Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Adjustment of carbon fluxes to light conditions regulates the daily turnover of starch in plants: a computational model†
Alexandra
Pokhilko
a,
Anna
Flis
b,
Ronan
Sulpice
c,
Mark
Stitt
b and
Oliver
Ebenhöh
*ad
aInstitute for Complex Systems and Mathematical Biology, University of Aberdeen, UK
bMax Planck Institute of Molecular Plant Physiology, Germany
cNational University of Galway, Plant Systems Biology Lab, Plant and AgriBiosciences Research Centre, Botany and Plant Science, Galway, Ireland
dCluster of Excellence on Plant Sciences (CEPLAS), Heinrich-Heine-University, Universitätsstraße 1, D-40225 Düsseldorf, Germany. E-mail: oliver.ebenhoeh@hhu.de
First published on 17th December 2013
Abstract
In the light, photosynthesis provides carbon for metabolism and growth. In the dark, plant growth depends on carbon reserves that were accumulated during previous light periods. Many plants accumulate part of their newly-fixed carbon as starch in their leaves in the day and remobilise it to support metabolism and growth at night. The daily rhythms of starch accumulation and degradation are dynamically adjusted to the changing light conditions such that starch is almost but not totally exhausted at dawn. This requires the allocation of a larger proportion of the newly fixed carbon to starch under low carbon conditions, and the use of information about the carbon status at the end of the light period and the length of the night to pace the rate of starch degradation. This regulation occurs in a circadian clock-dependent manner, through unknown mechanisms. We use mathematical modelling to explore possible diurnal mechanisms regulating the starch level. Our model combines the main reactions of carbon fixation, starch and sucrose synthesis, starch degradation and consumption of carbon by sink tissues. To describe the dynamic adjustment of starch to daily conditions, we introduce diurnal regulators of carbon fluxes, which modulate the activities of the key steps of starch metabolism. The sensing of the diurnal conditions is mediated in our model by the timer α and the “dark sensor” β, which integrate daily information about the light conditions and time of the day through the circadian clock. Our data identify the β subunit of SnRK1 kinase as a good candidate for the role of the dark-accumulated component β of our model. The developed novel approach for understanding starch kinetics through diurnal metabolic and circadian sensors allowed us to explain starch time-courses in plants and predict the kinetics of the proposed diurnal regulators under various genetic and environmental perturbations.
Introduction
Starch is the main carbohydrate transiently stored in the leaves of many plants. It is accumulated during the day and consumed at night, providing reduced carbon for plants in the absence of light. The daily rhythms of starch accumulation and degradation result from coordination of several metabolic pathways of carbon metabolism in plants. In the day-time plants fix carbon from atmospheric CO2 in the Calvin–Benson cycle (CBC) in chloroplasts. The resulting triose-phosphates (TP) can enter one of the two main pathways of carbon partitioning: either they remain in the chloroplast to be converted into starch or they are exported to the cytosol for the synthesis of sucrose, which can be used as an energy source or imported and consumed by growing sink tissues. Starch acts as a carbon reserve. In the night starch is degraded and the degradation products are exported to the cytosol, where they are used for sucrose synthesis, ensuring that plants have sufficient carbon and energy to keep growing.For many plant species, transitory starch is the main source of carbon at night. Therefore its synthesis and degradation are tightly regulated to avoid periods of starvation. On short days, carbon is partitioned mainly to starch, which is synthesized fast and degraded slowly at night, whereas on long days partitioning is shifted to sucrose, resulting in slow synthesis and fast degradation of starch.1–5 The diurnal dynamics of starch is also quickly adjusted to unexpected changes in environmental conditions. For example, starch degradation instantly slows down when plants are subjected to an unexpected “early dusk”.1 The rate of starch synthesis is also affected by changes in the light conditions, such as for example the introduction of several hours of darkness in the middle of the day.1 The shorter total period of light leads to an accelerated starch accumulation in the second light interval.1 Further, the circadian clock was shown to play an important role in the diurnal regulation of starch metabolism in plants, but the precise mechanisms are still unknown.1,6 These observations lead to several questions.5,6 How do plants sense changes in the environmental conditions such as photoperiod? What are the mechanisms that allow plants to quickly adjust the rates of starch accumulation and degradation to unpredictable changes in the conditions? What is the role of the circadian clock in the regulation of starch turnover?
Because the answers to these questions are mainly unknown, we decided to take a mathematical modelling approach to explore possible mechanisms governing the diurnal regulation of starch. The model therefore comprises three modules: (i) the circadian clock, (ii) metabolism, (iii) a regulatory module linking the clock and metabolism. We here consider the clock as an independent regulatory circuit providing regulatory input to the other modules and model the clock as described in ref. 7 (Fig. S1A, ESI†). A key requirement of the metabolic module is to describe the regulation of carbon partitioning correctly. Consequently it must contain carbon fixation, starch synthesis and degradation in the chloroplast, sucrose synthesis in the cytosol and sucrose consumption in sink tissues (Fig. 1). Starch and sucrose synthesis are directly linked because they compete for the same precursor substrate triose-phosphate.8 Another important experimental observation related to possible diurnal mechanisms is that carbohydrate metabolism in source tissues is regulated by demand from sinks.9,10 While some of the included metabolic pathways, in particular carbon fixation, sucrose synthesis and starch degradation were modelled before,11–13 other processes (such as consumption of sucrose by sinks and the diurnal mechanisms of the metabolic coordination) were not described by mathematical models. For the development of the metabolic module we therefore combined the essential pathways of carbon metabolism, using and simplifying the existing models whenever possible, to provide a correct description of the available data on the kinetics of starch metabolism in plants. A key question addressed by the model is how these metabolic processes are regulated and controlled. Therefore, the emphasis in this work lies on the third module describing diurnal regulatory mechanisms, linking metabolism to the clock and environmental cues (Fig. 2). The overall aim of the comprehensive model is to describe existing data on starch turnover under multiple genetic and environmental perturbations and explain the dynamic adjustment to changing daily light conditions. In the following, we explain in detail how the model assumptions, in particular those governing the regulatory module, were derived from experimental facts.
