Phage display libraries of differently sized bicyclic peptides†
Inmaculada Rentero
Rebollo
,
Alessandro
Angelini‡
and
Christian
Heinis
*
Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland. E-mail: christian.heinis@epfl.ch
First published on 17th August 2012
Abstract
Phage selections with combinatorial libraries of uniformly sized bicyclic peptides have recently yielded potent and selective binders of several protein targets. In this work we varied in a combinatorial fashion the ring sizes of bicyclic peptides in phage libraries, expecting that they would yield binders with higher affinities and/or more diverse binding motifs that could be affinity matured. 14 new phage peptide libraries of the format Cys-(Xaa)m-Cys-(Xaa)n-Cys (Xaa are random amino acids, m and n = 3, 4, 5 or 6) were generated and cyclized with tris-(bromomethyl)benzene. Affinity selections against the tumor-associated serine protease urokinase-type plasminogen activator yielded bicyclic peptideinhibitors with a large variety of consensus sequences. Several of the identified consensus sequences were exclusively found in bicyclic peptides having defined ring size combinations. Some of these peptides may bind in orientations that allow affinity maturation of non-conserved regions, while others do not. Having available multiple leads isolated from such bicyclic peptide libraries with variable ring sizes could therefore be a great asset for the generation of high affinity binders.
Introduction
Cyclic peptides are an enticing class of molecules for the development of therapeutics. They can act as protein domain mimics, reaching in some cases antibody-like affinity and specificity, while having favorable properties of small molecules such as high stability, access to chemical synthesis and good tissue penetration.1,2 Phage display has emerged as a powerful tool for the high-throughput screening of polypeptides including cyclic peptides.3 Since it relies on ribosomal synthesis, peptides are expressed as linear polymers and cyclization can be achieved by flanking the randomized segment with a pair of cysteine residues, which form a disulfide bridge upon oxidation. Recently, a new strategy has been developed to generate phage-encoded libraries of bicyclic peptides.4 In brief, random peptides containing three cysteine residues are displayed on phage and reacted with the trivalent thiol-reactive compound tris-(bromomethyl)benzene (TBMB) (Fig. 1A). TBMB reacts efficiently and selectively with the cysteine side chains under mild conditions and yields bicyclic peptide structures.5 Affinity selections against plasma kallikrein, cathepsin G and, more recently, urokinase-type plasminogen activator (uPA) yielded highly selective inhibitors in the nanomolar to sub-nanomolar range.4,6,7 These bicyclic inhibitors had higher binding affinities than monocyclic ones previously generated to the same targets.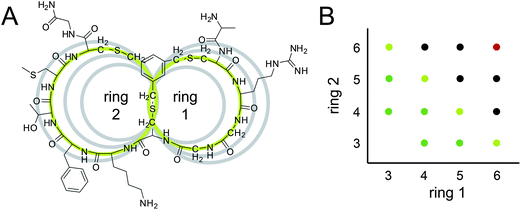 | ||
| Fig. 1 Bicyclic peptide phage libraries with different ring sizes. (A) Chemical structure of a representative bicyclic peptide (UK368) with green rings of 3 and 4 amino acids. For size comparison, the peptide rings with 3, 4, 5 and 6 variable amino acids are shown as grey circles. (B) Overview of the libraries. Indicated on the axes is the number of variable amino acids in the two rings of the bicyclic peptides. Libraries that were cloned in this work are indicated with light green (library A), green (library B) and black dots (library C). The 6 × 6 library indicated with a red dot was cloned previously. | ||
Loop length diversity is already an acknowledged component for the engineering of molecular recognition surfaces in protein scaffolds or cyclic peptides. Length plays an important role in the diversity of complementarity-determining regions of natural antibodies, and it is the primary determining factor for their conformations. Insertion of length diversity in synthetic antibody libraries allowed higher binding affinities to be reached.8 Similar results were obtained when the length of the binding loops was varied in other protein scaffolds such as the 10th type III domain of human fibronectin (Fn3).9 Furthermore, conformational diversity, given by loop length variability, was sufficient to compensate for restricted chemical diversity when screening Fn3 libraries having loops consisting of only Tyr and Ser residues.10 Studies on cyclic peptide libraries suggested that the tightest binders are more likely to be identified by screening multiple libraries with variable loop length since targets often have preferences for specific peptide constraints.11,12
So far, only macrocycle libraries having two rings of equal size have been screened by phage display.4,6,7 Most affinity selections have been performed with a library having two equal rings, each containing six random amino acids (abbreviated as 6 × 6 bicyclic peptides).4,6 Phage panning against the serine protease plasma kallikrein yielded potent inhibitors.4 Comparison of the peptide sequences revealed three consensus regions present in the first or second peptide loop. To affinity mature these inhibitors, three semi-randomized libraries were created, each having one of the three consensus sequences in one loop and six random amino acids in the other loop. Several improved inhibitors with Kis as low as 2 nM were obtained, wherein all improved clones were derived from only one of the three libraries.4 In selections against uPA, two classes of bicyclic peptides were isolated, the major one with a consensus sequence in the first peptide ring, and the minor one with a consensus sequence around the middle cysteine.6 The best inhibitor (UK18) showed a Ki of 53 nM. In this case, affinity maturation attempts with libraries based on either of the consensus sequences could not improve the potency beyond the one of the best inhibitor.6 These results suggested that some peptide leads are more suited for the affinity maturation than others and that it is of advantage to have several consensus sequences as starting points.
