9. Interestingly, the pH responsive nature of the dye could also be monitored by the naked eye. Whilst it was dark blue at acidic pH, an aqueous solution of 1 turned orange when the pH was raised above the physiological value of 7.4 (Fig. 2a). When observing the UV/Vis spectra of 1 in buffers of different pH, the characteristic absorbance of the monomethine cyanine dye centred at 578 nm was found to decrease with increasing pH. A similar trend was observed when looking at the second absorbance maximum of the dye centred at 396 nm. The disappearance of these two maxima occurred with the appearance of a new maximum at around 476 nm, although of much weaker intensity (see ESI†). It is noteworthy that under the conditions of the experiments, no aggregation was ever observed. The pH responsive nature of 1 can also be monitored by fluorescent spectroscopy, offering a significantly greater sensitivity when compared to UV/Vis spectroscopy (Fig. 2b). While in aqueous solution the fluorescence emitted upon excitation at 578 nm was below the detectable limit, a strong fluorescence was obtained when exciting the dye at the second absorption maximum wavelength (ca. 400 nm). When plotting the fluorescence intensity (λexc = 400 nm, λem = 488 nm) as a function of pH, a regular sigmoid response was obtained, from which a pKa value of 6.6 was estimated (Fig. 2c).
![(a) Colour change of 1 in potassium phosphate buffers (100 mM) at various pH values; (b) fluorescence emission spectra of dye 1 in potassium phosphate buffers (100 mM) at various pH values ranging from 4.2 to 10; (c) fluorescence emission intensities (I/Imax) of 1 as a function of pH recorded in potassium phosphate buffers (λexc = 400 nm, λem = 488 nm, [1] = 1 μM).](/image/article/2013/MD/c2md20173a/c2md20173a-f2.gif) | ||
| Fig. 2 (a) Colour change of 1 in potassium phosphate buffers (100 mM) at various pH values; (b) fluorescence emission spectra of dye 1 in potassium phosphate buffers (100 mM) at various pH values ranging from 4.2 to 10; (c) fluorescence emission intensities (I/Imax) of 1 as a function of pH recorded in potassium phosphate buffers (λexc = 400 nm, λem = 488 nm, [1] = 1 μM). | ||
Taken together, these results show that dye 1 can act as a fluorescent and colorimetric pH-sensitive probe, sensing pH below and above the physiological value of 7.4. pH sensitivity is introduced by the non-alkylated acridine moiety. When the acridine is protonated, a positive charge delocalised between the acridine and indole moieties restores the push–pull mechanism responsible for the fluorescence properties of traditional cyanine dyes. At higher pH values where acridine is non-protonated, the conjugated system is no longer charged and the absence of a push–pull mechanism between both heterocycles accounts for the observed fluorescence decrease.
A G-quadruplex binding ligand
Intramolecular G-quadruplexes are four-stranded structures formed by G-rich oligonucleotide sequences of the type G3+N1–7G3+N1–7G3+N1–7G3+ in the presence of physiological cations, notably K+ and Na+.14 Formation of intramolecular DNA quadruplexes was first proposed to occur at the 3′-end of telomeres, thus preventing telomere maintenance by the enzyme telomerase in tumour cells. Genome-wide analyses then showed that such structures were highly represented in key areas of the genome, in particular in the promoter region of genes.15 Despite the fact that, in order to form, promoter quadruplexes need to compete with a highly stable duplex conformation, several pieces of indirect evidence have been accumulated in the recent literature which suggest that such structures can influence DNA transcription efficiency, negatively or positively, when either deleted or stabilised. Since DNA quadruplexes have been linked to ageing and cancer, a large number of small molecules have appeared in the literature that bind to and stabilise these motifs in a structure-specific manner. Selected molecules that bind promoter quadruplexes in vitro were also shown to inhibit the transcription of the corresponding genes in vitro and also in vivo.16 Among them, poly-substituted acridines have been shown to bind DNA quadruplexes with both high affinity and high selectivity versus duplex DNA.7,17 In order to determine the potential of dye 1 for recognising DNA quadruplex structures, particularly those located in the promoter of oncogenes, we performed a series of fluorescence titration experiments using as targets the