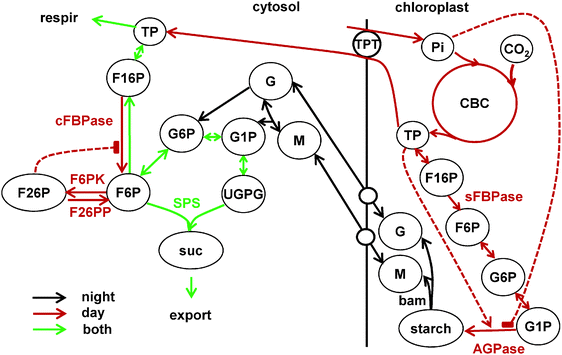 | ||
| Fig. 1 Main reactions of carbon partitioning between chloroplast and cytosol in source tissues, included in the model. Short names of the key enzymes, regulating the fluxes are shown near the arrows. Metabolite names are abbreviated in circles. Solid lines show reactions and dashed lines the key allosteric regulations. Reactions of carbon fixation in the Calvin–Benson cycle (CBC) leading to starch synthesis in the chloroplast during the day are shown by red lines with two key irreversible steps determined by stromal fructose bisphosphatase (sFBPase) and ADPglucose phosphatase (AGPase), which is allosterically regulated by triose-phosphates (TP) and inorganic phosphate (Pi). Day-time reactions also include TP export to the cytosol via TPT for sucrose synthesis. The reactions of the sucrose synthesis pathway are active in both day and night and are shown by green lines. The key irreversible steps in the sucrose synthesis pathway are conducted by cytosolic FBPase (cFBPase) and sucrose-phosphate synthase (SPS, combined with sucrose-phosphate phosphatase, SPP). cFBPase is involved in sucrose synthesis only during day, when TP is supplied by the CBC. It is allosterically inhibited by F26P, which is synthesised from F6P by the bisfunctional enzyme F6P kinase/F26P phosphatase (F6PK/F26PP). Main reactions of starch degradation during the night are shown by black lines. In the chloroplast only β amylase (bam) is shown for clarity. Isoamylase (ISA) and disproportionating enzyme 1 (DPE1) reactions are omitted from the scheme. Maltose (M) and glucose (G) are exported from chloroplasts to the cytosol through the respective transporters MEX1 and GLUT and converted to the glucose phosphates G1P and G6P by DPE2, α-glucan phosphorylase (PHS) and hexokinase. Reactions downstream of hexose phosphates in the cytosol (G1P, G6P, F6P) are active both during day and night, with SPS being a key irreversible step leading to generation of sucrose, which is further exported and consumed by sink tissues. Photosynthetically derived carbon in the form of TP is also consumed by respiration. Additional diurnal regulation of the above reactions is presented separately in Fig. 2. | ||
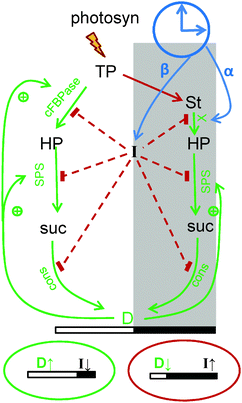 | ||
| Fig. 2 Schematic presentation of diurnal regulation of carbon metabolism in the model. Carbon fluxes are modulated via changes in the rates of the key steps of metabolic pathways, which are presented in more details in Fig. 1. During the day triose-phosphates (TP), produced by photosynthesis in source tissues are partitioned between starch (St) and sucrose (suc) (via hexose-phosphates, HP). In the night (shadowed area) sucrose is synthesized from degraded starch. Sucrose is consumed by growing sink tissues (cons), both in the light and the dark. The scheme shows the diurnal regulation of carbon fluxes by the sensor of carbon deficit I and by the sink's demand D. Decrease of I and increase of D on long days (green oval below the picture) slows down starch synthesis through accelerating sucrose synthesis by activation of the key enzymes cFBPase and SPS (SPS is regulated directly, and cFBPase through F26PP, see Fig. 1). Decrease of I also activates consumption of sucrose by sinks, thus increasing sink demand D and further accelerating carbon flux through cFBPase and SPS. On short days (red oval below the picture) the carbon deficit sensor I is increasing and D is decreasing, resulting in a redistribution of carbon flux away from sucrose towards starch. The induction of glucose-phosphate translocator 2 (GPT2) in the chloroplast membrane further accelerates starch synthesis under carbon-limited conditions (GPT2 is not shown for clarity). In the night starch degradation is set at dusk according to the levels of starch and timer α through the model component X, which reflects the activity of the starch-degrading enzymatic complex. In addition, I levels at dusk downregulate starch degradation by inhibiting X, thus providing a slow delayed feedback to adapt plants to the carbon deficit conditions experienced in the previous day. Green colored arrows on the scheme show processes related with stimulation of carbon utilization and red colored connections show processes related with reduction of carbon utilization. | ||
Building a mathematical model of such a complex molecular network, including metabolic, genetic and regulatory components, is a difficult task, in particular considering that our knowledge of the exact mechanisms of the diurnal regulation of starch kinetics is incomplete. Constructing a model based on limited knowledge necessarily involves setting up new hypotheses, making sophisticated guesses and a considerable degree of trial and error. The design process is naturally guided by the key experimental features which the model should be able to reproduce, while care must be taken that the hypotheses underlying the model formulation are based on experimental findings and supported by plausible arguments. The model we present here contains a number of hypothesised key components, which together generate the key properties of the diurnal regulation of starch metabolism, but for which conclusive experimental evidence does not always exists. Thus, we first integrated existing knowledge about the possible diurnal mechanisms and then followed a top-down approach to derive molecular hypotheses from general observations. In a following bottom-up approach, we derived properties of the hypothesised components, which allow experimental validation of our assumptions and will eventually support the future identification of the key regulatory elements.
The dynamic adjustment of carbon partitioning suggests the existence of regulatory mechanisms, which sense and respond to light conditions and the time of the day. The apparently co-regulated activity of key metabolic enzymes of the sucrose synthesis pathway8 further indicate the existence of universal regulators which can shift metabolic fluxes in a coordinated fashion according to external conditions. In our model, we include two sensors α and β, where α serves as a timer measuring the circadian time and β senses the carbon status and additionally underlies circadian regulation. These sensors integrate metabolic and circadian cues and control the activities of the global regulators D, I and X. Here, D is an activator responsible for the observed shift of carbon partitioning towards demand pathways9,10 and I is an inhibitor limiting carbon consumption when supply is low, with SnRK1/AMPK/SNF1-related kinase being a suitable candidate for I.14,15 We show by comparison of the modelled characteristics of β with published and novel data that the β subunit of SnRK1 (AKINβ1), which is regulated by both clock and light,16,17 is a suitable candidate for β. The data also suggest that the amount of stored transitory starch is measured by some means, probably through the diurnal modulation of enzymatic complexes on the surface of starch granules.6,18 This is described in our model through the regulator of starch breakdown X, which integrates information on the amount of stored starch and the time of the day.
In summary, we created a comprehensive model of the diurnal regulation of carbon metabolism in plants, combining the essential enzymatic steps with diurnal regulation of metabolism by light and the clock. After building the model and connecting it with the most recent model of the circadian clock,7 we demonstrated a good match of the model simulations to published and novel data on the response of the starch kinetics to various perturbations, such as the duration of the light period, changes in the illumination protocol and genetic manipulations. Additionally the model predicted the possible properties of the diurnal regulators of carbon metabolism, such as α and β, which are important for their further molecular characterization. The model also predicts the responses of the system to new perturbations of the daily light cycle such as decreased light intensities at dawn (“early dawn”) or mutations of the clock genes. Our results suggest possible ways to further explore the diurnal regulation of starch metabolism in future experimental studies.
Results
Model structure and regulatory principles
The clock module (Fig. S1A, ESI†) and a metabolic module (Fig. 1) of the model are connected by the regulatory module (Fig. 2) integrating the timed input from the clock and environmental cues to control starch metabolism. In the following, the regulatory principles are briefly outlined, a more detailed description of key features and experimental background are described in Material and methods and in the ESI.†A key model component integrating clock, light and metabolic signals is the global inhibitor I, which reflects the “carbon deficit” the plant is experiencing. Clock and light signals are perceived and processed by the light and dark sensors α and β, respectively. While levels of α are low at dawn and increasing during the day, levels of β are low at dusk and increase during the night. The inhibitor I is induced by high levels of β (short days – environmental cue) and further by carbon stress (metabolic signal), e.g. when carbon is prematurely depleted before the end of the night, as in the “early dawn” experiment (see Results).
The partitioning of newly fixed carbon between sucrose and starch is regulated by carbon availability and demand. In our model, the key enzymes of sucrose synthesis, cFBPase and SPS, as well as sucrose consumption are regulated by the diurnal regulators I and D, where the global inhibitor I reflects the amount of “carbon deficit” (see above) and the activator D reflects the carbon demand of sink tissues. When “carbon deficit” is high (e.g. on short days), I increases and inhibits cFBPase, SPS and sucrose consumption by sink tissues (Fig. 2). In contrast, D increases when carbon is abundant (e.g. on long days), stimulating sucrose synthesis through activation of cFBPase and SPS. Thus, the combined regulation of I and D affects sucrose synthesis (Fig. 2): on long days, activation by D and reduced inhibition by I partitions carbon towards sucrose synthesis whereas on short days inhibition by I and reduced activation by D changes partitioning towards starch synthesis. In the model, pronounced carbon deficit further stimulates partitioning towards starch by the induction of the glucose-phosphate translocator 2 (GPT2) in the chloroplast membrane.19,20 Through incorporation of several parallel mechanisms regulating sucrose synthesis, the model ensures the efficient adjustment of carbon partitioning and hence starch synthesis to the external conditions.