In this work, we aimed at generating libraries of bicyclic peptides with different combinations of ring sizes in order to find more potent inhibitors and/or a larger diversity of binding motifs that could be used as constant regions in affinity maturation libraries. In a recent study, the size of bicyclic peptides was reduced to modulate their specificity. These bicyclic peptides having slightly smaller rings (5 instead of 6 randomized amino acids per ring, termed 5 × 5 peptides) bound tightly to the serine protease plasma kallikrein, inhibiting the human and murine orthologs but not any human paralogous proteases.7 Herein, we generated bicyclic peptide phage libraries with combinations of differently sized macrocyclic rings. Specifically, we cloned 14 phage peptide libraries of the format Cys-(Xaa)m-Cys-(Xaa)n-Cys, wherein the number ‘m’ and ‘n’ of random amino acids between the cysteine residues was 3, 4, 5 or 6 (Fig. 1). The libraries were subjected to affinity selections either in groups or all together against the cancer-associated protease uPA.
Results and discussion
Phage-encoded bicyclic peptide libraries with variable ring sizes
The 14 bicyclic peptide phage libraries were cloned using degenerate primers that allow all 20 amino acids in the randomized positions. The libraries are termed according to the number of random amino acids per peptide ring. For example, the library 3 × 4 contains bicyclic peptides with 3 and 4 random amino acids in the first and second ring, respectively (Fig. 1A). The library 6 × 6 already existed and was not newly cloned (Fig. 1B, indicated as a red dot). Libraries with similar numbers of randomized amino acid positions were pooled as follows: bicyclic peptides of the format 3 × 4, 4 × 3, 4 × 4, 3 × 5 and 5 × 3 formed the library A (7 and 8 random amino acids; Fig. 1B, light green dots), those of the format 3 × 6, 6 × 3, 4 × 5 and 5 × 4 formed the library B (9 random amino acids; Fig. 1B, green dots) and those of the format 4 × 6, 6 × 4, 5 × 5, 5 × 6, 6 × 5 formed the library C (10 and 11 random amino acids; Fig. 1B, black dots). The diversities of the libraries were quantified by counting the number of bacterial colonies formed after transformation and were relatively small (between 107 and 5 × 108 cfu). Sequencing clones from libraries A, B and C showed that peptides with all possible combinations of ring sizes were represented (Table 1).| Library | Loop length | Before selection | 2nd round | 3rd round |
|---|---|---|---|---|
| A | 3 × 4 | 7 | 3 | 12 |
| 4 × 3 | 2 | 1 | 0 | |
| 4 × 4 | 8 | 4 | 7 | |
| 3 × 5 | 10 | 1 | 1 | |
| 5 × 3 | 6 | 3 | 1 | |
| B | 3 × 6 | 4 | 1 | 0 |
| 6 × 3 | 1 | 2 | 0 | |
| 4 × 5 | 15 | 12 | 8 | |
| 5 × 4 | 8 | 1 | 0 | |
| C | 4 × 6 | 10 | 1 | 5 |
| 6 × 4 | 4 | 4 | 6 | |
| 5 × 5 | 13 | 2 | 0 | |
| 5 × 6 | 3 | 1 | 0 | |
| 6 × 5 | 7 | 3 | 1 |
Phage selections of bicyclic peptides against a serine protease
Phage selections were performed against the human serine protease uPA. The three libraries A, B and C modified with TBMB5 were individually subjected to affinity selections. In parallel, a fourth selection was performed mixing the three libraries with the previously generated 6 × 6 library (size: 4 × 109 clones),4 in order to allow competition among all the bicyclic peptide formats. Biotinylated uPA was immobilized on magnetic streptavidin beads in the first round and on magnetic neutravidin beads in the second round to prevent the enrichment of streptavidin or neutravidin binders. Negative controls in which the libraries were panned against streptavidin- (first round) or neutravidin- (second round) coated beads allowed the quantification of phage that bound specifically to uPA. Already in the first round of selection, the number of phage isolated against uPA was 10-fold higher compared to the negative control selection (Fig. 2A). The enrichment over the negative control rose to 104-fold in the second round. Similar enrichments were observed for all libraries, suggesting that all contained a large portion of uPA-specific bicyclic peptide binders.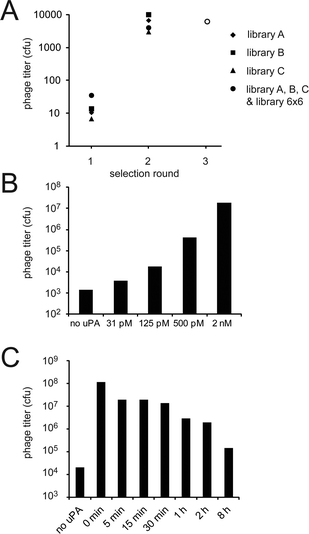 | ||