quadruplex motifs identified in the promoter regions of the genes coding for the proteins c-myc,18 c-kit,19 and K-ras.20 Binding to a duplex DNA was also assessed by the same technique in order to detect a potential quadruplex versus duplex selectivity. First, titration experiments were carried out on the c-myc quadruplex and a control duplex DNA. Briefly, a solution of dye 1 (0.5 μM in 100 mM potassium phosphate buffer of pH 7.4 also containing 100 mM KCl) was titrated with a solution of DNA (up to 2.5 equivalents). The fluorescence emission spectra were then recorded on a Jobin-Yvon fluorolog spectrofluorimeter (λexc = 400 nm, Fig. 3a and b). A strong fluorescence quenching was observed upon addition of both duplex and quadruplex DNA. However, a weaker quenching was observed with duplex DNA when compared to that observed with the c-myc quadruplex. Indeed, while the fluorescence of dye 1 is almost completely quenched after addition of 0.5 equivalent of the quadruplex, a significant amount of fluorescence remains (ca. 10–12%) even after addition of 1 equivalent of double-stranded DNA (Fig. 3c). The binding of dye 1 to other promoter quadruplexes was then assessed using a fluorescence plate reader. Experiments were carried out in a 384-well plate, using a fixed dye concentration of 10 μM and increasing concentrations of DNA (from 0 to 20 μM). Fluorescence emission was detected using a fluorescence plate reader (λexc = 400 nm, λem = 475 nm). Near identical profiles were obtained for the c-myc, c-kit2 and K-ras quadruplexes, while a weaker quenching was observed for the c-kit1 quadruplex, indicating a lower affinity (see ESI†).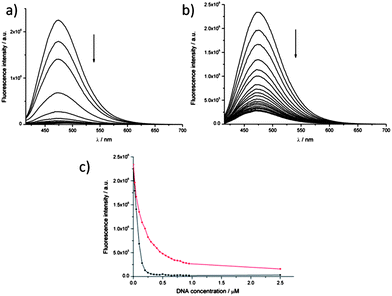 | ||
| Fig. 3 Fluorescence titration experiments: a solution of dye 1 (0.5 μM) was titrated with increasing concentrations of DNA (up to 1.25 μM, i.e. 2.5 equivalents). Emission spectra (λexc = 400 nm) were recorded for titration of either c-myc quadruplex (a) or double-stranded DNA (b). (c) shows a plot of the maximum fluorescence intensity (λexc = 400 nm, λem = 475 nm) as a function of DNA concentration: data for the c-myc quadruplex are shown in black and data for double-stranded DNA in red. | ||
Although these fluorescence titration experiments do not provide accurate information about the affinity and specificity of binding to either duplex or quadruplex DNA, they suggest that the cyanine dye 1 (1) has a strong affinity for every DNA tested, (2) binds tighter to the c-myc, c-kit2 and K-ras quadruplexes compared to the c-kit1 quadruplex and (3) also binds duplex DNA although likely via a different binding mode. Surface Plasmon Resonance (SPR) studies were also carried out to further characterise the DNA-binding properties of dye 1 to three selected DNA targets: the c-myc and c-kit2 quadruplexes as well as a 23-bp duplex DNA control.21 As already suggested by the results of the fluorescence titration experiments, 1 was shown to bind strongly and selectively to the c-myc quadruplex (Kd = 670 nM). Binding to the c-kit2 quadruplex appeared to be ca. 3.5-fold weaker (Kd = 2.75 μM) while unspecific binding to the double-stranded DNA control was detected at concentrations above 1 μM (Fig. 4).
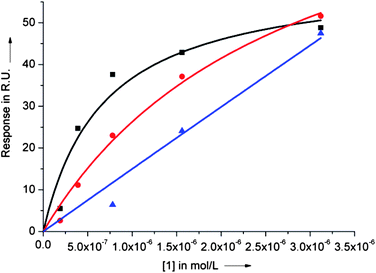 | ||
| Fig. 4 SPR binding curves for dye 1 binding to the c-myc (black squares) and ckit2 (red circles) quadruplexes and to a 23bp duplex DNA (blue triangles). Dissociation constants (Kd) were obtained by fitting the experimental values of response at equilibrium (Req) as a function of ligand concentration (0–3.125 μM) using the steady state affinity algorithm (Biaevaluation 3.0.2). | ||
Finally, the ability of dye 1 to stabilise the c-myc quadruplex was assessed by Circular Dichroism (CD). In 50 mM TRIS buffer (pH 7.4) also containing 10 mM KCl, a solution of 10 μM c-myc DNA was shown to form a highly stable, parallel-stranded, G-quadruplex that can be fully denatured by heating the solution up to 95 °C. Under such conditions of salt and pH, the c-myc quadruplex is characterised by a melting temperature (Tm) of ca. 78 °C. Upon addition of 1 equivalent of compound 1, a parallel G-quadruplex is still formed which cannot be fully denatured/unfolded even when raising the temperature up to 95 °C (Fig. 5). Although the absence of complete unfolding at high temperatures precluded the determination of a Tm value in the presence of ligand, this experiment clearly demonstrated the G-quadruplex-stabilising potential of 1.