Starch degradation rates are determined by dusk levels of starch and the circadian “timer” α, via the model variable X, which describes the activity of a starch-degrading complex. The level of X and hence the starch degradation rate is set at dusk and is not changing in the darkness. The timer α accumulates during the day and positively regulates the level of X. This leads to a fast starch degradation rate on long days and a slow rate on short days. In order to allow for a correction of degradation rates as a result of starvation in the previous night, starch degradation is additionally inhibited by high levels of I. This long-term adaptation represents a delayed feedback regulation of starch degradation by the previous day conditions. Thus, I integrates clock, light and metabolic signals to inhibit processes related to carbon consumption and stimulate accumulation of starch under “carbon deficit” conditions.
Although there might be several alternative scenarios for the diurnal regulation, our model is in good agreement with both literature data on the diurnal regulation of carbon fluxes (Materials and methods) and all available data on starch timecourses under various perturbations (Results). While the model is also capable of reproducing the experimentally observed levels of intermediate metabolites of carbohydrate metabolism (ESI†), we here put an emphasis on analysing starch turnover under various environmental and genetic perturbations to explore potential regulatory mechanisms underlying the adjustment of starch metabolism to diurnal conditions.
Regulation of starch metabolism by day length
The essential requirement of the model is to correctly predict the diurnal turnover of starch in plants when grown under different photoperiods. We therefore analysed the model with respect to the regulation of starch metabolism by day length.During the day, carbon is partitioned into starch or sucrose, depending on the photoperiod. During short photoperiods, the level of “carbon deficit” I increases (see above) through increase of the dark-accumulated circadian sensor β (Fig. 3C). The increase of I in turn accelerates starch synthesis (Fig. 3A) through reduction of carbon partitioning towards sucrose synthesis (Fig. 3B and Fig. S2C, ESI†). In parallel, the increase in I downregulates consumption of sucrose by growing sinks, thus decreasing demand D (Fig. S2A, ESI†), further increasing starch synthesis rates (Fig. 3A and B). An additional acceleration of starch synthesis is modelled by the induction of GPT2 under short day conditions (Fig. S2B and S6A, ESI†). In contrast, in long days the decrease of I (Fig. S2B, ESI†) and increase of D (Fig. S2A, ESI†) shift carbon partitioning towards sucrose synthesis and reduced starch accumulation (Fig. 3A and B) through the stimulation of SPS and cFBPase. Therefore, the parallel mechanisms via I, D and GPT2 adjust the starch synthesis rate to the duration of the day in our model.
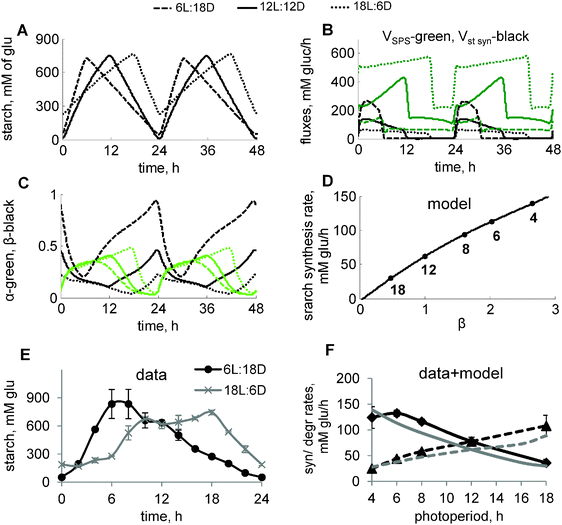 | ||
| Fig. 3 Diurnal regulation of starch kinetics under different photoperiods. (A–D) Model simulations of carbon metabolism in plants grown under varying photoperiods. Diurnal profiles of starch accumulation (A) and carbon fluxes through starch and sucrose synthesis pathways (B), and circadian components α and β (C) under 6L:18D, 12L:12D and 18L:6D photoperiods are shown by dashed, solid and dotted lines, respectively. Concentration of starch is expressed in mM of glucosyl units. Carbon fluxes in B are represented by the rates of SPS (green) and total rate of starch synthesis (black). α and β in C are shown by green and black lines, respectively. (D) Simulated dependence of the starch synthesis rate on the peak level of β at dawn. Data points correspond to different photoperiods, indicated by a number near each data point. β levels for each photoperiod were normalized to the value for 12L:12D. (E) Experimental time-courses of starch in plants, grown under short (6L:18D) and long (18L:6D) photoperiods. Starch levels in chloroplasts were recalculated from mg g−1 FW units to mM using a value of 65 μl g−1 FW for the chloroplast volume.51 (F) Dependence of starch synthesis (solid lines) and degradation (dashed lines) rates on the duration of the day. Rates were calculated based on the difference between end-of-the-day and end-of-the-night levels of starch under various photoperiods. Experimental data and model simulations are shown by black and grey lines respectively. Panels with data points have an additional title (“model” or “data”). | ||
At night, starch is degraded with a rate which increases on long photoperiods.1,2 In our model, this is described through the acceleration of starch degradation by the clock-related “timer” α, which increases with the duration of time spent in the light (Fig. S1B, ESI,† and Fig. 3C). This mechanism, together with regulation of starch synthesis via carbon partitioning, described above, resulted in the adjustment of the starch kinetics to the various photoperiods (Fig. 3A). The model simulations correctly predicted our data on starch turnover under short (6 h light/18 h dark; 6L:18D) and long (18L:6D) days, which demonstrate a drastic change in the starch synthesis and degradation rates under these extremely different durations (3-fold) of the light period (Fig. 3E). Our further experimental measurements across multiple photoperiods demonstrate an almost linear dependence of starch synthesis and degradation rates on the duration of the light period (Fig. 3F). These data are quantitatively reproduced by our model (Fig. 3F). Moreover, our simulations suggest that starch synthesis rates correlate with the level of the circadian sensor of darkness β, which regulates carbon partitioning during the day (Fig. 3D).
Starch kinetics in clock mutants
The clock is known to affect the turnover of starch.1 In particular, mutation of the key clock genes LHY and CCA1 disrupts the normal starch time-course by accelerating the degradation of starch at night in the lhy/cca1 mutant1 (Fig. 4A). The model is in good agreement with these data (Fig. 4B) and explains the observed acceleration of starch degradation in the lhy/cca1 mutant through an increase in the clock-related regulator of starch degradation α (Fig. 4C, grey lines). The model also reproduces the experimentally observed decrease of the starch synthesis rate in the lhy/cca1 mutant (Fig. 4B).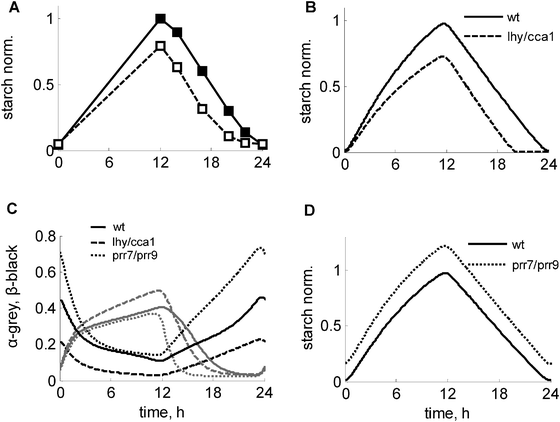 | ||
| Fig. 4 Modulation of the starch kinetics in clock mutants under 12L:12D conditions. (A) Experimental data of starch turnover in wild type and lhy/cca1 mutant, redrawn from ref. 1. (B–D) Simulated kinetics of starch, α and β in the wild type (solid lines), lhy/cca1 (dashed lines) and prr7/prr9 (dotted lines) mutants. C. Simulated profiles of α and β are shown by grey and black lines respectively. Starch levels were normalized to peak starch level in wild type under 12L:12D. Clock mutants were simulated by setting the transcription rates of the corresponding clock genes to 0. | ||
The model suggests an explanation for this counterintuitive experimental observation of a decreased starch synthesis rate in the lhy/cca1 mutant. Intuitively, the premature exhaustion of starch and concomitant carbon starvation at the end of the night in this mutant (ref. 1 and 21, Fig. S2F, ESI†) would be expected to lead to a decrease of sucrose flux and an increase of starch synthesis in the next day (Fig. 3). However, the model suggests that the impairment of the circadian regulation of the dark sensor β in the lhy/cca1 mutant counteracts the effect of the nightly starvation on starch synthesis. Fig. 4C and Fig. S2E (ESI†) illustrate the reduction of β (and hence I) in the lhy/cca1 mutant caused by the impairment of the clock, which makes this mutant less responsive to night starvation. As a consequence, it maintains a relatively high flux towards sucrose synthesis during the day despite starvation during the previous night. This demonstrates that both metabolic and circadian factors are important for a proper adjustment of starch kinetics to environmental conditions.