| Fig. 2 Number of phage clones isolated from the various libraries in the 3 rounds of selection. (A) Enrichment factors found for the different selections (number of phage captured in the presence of uPA divided by the number of phage isolated in the absence of uPA). The enrichment factors of 103 to 104 obtained for all libraries indicate that uPA-specific peptides were isolated. In the third round, phage isolated in all the four individual second round selections were pooled, amplified and together subjected to a third round of panning towards uPA. (B) Phage selections performed with different concentrations of uPA (third round of panning). The numbers of isolated infective phage particles are indicated. (C) Phage selections with competitive ligand (UK18) (third round of panning). After binding of phage to immobilized uPA, the competitive inhibitor UK18 (Ki = 53 nM)6 was added for different incubation time periods (indicated on the abscissa in the graph) to prevent re-binding of weak ligands. The numbers of isolated infective phage particles are indicated. | ||
Consensus sequences of bicyclic peptides isolated after two rounds of selection
Sequencing of 48 peptides isolated from the three libraries revealed a total of six different consensus sequences (Fig. 3). One of the consensus sequences (S/TAR) was found in all the three libraries and in a total of 22 different peptides. This sequence appeared in either the first or second peptide ring, and was found in peptide rings with 4, 5 and 6 but not 3 amino acids length (Fig. 3). The conformational constraint imposed in a loop with 3 amino acids might prevent the tri-peptide from binding to uPA. For the other five consensus sequences, bicyclic peptides within each group had in most cases the same or similar ring sizes. In peptides isolated from library A, two consensus sequences were identified, the first one being present in the second 4-amino acid ring of 3 × 4 and 4 × 4 peptides (K/RFSX; ‘X’ represents any amino acid). The second consensus sequence was found exclusively in bicyclic peptides having the 5 × 3 format (S/TI/LR![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) PSF
PSF![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ). In peptides isolated from library B, a consensus sequence was found in the second ring of 4 × 5 bicyclic peptides (TKY/FTL/M). In selections with library C, a consensus sequence was found in bicyclic peptides of the 6 × 4 format (
). In peptides isolated from library B, a consensus sequence was found in the second ring of 4 × 5 bicyclic peptides (TKY/FTL/M). In selections with library C, a consensus sequence was found in bicyclic peptides of the 6 × 4 format (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) NXYYSX
NXYYSX![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) T/S). In the fourth experiment in which all libraries as well as the library 6 × 6 were mixed and subjected to selections, none of the bicyclic peptide formats was enriched over the others. However, the more ubiquitous S/TAR motif, which can be accommodated in different ring sizes, predominated in this selection.