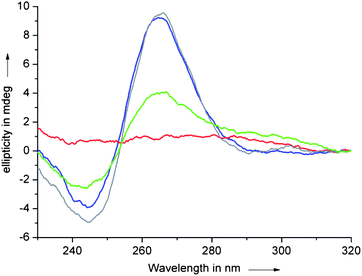 | ||
| Fig. 5 CD spectra of a solution of c-myc quadruplex (10 μM) in TRIS·HCl buffer (50 mM, pH 7.4) also containing 10 mM KCl recorded at 10 °C (blue) and 95 °C (red) and in the presence of 1 eq. of ligand 1 (10 μM) at 10 °C (grey) and 95 °C (green). | ||
The sub-micromolar affinity of 1 for the c-myc quadruplex, its ability to strongly stabilise the c-myc quadruplex and to discriminate between quadruplex and duplex DNA make it a suitable candidate for the development of original quadruplex-targeting drugs based on this scaffold. Moreover, we recently reported that 9-amino acridines could undergo a reversible amine exchange reaction under near-physiological conditions, thus potentially reducing their bioavailability in vivo.22 Substitution of this labile 9-amino linkage with a more chemically stable carbon–carbon bond, while keeping the DNA-binding properties of the molecules intact, could therefore become an interesting way to overcome this drawback intrinsic to 9-aminoacridines.
Conclusions
We described the dual properties of an engineered cyanine dye as a pH sensor and a quadruplex binding ligand. We recently reported the fluorogenic and DNA-binding properties of the first family of acridine-containing cyanine dyes.10 When compared to these previously reported dyes, compound 1 has a non-N-alkylated acridine moiety, thus making it responsive to pH. While in its protonated form dye 1 behaves like a cyanine dye and emits a strong fluorescence at ca. 475 nm, its non-protonated form is characterised by a much weaker and blue-shifted fluorescence. A clear transition occurs at pH around the pKa value of the acridine (ca. 6.6) that can be monitored by fluorescence spectroscopy with high sensitivity but also with the naked eye since raising the pH also leads to a strong colour change from dark blue to yellow. We therefore demonstrated that dye 1 could be used as either a pH colorimetric indicator or fluorosensor and is capable of sensing pH values ranging from ca. 5 to 8.Like its parent molecule BRACO-19, this 3,6,9-trisubstituted acridine also binds strongly to promoter quadruplexes and is capable of discriminating between quadruplex and duplex DNA. We have shown that compound 1 could bind with a sub-micromolar affinity to the c-myc quadruplex and with low micromolar affinity to the c-kit2 quadruplex. Although it may not be quite as specific for quadruplex DNA (with respect to the double-stranded form of DNA) as other ligands recently reported in the literature, molecules based of this scaffold may still have potential as quadruplex-targeting therapeutic agents and analogues of 1 are currently being developed by our group.
Notes and references
- For recent reviews see M. Schäferling, Angew. Chem., Int. Ed., 2012, 51, 3532 Search PubMed; L. M. Wysocki and L. D. Lavis, Curr. Opin. Chem. Biol., 2011, 15, 752 CrossRef; M. S. Gonçalves, Chem. Rev., 2009, 109, 190 CrossRef CAS; L. D. Lavis and R. T. Raines, ACS Chem. Biol., 2008, 3, 142 CrossRef CAS.
- A. Mishra, R. K. Behera, P. K. Behera, B. K. Mishra and G. B. Behera, Chem. Rev., 2000, 100, 1973 CrossRef CAS; F. M. Hamer, The Cyanine Dyes and Related Compounds, ed. A. Weissberger, Interscience, London, UK, 1964 Search PubMed.
- P. Yang, A. De Cian, M.-P. Teulade-Fichou, J.-L. Mergny and D. Monchaud, Angew. Chem., Int. Ed., 2009, 48, 2188 CrossRef CAS; H. Ozhalici-Unal, C. L. Pow, S. A. Marks, L. D. Jesper, G. L. Silva, N. I. Shank, E. W. Jones, J. M. Burnette 3rd, P. B. Berget and B. A. Armitage, J. Am. Chem. Soc., 2008, 130, 12620 CrossRef CAS; H. J. Karlsson, M. Eriksson, E. Perzon, B. Akerman, P. Lincoln and G. Westman, Nucleic Acids Res., 2003, 31, 6227 CrossRef CAS.