To explore the influence of the clock on the starch kinetics further, we simulated the effect of the impairment of other clock genes. Another mutant for which diurnal data on starch is available is toc1. The model simulations agree with experimental data, in which a minor effect on the starch kinetics was observed (Fig. S4A, ESI;†ref. 1). Since the mutation of the LHY and CCA1 genes has a strong effect on the diurnal kinetics of starch, we next predicted starch turnover kinetics for the prr7/prr9 double mutant, which is characterized by up-regulated expression levels of both LHY and CCA1 due to the absence of their inhibitors PRR9 and PRR7.22,23 Our model predicts that the prr7/prr9 mutant should have a mild starch-excess phenotype (Fig. 4D), caused by a minor reduction of the starch degradation rate when compared to the wild type (Fig. 4C and D). Also, the model predicts some increase of the starch synthesis rate due to the higher levels of the dark sensor β (Fig. 4C) and hence I (Fig. S2E, ESI†), suggesting slightly higher sensitivity of this mutant to starvation relative to the wild type.
Response of starch metabolism to environmental perturbations
Next we tested the response of the model to sudden perturbations in the light conditions. We first simulated a key experiment, in which plants were subjected to an unexpected early dusk.1Fig. 5A and B show the comparison between starch degradation profiles in plants entrained to normal 12L:12D cycles with plants subjected to a sudden unexpected early dusk. The model simulation reproduced the immediate response of a reduced starch degradation rate. To explore the mechanisms responsible for the adaptation of the starch degradation rate, we compared the simulations of an unexpected “early dusk” with long-term adapted plants grown continuously under short days. Fig. 5A demonstrates (compare black and grey dashed lines) that the end-of-the-night (EN) starch level is predicted to be reduced in unexpected short days compared to adapted plants, which qualitatively agrees with the experimentally observed decrease of EN starch levels.24 The model explains this difference through the higher levels of I and thus higher inhibition of starch degradation in adapted plants compared to plants that are subjected to a sudden “early dusk”. It is worthwhile noting that this reduction in EN starch levels cannot be reproduced by a model without the feedback regulation of the starch degradation by I (Fig. S3, ESI†).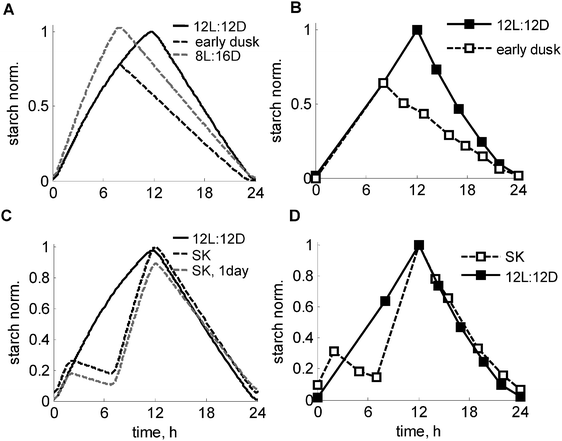 | ||
| Fig. 5 Simulated and experimental time-courses of starch turnover under various light conditions. (A and B) Adjustment of starch degradation to unexpected “early dusk”. Simulated (A) and measured (B) kinetics of starch in the “early dusk” experiment, where control plants (solid line), grown under 12L:12D photoperiod were suddenly exposed to preliminary darkness at 8ZT, 4 h before normal dusk (dashed black line). The grey dashed line in A shows the starch profile in plants entrained to short days (8L:16D) for comparison. The experimental data on panels B are re-plotted from ref. 1. (C and D) Time-courses of starch turnover under a skeleton photoperiod. Simulated (C) and measured (D) profiles of starch in plants, grown under skeleton 2L:5D:5L:12D conditions are shown by black dashed lines, with 12L:12D kinetics shown by solid lines. The experimental data points on F are re-plotted from ref. 1. Simulated kinetics of starch on the first day of the skeleton photoperiod after the transition from 12L:12D condition is shown by the grey dashed line. Starch levels were normalized to the peak level in wild type under 12L:12D. | ||
Another classical perturbation of the daily light cycle is related to applying “skeleton” (SK) photoperiods, where a period of darkness is introduced during the light period.25 The circadian clock was shown to be quite robust under SK photoperiods, even if the period of darkness extends over several hours.25,26 Similarly, the starch degradation rate was shown to be set properly under SK photoperiods.1 Our model was able to reproduce the key observation that starch was exhausted only at the end of the night in the both SK and normal 12L:12D photoperiods (Fig. 5C), in agreement with the data1 (Fig. 5D). The model explains this through comparable levels of α at dusk (12ZT) under both 12L:12D and SK photoperiods (Fig. S2D, ESI†) as a consequence of an accelerated accumulation of α upon dark-to-light transitions, which compensates for a fall in α in the midday dark period. The simulated kinetics of starch synthesis under SK photoperiods also closely match the experimental data showing an acceleration of the rate of starch synthesis after 5 h of midday darkness compared to a normal 12L:12D day (Fig. 5C and D). The accelerated synthesis rate is explained by an increased carbon deficit under SK photoperiods due to a prolonged darkness, which leads to elevated β and thus I (Fig. S4B, ESI†). This in turn redirects carbon partitioning towards starch by inhibiting sucrose synthesis. An induction of GPT2 after midday darkness (Fig. S4B, ESI†) further increases the rate of starch synthesis under SK photoperiods (Fig. 5C). The model also suggests that an adaptation to the skeleton photoperiods should take several days in plants grown in a 12L:12D photoperiod. Fig. 5C illustrates this by showing that plants have a lower starch level on the first day of the skeleton photoperiod, which is related to lower I immediately after transfer from 12L:12D conditions. This example further illustrates the diurnal mechanisms of starch synthesis and carbon partitioning, related to upregulation of β and hence I under carbon-limited conditions, which are caused by the shorter total duration of the light period in the SK photoperiods.
In summary, the simulation results shown in Fig. 3–5 demonstrated that the proposed diurnal mechanism of the regulation of starch synthesis and degradation by the light and the clock through α and β is capable of describing correctly a large range of different experimental data and further suggests explanations for these observations.