T/S). In the fourth experiment in which all libraries as well as the library 6 × 6 were mixed and subjected to selections, none of the bicyclic peptide formats was enriched over the others. However, the more ubiquitous S/TAR motif, which can be accommodated in different ring sizes, predominated in this selection.
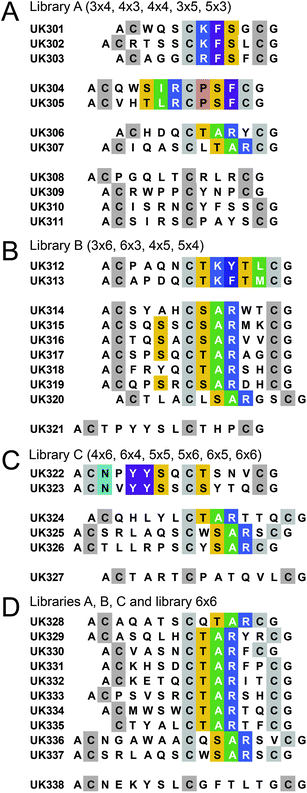 | ||
| Fig. 3 Bicyclic peptides isolated after two rounds of affinity selection. Libraries with similar numbers of randomized amino acids were pooled and subjected to selections with uPA. (A) Library A (7 or 8 randomized amino acids). (B) Library B (9 randomized amino acids). (C) Library C (10 or 11 randomized amino acids). (D) Library A, B and C as well as library 6 × 6. Sequence similarities are highlighted with colors. | ||
Third round of panning applying more stringent selection conditions
Phage clones isolated after two rounds of panning in the four independent selections were mixed together and subjected to a third round (Fig. 2A). To isolate only the tightest-binding peptides, we applied more stringent panning conditions. In one set of experiments, phage was incubated with lower concentrations of biotinylated uPA (ranging from 2 nM to 31 pM) before capturing them on streptavidin-coated beads. At lower concentrations, the number of captured phage was gradually reduced (Fig. 2B). In the second set of experiments, the bicyclic peptide UK18, which binds to the active site of uPA with nanomolar affinity, was used to compete off weak binders. As anticipated, fewer phage remained on the beads at longer incubation times (Fig. 2C). This latter experiment indicated that a large portion of the bicyclic peptides isolated in the second round of selection was binding to the active site of uPA.Consensus sequences of bicyclic peptides isolated after three rounds of selection
Sequencing of 149 clones isolated in the third selection round showed a smaller but still high sequence diversity (50 different peptides) and significantly stronger consensus sequences (Fig. 4). Two of the consensus sequences derived from the 6 × 6 library were exactly the same as found in previous selections against uPA (Fig. 4, top two consensus sequences). Several peptides contained again the tri-peptide motif (S/TAR) that was already found in different bicyclic peptide formats after the second round of panning. This consensus sequence was flanked by specific amino acids that likely increase the binding affinity. A strong consensus covering 9 amino acid positions (CTA/VRTCPAT/SXVL/M![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ) and containing the TAR motif was preferentially found in the 4 × 6 bicyclic peptide format with one exception, a 4 × 4 bicyclic peptide (UK364). Another consensus sequence also containing the TAR motif covered 6 amino acid positions (TAR
) and containing the TAR motif was preferentially found in the 4 × 6 bicyclic peptide format with one exception, a 4 × 4 bicyclic peptide (UK364). Another consensus sequence also containing the TAR motif covered 6 amino acid positions (TAR![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) PQS) and was found in bicyclic peptides of the 4 × 4 and 6 × 4 formats. A strong consensus sequence not containing the TAR motif was found in 10 different peptides all having the 3 × 4 format (
PQS) and was found in bicyclic peptides of the 4 × 4 and 6 × 4 formats. A strong consensus sequence not containing the TAR motif was found in 10 different peptides all having the 3 × 4 format (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) XGG
XGG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) K/RF/YT/SL/M
K/RF/YT/SL/M![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ).
).