- T. Mahmood, A. Paul and S. Ladame, J. Org. Chem., 2010, 75, 204 CrossRef CAS.
- P. Belmont, J. Bosson, T. Godet and M. Tiano, Anti-Cancer Agents Med. Chem., 2007, 7, 139 CrossRef CAS; M. Wainwright, J. Antimicrob. Chemother., 2001, 47, 1 CrossRef CAS; S. A. Gamage, D. P. Figgitt, S. J. Wojcik, R. K. Ralph, A. Ransijn, J. Mauel, V. Yardley, D. Snowdon, S. L. Croft and W. A. Denny, J. Med. Chem., 1997, 40, 2634 Search PubMed; P. M. Chauhan and S. K. Srivastava, Curr. Med. Chem., 2001, 8, 1535 CAS.
- B. Rene, P. Fosse, T. Khelifa, A. Jacquemin-Sablon and C. Bailly, Mol. Pharmacol., 1996, 49, 343 Search PubMed.
- N. H. Campbell, G. N. Parkinson, A. P. Reszka and S. Neidle, J. Am. Chem. Soc., 2008, 130, 6722 CrossRef CAS.
- M. J. Moore, C. M. Schultes, J. Cuesta, F. Cuenca, M. Gunaratnam, F. A. Tanious, W. D. Wilson and S. Neidle, J. Med. Chem., 2006, 49, 582 CrossRef CAS.
- A. M. Burger, F. Dai, C. M. Schultes, A. P. Reszka, M. J. Moore, J. A. Double and S. Neidle, Cancer Res., 2005, 65, 1489 CrossRef CAS; C. M. Incles, C. M. Schultes, H. Kempski, H. Koehler, L. R. Kelland and S. Neidle, Mol. Cancer Ther., 2004, 3, 1201 CAS; S. M. Gowan, J. R. Harrison, L. Patterson, M. Valenti, M. A. Read, S. Neidle and L. R. Kelland, Mol. Pharmacol., 2002, 61, 1154 CrossRef CAS.
- R. J. Harrison, A. P. Reszka, S. M. Haider, B. Romagnoli, J. Morrell, M. A. Read, S. M. Gowan, C. M. Incles, L. R. Kelland and S. Neidle, Bioorg. Med. Chem. Lett., 2004, 14, 5845 CrossRef CAS.
- M. A. Barry, J. E. Reynolds and A. Eastman, Cancer Res., 1993, 53, 2349 Search PubMed.
- J. Han and K. Burgess, Chem. Rev., 2010, 110, 2709 CrossRef CAS.
- For selected examples, see (a) W. Shi, X. Li and H. Ma, Angew. Chem., Int. Ed., 2012, 51, 6432 CrossRef CAS; (b) F. Doria, M. Nadai, G. Sattin, L. Pasotti, S. N. Richter and M. Freccero, Org. Biomol. Chem., 2012, 10, 3830 RSC; (c) X. Zhang, S. Rehm, M. M. Safont-Sempere and F. Würthner, Nat. Chem., 2009, 1, 623 CrossRef CAS; (d) M. S. Briggs, D. D. Burns, M. E. Cooper and S. J. Gregory, Chem. Commun., 2000, 2323 RSC.
- S. Neidle and S. Balasubramanian, in Quadruplex Nucleic Acids, Royal Society of Chemistry, Cambridge, 2006 Search PubMed.
- For recent reviews or examples see (a) Y. Qin and L. H. Hurley, Biochimie, 2008, 90, 1149 CrossRef CAS; (b) J. L. Huppert, Biochimie, 2008, 90, 1140 CrossRef CAS; (c) J. Eddy and N. Maizels, Nucleic Acids Res., 2008, 36, 1321 CAS.
- (a) M. Bejugam, S. Sewitz, P. S. Shirude, R. Rodriguez, R. Shahid and S. Balasubramanian, J. Am. Chem. Soc., 2007, 129, 12926 CrossRef CAS; (b) K. I. McLuckie, Z. A. Waller, D. A. Sanders, D. Alves, R. Rodriguez, J. Dash, G. J. McKenzie, A. R. Venkitaraman and S. Balasubramanian, J. Am. Chem. Soc., 2011, 133, 2658 CrossRef CAS.