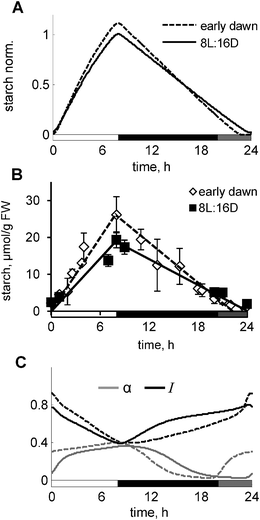
Decoupling of the metabolism from the clock in the wild type plants – “early dawn”
Light acts to entrain the clock, and also provides a source of energy to drive photosynthesis and generate metabolites that are used for growth. Normally, these actions occur in a close temporal sequence after illuminating plants. To separate clock-related from the metabolic-related effects of light we investigated the effect of a treatment in which weak light (for example, weak red and blue light) was given to plants 4 h before the normal dawn. This light was shown to be sufficient to entrain the clock to the new, early dawn,27 but is below the light compensation point for photosynthesis, i.e. the irradiance is too low to support net photosynthetic carbon gain. In this way, the plants should experience two dawns: a “circadian dawn” corresponding to the beginning of the weak light pulse and a “photosynthetic dawn” at which carbon assimilation commences (at time 0). The model predicts that starch is prematurely exhausted before the photosynthetic dawn, resulting in a period of carbon starvation in the interim period until high light is provided to support photosynthesis. This results in a substantial change in the simulated kinetics of starch turnover (Fig. 6A). Our experimental data confirmed this model prediction, demonstrating preliminary depletion of starch and higher rate of starch synthesis in “early dawn” plants compared to control plants, grown under 8L:16D conditions (Fig. 6B). The model explains the depletion of starch through the higher level of α due to the resetting the clock and hence, the clock-driven component α to the circadian dawn (Fig. 6C). The model also explains the increase of starch synthesis by the increased level of the “starvation marker” I, related to the drop of sugar levels during the very low light period. Again, increased I reduces sucrose synthesis rates, thus accelerating starch synthesis in the “early dawn” plants, which is also enhanced by the increased activity of GPT2. Therefore, the model suggests a new way for identifying the unknown regulator of starch degradation α among the compounds, which are upregulated during the low light period in the “early dawn” plants (Fig. 6C).AKINβ1 – a good candidate for the diurnal and circadian “dark sensor” β
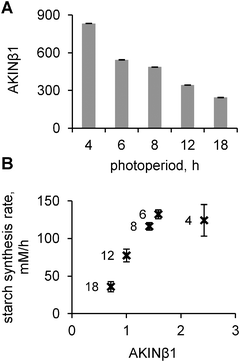
As discussed above, a key model component is the “dark sensor” β, which activates I and thus provides the diurnal regulation of carbon fluxes under multiple conditions. We searched for a possible candidate for this hypothesised “dark sensor” β. The experimental data on the inhibition of both SPS and cFBPase through SnRK1-related mechanisms28,29 and the regulation of carbon consumption and growth by SnRK114 suggest that this kinase might be a good candidate for I. Thus we first analysed available microarray data for the expression profiles of known regulators of SnRK1. We found that the β subunit of SnRK1 (AKINβ1) possesses all the necessary properties for our model component β: its mRNA accumulates in darkness and quickly decays in light. Besides, AKINβ1 is a necessary element for SnRK1 activity.16,17 To obtain further support for our assumption, we measured the expression levels of AKINβ1 under various photoperiods by microarray analysis.The expression levels of AKINβ1 showed an increase at the end-of-the-night on short days compared to longer days (Fig. 7A). There was an almost linear dependence of peak AKINβ1 levels at dawn and the duration of the night (Fig. 7A), which resulted in a strong correlation between measured rates of starch synthesis and AKINβ1 levels under all photoperiods except 4 h. In this extremely short photoperiod, starch synthesis was probably saturated (Fig. 7B). This matches the model's predictions for β (Fig. 3F) and points to AKINβ1 as a good candidate for being the dark sensor β in our model.
This view is further supported by published microarray data available for diurnal and constant light conditions (http://diurnal.mocklerlab.org), showing that AKINβ1 expression (At5g21170) is strongly regulated by the clock. The promoter of AKINβ1 has 7 copies of the CCA1 binding site,30 and also 3 copies of the G-box-expanded and 2 copies of EE-like expanded binding sites for TOC1.31 This suggests a possible molecular basis for the regulation of β by the clock, which is implemented in our model by activation of β by CCA1 and LHY proteins and inhibition by TOC1 protein (part E of the ESI†). Moreover, our simulations of the lhy/cca1 and toc1 mutants (Fig. 4B and Fig. S4A, ESI†) demonstrated that such a regulation agrees with the kinetics of starch observed in these clock mutants, further suggesting AKINβ1 to be a good candidate for the diurnal and circadian regulation of carbon partitioning in plants. The model suggests that AKINβ1 levels should change dramatically under various genetic and environmental perturbations, such as mutations of the clock genes or skeleton and normal photoperiods (Fig. 3C and 4C), thus outlining experimental strategies to further consolidate the possible regulatory role of AKINβ1 in carbon metabolism.
In summary, our results showed that the model is able to describe and explain all existing data on the diurnal changes of starch under genetic and environmental perturbations, including various T cycles, starch-excess mutants and “night pulse” experiments, presented in the ESI† (Fig. S4C–F and S5). Moreover, the model is robust to the variation of the unknown parameters of the diurnal regulation (Fig. S7 and chapter “Parameter stability analysis” in the ESI†). This suggests that the proposed structure of the diurnal regulation is capable of catching the most essential regulatory features of starch metabolism in plants.
Discussion
The diurnal kinetics of starch accumulation in plants adapts to long term differences in the light regime and also has a striking ability to quickly adjust to unpredictable changes in the light conditions. Such changes will occur continually in a natural environment. Somehow plants “know” how quickly they should accumulate starch to satisfy their needs of carbon during the coming night, and can also adjust the rate of starch degradation according to the length of the night and the amount of starch accumulated at the end of the light period. This response ensures that plants have enough carbon to survive and even grow until the next dawn and avoid starvation. These unique properties of the daily regulation of the starch kinetics imply that there should be some sensor mechanisms that regulate the fluxes of carbon into and out of starch. Experimental data demonstrated that these mechanisms are regulated by the circadian clock.1,21,24 Firstly, starch kinetics is severely impaired in the lhy/cca1 clock mutant, which prematurely depletes its starch reserve at the end of the night (Fig. 4A). Secondly, wild type plants cannot adjust their rate of starch degradation when grown in an environmental period different from their natural 24 h clock period. Thirdly, plants can easily adjust their starch kinetics to skeleton photoperiods (Fig. 5D), which do not impair the functioning of the clock.25,26The importance of the circadian clock in the regulation of starch turnover and other physiological processes, such as growth, flowering and opening of the plant pores (stomata) is well acknowledged, however the molecular mechanism of this regulation is only understood in a few cases.32,33 The circadian clock regulates transcription of a large number of plant genes – about 30% of the whole genome is rhythmic under constant light conditions.34 The rhythmically expressed genes include some of the important enzymes of starch metabolism, such as DPE2, PHS2, glucan water dikinase (GWD1) and β-amylase.24 However, protein abundance of these enzymes does not seem to follow transcription and remains nearly constant during the day.24 The absence of the diurnal fluctuation of protein abundance was recently shown for a large number of other plant genes, which are rhythmically expressed.35 As discussed elsewhere,36 the diurnal changes in gene expression allow the plant to continuously monitor physiological and environmental conditions, but the changes of transcripts are often rectified by a slower rate of protein turnover. They do not lead to large changes in protein levels within a 24 h cycle, but instead allow a gradual change in protein levels in response to a sustained change in environmental conditions or developmental program.35,37 Therefore, a fast adjustment of starch kinetics to fluctuating environmental conditions requires the presence of some other circadian mechanisms, which are not reduced to changes in abundance of metabolic enzymes.