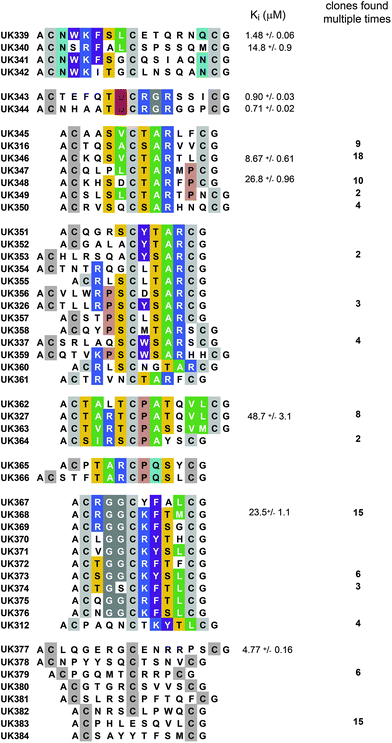 | ||
| Fig. 4 Bicyclic peptides isolated after three rounds of affinity selection. Sequence similarities are highlighted in color. The inhibitory activities (Kis) of several TBMB-cyclized peptides are indicated (average values of at least three measurements). | ||
Synthetic bicyclic peptides of several clones were chemically synthesized. All of them inhibited uPA but none was more potent than the previously isolated UK18 (Fig. 4). The most potent inhibitors isolated here, UK343 and UK344, were derived from the 6 × 6 library and showed Kis of 0.90 and 0.71 μM, respectively. It is likely that the 6 × 6 library had yielded the best binders because of the larger number of randomized amino acid positions and/or the larger number of different clones in the library (4 × 109 clones versus 107 to 5 × 108 clones in the libraries A, B and C). Although more potent inhibitors could not be isolated from the naïve libraries, the numerous binding motifs identified provide more starting points for future affinity maturation attempts. Of particular interest are the bicyclic peptide formats with large rings that appear, based on the consensus sequences, not to be optimized in all of the amino acid positions.
Conclusion
Phage panning experiments with bicyclic peptides having different ring sizes yielded more diverse consensus sequences than previously found in selections with a 6 × 6 bicyclic peptide phage library. The bicyclic peptideinhibitors with unrelated consensus sequences are presumably interacting differently with the active site of uPA. Some of these peptides may bind in orientations that allow affinity maturation of non-conserved regions while others do not. Having available multiple leads isolated from such bicyclic peptide libraries with variable ring sizes could therefore be a great asset for the generation of high affinity binders. It is likely that other multicyclic peptide structures evolved by phage display such as cysteine knots13 or other disulfide-constrained mini-proteins14–16 would similarly benefit from variation of the peptide ring size.Experimental section
Library generation
All primers used for library cloning, as well as the vector 21tet(5), are described in the ESI†. Phage libraries were created by inserting DNA sequences encoding the semi-random peptide sequences (Ala-Cys-(Xaa)m-Cys-(Xaa)n-Cys, m and n ranged from 3 to 6 amino acids), the linker Gly-Gly-Ser-Gly, and the disulfide-free domains D1 and D2 into the phage vector 21tet(5). The insert was step-wise created in two consecutive PCR reactions. First, the genes of D1 and D2 were PCR amplified with the two primers prepcr and sfi2notfo using the vector fdg3p0ss21 (ref. 17) as a template. Second, the DNA encoding the random peptides was appended in a PCR reaction using primers of the type 5′-TATGC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ATGGCAGCATGC(NNK)mTGC(NNK)nTGTGGCGGTTCTGGCGCTG-3′ (m, n = 3, 4, 5 or 6) and the primer sfi2notfo. The PCR products were digested with SfiI (underlined in primers) and ligated into SfiI-digested vector 21tet(5). For each set of libraries (A, B and C), 23 μg and 7 μg of SfiI-digested vector and PCR products respectively were ligated and electroporated into E. coli TG1 cells. An equal amount of each insert was added into the ligation reaction to a total of 7 μg. After electroporation, cells were incubated for 1 hour in 2YT at 37 °C and plated on large (20 cm diameter) chloramphenicol (30 μg ml−1) 2YT plates. Colonies were scraped off the plates with 2YT media, supplemented with 10% v/v glycerol and stored at −80 °C.
ATGGCAGCATGC(NNK)mTGC(NNK)nTGTGGCGGTTCTGGCGCTG-3′ (m, n = 3, 4, 5 or 6) and the primer sfi2notfo. The PCR products were digested with SfiI (underlined in primers) and ligated into SfiI-digested vector 21tet(5). For each set of libraries (A, B and C), 23 μg and 7 μg of SfiI-digested vector and PCR products respectively were ligated and electroporated into E. coli TG1 cells. An equal amount of each insert was added into the ligation reaction to a total of 7 μg. After electroporation, cells were incubated for 1 hour in 2YT at 37 °C and plated on large (20 cm diameter) chloramphenicol (30 μg ml−1) 2YT plates. Colonies were scraped off the plates with 2YT media, supplemented with 10% v/v glycerol and stored at −80 °C.