- For recent examples see (a) C. Bazzicalupi, M. Chioccioli, C. Sissi, E. Porcù, C. Bonaccini, C. Pivetta, A. Bencini, C. Giorgi, B. Valtancoli, F. Melani and P. Gratteri, ChemMedChem, 2010, 5, 1995 Search PubMed; (b) J. Debray, W. Zeghida, M. Jourdan, D. Monchaud, M. L. Dheu-Andries, P. Dumy, M. P. Teulade-Fichou and M. Demeunynck, Org. Biomol. Chem., 2009, 7, 5219 RSC.
- V. Gonzalez and L. H. Hurley, Annu. Rev. Pharmacol. Toxicol., 2009, 50, 111 Search PubMed.
- (a) S. Rankin, A. P. Reszka, J. Huppert, M. Zloh, G. N. Parkinson, A. K. Todd, S. Ladame, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2005, 127, 10584 CrossRef CAS; (b) H. Fernando, A. P. Reszka, J. Huppert, S. Ladame, S. Rankin, A. R. Venkitaraman, S. Neidle and S. Balasubramanian, Biochemistry, 2006, 45, 7854 CrossRef CAS; (c) S. T. Hsu, P. Varnai, A. Bugaut, A. P. Reszka, S. Neidle and S. Balasubramanian, J. Am. Chem. Soc., 2009, 131, 13399 CrossRef CAS.
- S. Cogoi and L. E. Xodo, Nucleic Acids Res., 2006, 34, 2536 CrossRef CAS.
- (a) J. E. Redman, Methods, 2007, 43, 302 CrossRef CAS; (b) J. Dash, Z. A. Waller, G. D. Pantos and S. Balasubramanian, Chem.–Eur. J., 2011, 17, 4571 CrossRef CAS.
- A. Paul and S. Ladame, Org. Lett., 2009, 11, 4894 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H and 13C NMR spectra of 1, UV/Vis absorption spectra of 1 at various pH values, oligonucleotide sequences, fluorescence titration experiments of 1 with the c-myc quadruplex and with double-stranded DNA. See DOI: 10.1039/c2md20173a |
| ‡ Current address: COMSATS, Institute of Information Technology, Abbottabad Campus, Pakistan. |
| § Synthesis of dye 1: 150 mg of acridone BRACO-1410 (0.31 mmol) were suspended in POCl3 (10 ml) and the brown yellow mixture was heated to reflux under N2. After 2 h the yellow suspension was cooled down to room temperature and then to 0 °C. Cold diethyl ether (10 ml) was added and a yellow/brown solid separated. The precipitate was collected by filtration, washed with diethyl ether (3 × 10 ml), and dried under vacuum affording a yellow/brown solid. The crude 9-chloroacridine product was used in the next step without any further purification. To the crude product was added 2-methylene-1,3,3-trimethylindoline (2.80 ml, 15.8 mmol, 50 eq.) followed by chloroform (10 ml). The dark mixture was heated at 60 °C for six days under a N2 atmosphere. After cooling to room temperature the solvent was removed under reduced pressure to afford a dark oil which was triturated in diethyl ether (10 ml). The mixture was cooled at 0 °C for 30 min and a dark precipitate formed. The precipitate was collected by filtration affording a crude violet solid. The crude product was purified by HPLC on a Phenomenex C18 Gemini-NX 110 Å column (250 × 10.0 mm) using a water–acetonitrile gradient (both containing 0.1% of TFA). Lyophilization afforded the product as a dark violet solid (25 mg as a TFA salt, yield 12%). 1H-NMR (400 MHz, CD3OD) δ (ppm): 8.69 (2H, bs), 8.19 (2H, d, J = 8 Hz), 7.57–7.59 (2H, m), 7.46 (1H, d, J = 8 Hz), 7.33 (1H, t, J = 8 Hz), 7.16 (1H, t, J = 8 Hz), 6.93 (1H, d, J = 8 Hz), 6.42 (1H, s), 3.76 (4H, bs), 3.64 (4H, bs), 3.19 (4H, bs), 3.09 (4H, bs), 2.63 (3H, s), 2.20 (4H, bs), 2.09 (4H, bs), 1.75 (6H, s). 13C-NMR (100 MHz, CD3OD) δ (ppm): 172.6, 170.9, 157.6, 146.4, 146.2, 141.9, 139.7, 130.1, 129.5, 124.3, 123.5, 120.9, 120.3, 110.3, 106.5, 86.4, 55.6, 51.7, 50.1, 35.8, 33.5, 29.2, 24.08. TFA signals: 163.0 (q, J = 136 Hz), 118.2 (q, J = 1164 Hz). LC-MS (ESI) m/z calcd for C39H47N6O2 [M]+ 631.38, found 631.43. |
| This journal is © The Royal Society of Chemistry 2013 |