An important alternative strategy for circadian regulation is the control of enzyme activities by post-translational regulation through some global regulatory proteins like kinases or phosphatases. Indeed, early studies highlighted the diurnal and circadian rhythms of the activity of the key enzyme of the sucrose synthesis pathway, SPS.8 Similarly, F26PP activity was shown to be regulated during the day and coordinated with SPS, which was suggested to be important for the diurnal adjustments of the carbon flux through the sucrose synthesis pathway.8 Later on it was shown that indeed SPS and F26PP share common regulatory mechanisms, for example by inactivation through phosphorylation by SnRK1 kinase.28,29 Interestingly SnRK1 not only modulates enzyme activities, but is also involved in the regulation of the consumption of sugars and plant growth.14,38
SnRK1 is known as a global regulator of plant metabolism under stress conditions.14,39 However, also under normal conditions SnRK1 is important for plant growth and survival,38,39 which implies that it might work as a measure of the relative stress or carbon deficit to regulate metabolic fluxes according to the continuously changing environmental conditions. Thus, SnRK1 possesses all important features of a global metabolic regulator, making it a good candidate for the model component I regulating carbon partitioning. The model further includes sensing mechanisms which transduce environmental and circadian cues to control the global regulators. In the model, daily information about light conditions and time of the day is perceived through the clock- and light-related “dark sensor” of carbon deficit β and the “timer” of starch degradation α. Although the identity of α is not clear yet, our present work suggests one plausible candidate for β. Based on the functional properties of β, such as its accumulation in darkness and activation of I by β we proposed the β subunit of SnRK1 (AKINβ1) as a possible candidate for β. AKINβ1 is transcriptionally induced in darkness and also strongly regulated by the clock. Our new experimental data also demonstrate a strong correlation between peak levels of AKINβ1 and the starch synthesis rates, which agrees with the model predictions related to the key role of β in the diurnal regulation of carbon partitioning. Further experimental validation of the role of AKINβ1 in carbon partitioning might include transient overexpression of AKINβ1 in plants, which is predicted to increase starch synthesis rate (Fig. S6B, ESI†).
The circadian regulation of β allowed us to explain the counter-intuitive experimental observation that starch synthesis rates in the lhy/cca1 mutant are reduced compared to the wild-type. The model predicts some reduction of the sensitivity of this mutant to starvation due to the clock impairment and reduced levels of β. We propose that the activation of the expression of AKINβ1 by the clock proteins LHY and CCA1 explains these results. In summary, our combined modelling and experimental results suggest a new molecular mechanism of the circadian regulation of the important physiological process of starch accumulation through β (AKINβ1). Together with ABAR,32 which participates in the circadian regulation of the stomata aperture, and FKF1, a circadian regulator of flowering time,33 AKINβ1 extends the list of potential target genes which mediate clock signals to downstream physiological processes in plants.
In addition to the diurnal regulation of the level of “carbon deficit” I through β, the model describes activation of I by carbon starvation when sugars are depleted, which is observed for example in starchless mutants2 or the lhy/cca1 mutant1 at the end of the night. Carbon starvation is normally avoided in wild type plants by means of the diurnal regulation described above. However under extreme conditions starch might be depleted, which results in a drop of sugar levels and induction of starvation genes.1,2 Thus we describe two signalling mechanisms of the “carbon deficit”: (i) diurnal via β under normal conditions and (ii) through metabolic changes, such as low sugars level, under starvation conditions. The “early dawn” experiment (Fig. 6) gives an example of the starvation-induced increase of I levels at the end of the night. In contrast, in normal short days, the increase of I is induced by β. These two modes of regulation – immediate and delayed – may interact under carbon-limited conditions. In particular, depletion of sugars at the end of the night might be sensed together with the light conditions and be transduced to regulate fluxes in the following 24 hour cycles, affecting both synthesis and degradation of starch. An additional level of slow adaptation to carbon deficit is mediated by the temporal induction of GPT2 in the chloroplast membrane, which redirects some of the cytosolic G6P back to the chloroplast, leading to a further acceleration of starch synthesis. This mechanism might contribute to the observed increase of the starch synthesis rate under short days and skeleton photoperiods. Additional mechanisms of the diurnal regulation of starch turnover might include post-translational activation of AGPase and regulation of carbon consumption by trehalose-6-phosphate,40,41 which are the important directions for further extension of the proposed model.
Moreover, the model allowed us to explore possible mechanisms of the regulation of starch degradation. We concluded that starch breakdown might be regulated by a combination of factors, such as the levels of the clock-related timer α and the global sensor of carbon deficit I, which provide a delayed metabolic feedback to starch degradation. At this stage, molecular mechanisms providing this feedback are speculative, but there exist indications that SnRK1 might again play a role. The fact that SnRK1 was found to be present in chloroplasts42 and the mammalian analogue of SnRK1 – AMPK – can bind to the surface of glycogen molecules via its β subunit43 indicate that SnRK1 might bind to the surface of starch granules and participate in starch degradation.
The rate of starch degradation is modelled to also depend on the starch level at dusk, as is suggested by various experimental findings.1–3 This is described through the proportional dependence of the starch degradation rate on the starch level at dusk through the variable X. This implies that the plant is able to somehow ‘measure’ the content of the stored starch. However, since starch is stored as granules and only the surface is accessible to enzymatic attack,44 a direct sensing of starch level is hard to conceive. However, as suggested in ref. 45, a soluble molecule whose concentration is proportional to the amount of starch might serve as a proxy for the starch content, allowing the plant to measure starch in the chloroplast. Although some hypothetical mechanisms of the regulation of starch-degrading enzymes on the surface of the starch granule were recently proposed,45 experimental evidence of the molecular details of the regulation await further research. Finally, the model correctly describes the starch kinetics for all the different experimental settings tested, such as different photoperiods, an “early dusk”, skeleton photoperiods, various T cycles (Fig. S5, ESI†) starch-excess mutants (Fig. S4C and D, ESI†), “night pulse” (Fig. S4E and F, ESI†) and new “early dawn” experiments (Fig. 6).
In conclusion, the model we present here embodies a novel approach to study metabolic regulation in the context of diurnal modulation of fluxes by global regulators. We provide theoretical and experimental results supporting the hypotheses that SnRK1 kinase and its β subunit are suitable candidates for the model components I (global regulator) and β (dark sensor). Although the experimental characterisation of other model components, such as D and α and their connections to metabolism and the clock need further research, we believe that our approach provides a valuable resource to investigate diurnal regulation of carbohydrate metabolism and thus allows to broaden the range of possible experiments, which now can be interpreted in a theoretical context. In particular the proposed properties of the diurnal and circadian regulators β and α under various perturbations of the daily light/dark cycle and the clock provide a good starting point for further studies to identify still unknown molecular details of the proposed interaction between diurnal regulatory systems and carbon metabolism.
Materials and methods
Model description
Therefore, we developed an additional block of reactions that mediates diurnal regulation of carbon metabolism through clock- and light-dependent processes. This is described in the model through coordination of metabolic reactions (Fig. 1) by key regulators, which sense the amount of available light and the time of day through the circadian clock (Fig. 2, part E of the ESI†). Some of these diurnal regulators and connections between them were derived from the existing literature, others were assumed based on the indirect observations, as described below. The model consists of 28 ordinary differential equations connected to the most recent model of the circadian clock (part F of the ESI,† Fig. S1A). The main principles of diurnal regulation of starch turnover in the model are schematically illustrated in Fig. 8.