Phage selections
Phage were produced and the peptides modified with TBMB as described previously.4 Chemically modified phage were dissolved in 3 ml washing buffer (10 mM Tris–Cl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM CaCl2), and blocked by addition of 1.5 ml washing buffer containing 3% w/v BSA and 0.3% v/v Tween 20, for 30 min. In parallel, 5 μg of biotinylated uPA was immobilized by incubation with 40 μl magnetic streptavidin beads (Dynabeads M-280 Streptavidin, Invitrogen, Lucerne, Switzerland). The beads with uPA as well as 40 μl magnetic streptavidin beads without uPA were blocked for 30 minutes in separate tubes with blocking buffer (washing buffer containing 1% w/v BSA and 0.1% v/v Tween 20). The 4.5 ml blocked phage were distributed equally to the two tubes and incubated for 30 minutes on a rotating wheel. The beads were washed eight times with washing buffer containing 0.1% v/v Tween 20 and twice with washing buffer. The bound phage were eluted by incubation with 100 μl of 50 mM glycine pH 2.2 for 5 min. Eluted phage were transferred to 50 μl of 1 M Tris–Cl, pH 8.0 for neutralization, and incubated with 30 ml of exponentially growing TG1 cells (OD600 = 0.4) for 90 min at 37 °C. The cells were pelleted by centrifugation, dissolved in 1 ml 2YT, spread on 2YT plates containing chloramphenicol (30 μg ml−1) and incubated at 37 °C overnight. The colonies were scraped off the plates with 2YT media, supplemented with 10% v/v glycerol and stored at −80 °C. In the second round of panning, uPA was immobilized on magnetic neutravidin beads. The beads were prepared by coating tosyl-activated beads (Dynabeads M-280 Tosylactivated, Invitrogen) with NeutrAvidin (Pierce, Rockford, IL, USA) following the manual of the manufacturer. In the third selection round, phage isolated in the second round of the four independent selections were produced separately, mixed at equal numbers, blocked as described above and panned together against uPA. In the in-solution capture procedure, biotinylated uPA (10 nM) was blocked, and incubated at final concentrations ranging from 31 pM to 2 nM with 2 ml of blocked phage. After incubation for 45 min at room temperature with rotation, the phage/uPA–biotin complexes were captured by incubation for 7 min with 40 μl blocked streptavidin beads. The washes and the elution were performed as described above. In the competitive capture procedure, 120 ng biotinylated uPA was immobilized on 30 μl streptavidin beads, washed twice and incubated in 300 μl blocking buffer for 30 min. They were subsequently added to 2 ml blocked phage and incubated for a minimum of 30 minutes at room temperature with rotation. Bicyclic peptide UK18 was added at different time points to a concentration of 9 μM. The phage were washed, eluted and propagated as described above.Chemical synthesis of bicyclic peptides
Peptides were synthesized by standard solid-phase peptide synthesis using Fmoc-protected amino acids and Rink amide AM resin (see Experimental procedures, ESI†). Peptides were eluted under reducing conditions and partially purified by precipitation. Crude peptide (0.5 mM) was reacted with TBMB (1 mM) in 80% aqueous buffer (20 mM NH4HCO3, 5 mM EDTA, pH 8.0) and 20% CH3CN for 1 h at 30 °C. The product was purified by reversed-phase chromatography on a C18 column (XBridge BEH300 Prep 5 μm, Waters, Milford, MA, USA) using a linear gradient with a mobile phase composed of eluant A (99.9% v/v H2O, 0.1% v/v TFA) and eluant B (94.9% v/v CH3CN, 5% v/v H2O and 0.1% v/v TFA) and a flow rate of 20 ml min−1. Pure bicyclic peptides were lyophilized and dissolved in H2O. The molecular mass was confirmed by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry.Determination of inhibitory activity of selected bicyclic peptides
Human uPA at a final concentration of 4 nM was incubated with different concentrations of bicyclic peptides and 100 μM fluorogenic substrate (Z-Gly-Gly-Arg-AMC, Bachem, Bubendorf, Switzerland) and residual activity measured at 25 °C for 30 minutes. The reactions were performed in volumes of 150 μl and a buffer containing 10 mM Tris–Cl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM CaCl2, 0.1% w/v BSA, 0.01% v/v Triton-X100 and 5% v/v DMSO at 25 °C. Fluorescence intensity was measured with a Spectramax Gemini fluorescence plate reader (excitation 355 nm, emission 480 nm, Molecular Devices). The reactions were performed in triplicate. The final Ki was calculated using the Cheng–Prusoff equation,18 wherein the kinetic constant Km for the hydrolysis of Z-Gly-Gly-Arg-AMC by human uPA was determined by the standard Michaelis–Menten equation as previously described.6Acknowledgements
The financial contribution from the Swiss National Science Foundation (SNSF Professorship PP00P3_123524/1 to C.H.) is gratefully acknowledged. The authors would like to thank all the members of the laboratory for helpful discussions.References
- R. C. Ladner, A. K. Sato, J. Gorzelany and M. de Souza, Drug Discovery Today, 2004, 9, 525–529 CrossRef CAS.