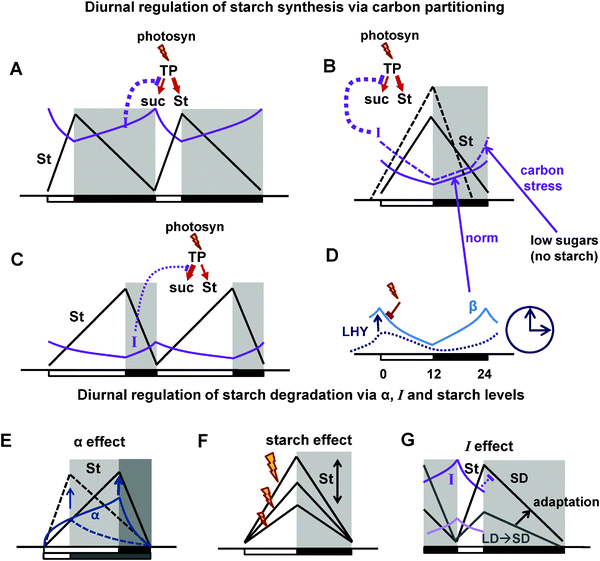 | ||
| Fig. 8 Schematic figures summarizing the main principle mechanisms of diurnal regulation of starch turnover in the model. (A–C) Diurnal regulation of starch synthesis via carbon partitioning under normal conditions (A and C) and under carbon stress, which is observed when starch is prematurely depleted at the end of the night due to mutations or specific perturbations of the light cycle (B; Fig. 6). The flux of newly-synthesised trioso-phosphate (TP) to starch (St) is regulated through changes in the flux of TP to sucrose (suc) synthesis during the day (red arrow schemes on the top of the pictures). The flux to sucrose is regulated through the inhibition by the global inhibitor I (SnRK1 is a candidate). Thus, high I results in the decrease of sucrose and increase of starch synthesis on short days (A) or under carbon stress (B), whereas low I increases sucrose and decreases starch synthesis under long days (C). (B and D) Diurnal regulation of I and β. B. Under normal conditions (solid lines) I is activated by the “dark sensor” β (β subunit of SnRK1 is a candidate). Under carbon stress (dashed lines) I is additionally activated by low sugar levels at the end of the night. (D) Diurnal regulation of β via its accumulation in the dark and activation by the clock component LHY. (E–G) Diurnal regulation of starch degradation. (E) The timer α increases during the day and stimulates starch degradation on long days (solid lines) compared to short days (dashed lines). (F) Starch degradation rate is proportional to starch level at dusk, which might fluctuate, for example under various intensities of light (indicated by flashes). (G) Inhibition of starch degradation by I results in faster starch degradation in plants transferred from long to short days (light coloured lines) compared to plants adapted to short days (darker lines). This provides adaptation of starch degradation to the metabolic conditions of the previous day. Days and nights are indicated on the pictures by white and black boxes on the x axis. Night is also shown by shadowed areas. | ||
Partitioning of carbon between starch and sucrose during the day depends on the activities of the key enzymes of sucrose synthesis cFBPase and SPS, which control the flux of photosynthetically derived triose-phosphates to sucrose (Fig. 1 and 2; ref. 8 and 40; see also part C of the ESI† for more details). The activities of SPS and cFBPase are regulated in a coordinated way during the day.8 cFBPase is controlled through allosteric inhibition by fructose-2,6-phosphate (F26P), the level of which is regulated by the bisfunctional enzyme fructose-6-phosphate kinase/F26P phosphatase (F6PK/F26PP; Fig. 1). The inactivation of both SPS and F26PP through phosphorylation by SnRK1 (SNF1-related kinase 1)28,29 suggests common regulatory mechanisms, which might coordinate SPS and cFBPase activities and thus the flux through the sucrose synthesis pathway during the day. SnRK1 belongs to the family of stress-related SnRK1/SNF1/AMPK kinases, which are global regulators of carbohydrate utilisation and other metabolic processes in plants and other organisms.14,15 It is plausible that SnRK1-related kinases modulate activities of the key metabolic enzymes and thus adjust fluxes to changing conditions. Therefore, in our model we assumed that a diurnal regulator I (for which SnRK1 is a good candidate) inhibits SPS and cFBPase (via F26PP) and thus regulates the flux through sucrose synthesis in source tissues (Fig. 2). This affects starch synthesis so that it is accelerated on shorter days, when sucrose flux is low (Fig. 8A and C). Moreover, consumption of sucrose in sink tissues is quickly adjusted to changing metabolic conditions.9,10 Interestingly, SnRK1 kinase is also involved in the regulation of consumption of carbon by growing tissues through downregulation under “carbon stress”.14,46 Therefore we assumed that I also inhibits sucrose consumption by sinks and thus provides a global regulation of the sucrose fluxes in plants (Fig. 2). Thus, in our model the partitioning of carbon between starch and sucrose is mainly determined by the level of the global inhibitor I.
Additional to inactivation, both SPS and F26PP might be activated through discrete phosphorylation sites.47 Both SPS and cFBPase activities were shown to be quickly upregulated “on demand” from sink tissues in experiments with source/sink manipulations, resulting in an increased partitioning of carbon to sucrose.9,10 Even though the identity of the diurnally-regulated kinase upregulating SPS and cFBPase needs to be clarified, their increased activities in source leaves “on demand” from sinks provides another mechanism for the diurnal regulation of sucrose flux. In our model the variable D reflects this demand.
Moreover, the existing data suggest that carbon partitioning might be regulated by induction of the glucose-6-phosphate (G6P) translocator (GPT2) in the chloroplast membrane under carbon-limited conditions.19 GPT2 increases the partitioning of carbon into starch by re-directing G6P back from the sucrose pathway towards starch synthesis. Although the exact molecular details of GPT2 induction are unknown, GPT2 expression is upregulated under limited carbon conditions, which are observed for example in starchless and starch-excess mutants,19,20 so we assumed in our model that a high level of I stimulates GPT2 induction.
The next important question in the modelling process is how the observed diurnal changes in carbon fluxes are regulated by light and the clock. To describe the observed sensing of diurnal conditions, we introduced in the model the clock- and light-modulated components β and α, which control the global regulators I and X and thus indirectly control starch synthesis and degradation (Fig. 2). The diurnal sensor β was introduced to provide the clock-modulated perception of light conditions for the marker of “carbon deficit” I. We assumed that β is accumulated in darkness, in the absence of photosynthesis (Fig. 3C and 8D; part E of the ESI†). The sensor β activates the regulator I to dynamically re-inform the system of carbon consumption on the amount of time spent in the dark. Experimental data show that the clock is affecting starch synthesis.1 Therefore we additionally assumed a modulation of β by the clock. This assumption allowed us to correctly describe the available data on the starch kinetics in the clock mutants (see Results). The predicted properties of β closely match the experimental observations for AKINβ1 (see Results). In the model, I is further activated under starvation through low sugar levels, which is in accordance to existing data.39,46 This additional level of regulation was introduced to describe conditions in which starch is prematurely exhausted at the end of the night, which activates starvation responses in some mutants (e.g., lhy/cca1 or mutants that are deficient in starch synthesis or breakdown) or in wild type plants during an extended night.1,2 Thus I is activated in our model by both metabolism (under the starvation through the fall in sugars level) and by the clock and light through β (under normal conditions) (Fig. 2 and 8B) and therefore integrates metabolic and circadian signals.
The second sensor of the diurnal conditions in our model is a “timer” α. This component is activated by light and acts to set the rate of starch degradation according to the time of the day.6 The presence of this hypothetical component in the model was necessary to describe the observed dependence of starch degradation on the clock and light signals,1,6 as discussed in Results. The timer α accumulates during the light period (Fig. 8E), therefore a higher accumulation of α on long days increases the rate of starch degradation compared to shorter days.2 It should be noted that α reflects the time spent in light, but does not itself require an input from the rate of photosynthesis. Multiple data suggest that the rate of starch degradation is set soon after dusk, since it remains almost constant in most of the conditions tested.1,2,24 This “dusk setting” might be related to the formation of multi-protein starch-degrading enzymatic complexes soon after lights-off.18 To model the dusk setting, we introduced a variable X, which changes in the light according to levels of α, but remains constant in darkness (Fig. S2D, ESI†). The data also suggest that, additional to the light and clock sensing, starch degradation depends on the starch level at dusk (Fig. 8F, Fig. S1B, ESI,†ref. 1, 5 and 6). Therefore X corresponds to the activity of some enzymatic complexes on the surface of starch granules, modulating starch degradation in dependence on α and the starch level. Although the molecular mechanisms of α–X interactions are still unknown, the presence of these two components in our model is sufficient to provide a good description of the large amount of data on starch degradation under various conditions as presented in Results. In summary, the sensors α and β are the only two components of our model, which directly perceive light- and clock-related information, transducing it to carbon metabolism through the model components I, D and X.