- M. Katsara, T. Tselios, S. Deraos, G. Deraos, M.-T. Matsoukas, E. Lazoura, J. Matsoukas and V. Apostolopoulos, Curr. Med. Chem., 2006, 13, 2221–2232 CrossRef CAS.
- F. Uchiyama, Y. Tanaka, Y. Minari and N. Tokui, J. Biosci. Bioeng., 2005, 99, 448–456 CrossRef CAS.
- C. Heinis, T. Rutherford, S. Freund and G. Winter, Nat. Chem. Biol., 2009, 5, 502–507 CrossRef CAS.
- P. Timmerman, J. Beld, W. C. Puijk and R. H. Meloen, ChemBioChem, 2005, 6, 821–824 CrossRef CAS.
- A. Angelini, L. Cendron, S. Chen, J. Touati, G. Winter, G. Zanotti and C. Heinis, ACS Chem. Biol., 2012, 7, 817–821 CrossRef CAS.
- V. Baeriswyl, H. Rapley, L. Pollaro, C. Stace, D. Teufel, E. Walker, S. Chen, G. Winter, J. Tite and C. Heinis, ChemMedChem, 2012, 7, 1173–1176 CrossRef CAS.
- C. V. Lee, W. C. Liang, M. S. Dennis, C. Eigenbrot, S. S. Sidhu and G. Fuh, J. Mol. Biol., 2004, 340, 1073–1093 CrossRef CAS.
- B. J. Hackel, A. Kapila and K. D. Wittrup, J. Mol. Biol., 2008, 381, 1238–1252 CrossRef CAS.
- A. Koide, R. N. Gilbreth, K. Esaki, V. Tereshko and S. Koide, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6632–6637 CrossRef CAS.
- L. L. Bonnycastle, J. S. Mehroke, M. Rashed, X. Gong and J. K. Scott, J. Mol. Biol., 1996, 258, 747–762 CrossRef CAS.
- E. Koivunen, B. Wang and E. Ruoslahti, Biotechnology, 1995, 13, 265–270 CrossRef CAS.
- G. P. Smith, S. U. Patel, J. D. Windass, J. M. Thornton, G. Winter and A. D. Griffiths, J. Mol. Biol., 1998, 277, 317–332 CrossRef CAS.
- H. J. Chang, H. J. Hsu, C. F. Chang, H. P. Peng, Y. K. Sun, H. M. Yu, H. C. Shih, C. Y. Song, Y. T. Lin, C. C. Chen, C. H. Wang and A. S. Yang, Structure, 2009, 17, 620–631 CrossRef CAS.
- F. Zoller, U. Haberkorn and W. Mier, Molecules, 2011, 16, 2467–2485 CrossRef CAS.
- T. Phan, H. D. Nguyen, H. Goksel, S. Mocklinghoff and L. Brunsveld, Chem. Commun., 2010, 46, 8207–8209 RSC.
- I. Kather, C. A. Bippes and F. X. Schmid, J. Mol. Biol., 2005, 354, 666–678 CrossRef CAS.
- Y. Cheng and W. H. Prusoff, Biochem. Pharmacol., 1973, 22, 3099–3108 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2md20171b |
| ‡ Present address: David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA. |
| This journal is © The Royal Society of Chemistry 2013 |