In addition to the immediate perception of the current day conditions, existing data suggest that starch degradation is affected by the metabolic state of the plant in previous days. Thus, starch degradation rate is slowed down over several days after transferring plants from long to short day conditions.24 To account for the long-term diurnal modulation of starch degradation, we included a delayed feedback, which provides an adaptation of plants to the higher level of carbon deficit experienced during the previous night (higher I) by inhibiting starch degradation on the next day by I. Experimental support for this assumption is provided by indications that SnRK1-related kinases in plants and other organisms bind to the surface of starch or glycogen.42,43 The long-term regulation of starch degradation by I was described in our model through inhibition of X by I. Since I is a relatively slow-changing model variable (Fig. S2B and E, ESI†) and starch degradation is set only once at dusk, an increase of I at night inhibits starch degradation only in the next day (Fig. 8G). Thus, regulation by starch levels and α is responsible for the dynamic control of starch degradation according to the present conditions, while the slow response through I provides a long-term adaptation of starch to conditions experienced during the previous day. This combined regulation of starch degradation by starch, α and I allowed to correctly describe multiple experimental time-courses of starch under various perturbations (see Results).
The model equations are described in details in the ESI.† Model parameters are presented in Table S1 (ESI†). Whenever possible, parameters of the metabolic reactions were taken from existing literature (Table S1, ESI†). Non-accessible parameters were estimated to achieve reported values of steady state metabolite concentrations (see the ESI† for more details). Unknown parameters of the diurnal regulation were fitted manually to achieve agreement with published data on starch time-courses in wild type plants grown under different photoperiods.2,24,48 Parameters were kept the same in all simulations, except mutant simulations, where rates of production of the corresponding mutated components were set to 0, as indicated in the respective figure legends. Photoperiod simulations were performed by varying the corresponding time of dusk (parameter Tdusk in the light function L(t), described in part E of the ESI†). The system of ordinary differential equations was solved using MATLAB, integrated with the stiff solver ode15s (The MathWorks UK, Cambridge). A MATLAB version of the model supplied as an ESI† model file.
Experimental conditions
For the starch time-course measurements, Arabidopsis thaliana seeds were sown on a wet soil in 10 cm diameter pots, covered with transparent lids and transferred to growth chambers (Percival Scientific Inc., Perry, IA). There was no stratification step. Plants were grown under 6L:18D and 18L:6D photoperiods. A light intensity was adjusted to 160 μmol m−2 s−1 and the temperature was maintained at 20 °C during the light phase and 18 °C during the dark phase. Pots were randomized to decrease positional effect. After a week lids were removed and the excessed plants were thinned. After another seven days plants were treated with Nematodes as a biological pest control. Plats were watered daily with tap water.
On the 21st day after sowing on soil two biological replicates per time point were harvested. Each replicate consisted of a pool of 7–13 plants. Sampling was performed with 2 h intervals, starting just before dawn (ZT0), within 5–10 minutes before a given time point. Standard deviation is shown on all figures in Results. During the dark phase plants were sampled in the presence of low-intensity green lamp. Rosettes were cut at the ground level, placed in plastic scintillation vials and frozen in liquid nitrogen.
Acknowledgements
This work was supported by the European Commission FP7 Collaborative Project TiMet (contract no. 245143).References
- A. Graf, A. Schlereth, M. Stitt and A. M. Smith, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9458–9463 CrossRef CAS PubMed.
- Y. Gibon, O. E. Bläsing, N. Palacios-Rojas, D. Pankovic, J. H. Hendriks, J. Fisahn, M. Höhne, M. Gunther and M. Stitt, Plant J., 2004, 39, 847–862 CrossRef CAS PubMed.
- Y. Gibon, E. T. Pyl, R. Sulpice, J. E. Lunn, M. Höhne, M. Gunther and M. Stitt, Plant, Cell Environ., 2009, 32, 859–874 CrossRef CAS PubMed.
- A. M. Smith and M. Stitt, Plant, Cell Environ., 2007, 30, 1126–1149 CrossRef CAS PubMed.
- M. Stitt and S. C. Zeeman, Curr. Opin. Plant Biol., 2012, 15, 282–292 CrossRef CAS PubMed.
- A. Graf and A. M. Smith, Trends Plant Sci., 2011, 16, 169–175 CrossRef CAS PubMed.
- A. Pokhilko, P. Mas and A. J. Millar, BMC Syst. Biol., 2013, 7, 23 CrossRef CAS PubMed.
- M. Stitt, S. Huber and P. Kerr, in The Biochemistry of Plants, ed. M. D. Hatch and N. K. Boardman, Academic Press, New York, 1987, vol. 10, pp. 327–409 Search PubMed.
- T. W. Rufty and S. C. Huber, Plant Physiol., 1983, 72, 474–480 CrossRef CAS PubMed.
- T. W. Rufty, S. C. Huber and P. S. Kerr, Plant Sci. Lett., 1984, 34, 247–252 CrossRef CAS.
- G. Pettersson and U. Ryde-Pettersson, Eur. J. Biochem., 1988, 175, 661–672 CrossRef CAS.
- X. G. Zhu, E. de Sturler and S. P. Long, Plant Physiol., 2007, 145, 513–526 CrossRef CAS PubMed.
- A. Nag, M. Lunacek, P. A. Graf and C. H. Chang, BMC Syst. Biol., 2011, 5, 94 CrossRef CAS PubMed.
- R. Ghillebert, E. Swinnen, J. Wen, L. Vandesteene, M. Ramon, K. Norga, F. Rolland and J. Winderickx, FEBS J., 2011, 278, 3978–3990 CrossRef CAS PubMed.
- S. J. Hey, E. Byrne and N. G. Halford, Ann. Bot., 2010, 105, 197–203 CrossRef CAS PubMed.
- C. Polge and M. Thomas, Trends Plant Sci., 2007, 12, 20–28 CrossRef CAS PubMed.
- C. Polge, M. Jossier, P. Crozet, L. Gissot and M. Thomas, Plant Physiol., 2008, 148, 1570–1582 CrossRef CAS PubMed.
- S. Streb and S. C. Zeeman, Arabidopsis, 2012, 10, e0160 CrossRef PubMed.
- H. H. Kunz, R. E. Häusler, J. Fettke, K. Herbst, P. Niewiadomski, M. Gierth, K. Bell, M. Steup, U. I. Flügge and A. Schneider, Plant Biol., 2010, 12(Suppl 1), 115–128 CrossRef CAS PubMed.
- M. Stettler, S. Eicke, T. Mettler, G. Messerli, S. Hörtensteiner and S. C. Zeeman, Mol. Plant, 2009, 2, 1233–1246 CrossRef CAS PubMed.
- N. Yazdanbakhsh, R. Sulpice, A. Graf, M. Stitt and J. Fisahn, Plant, Cell Environ., 2011, 34, 877–894 CrossRef PubMed.
- E. M. Farre, S. L. Harmer, F. G. Harmon, M. J. Yanovsky and S. A. Kay, Curr. Biol., 2005, 15, 47–54 CrossRef CAS PubMed.
- N. Nakamichi, T. Kiba, R. Henriques, T. Mizuno, N. H. Chua and H. Sakakibara, Plant Cell, 2010, 22, 594–605 CrossRef CAS PubMed.
- Y. Lu, J. P. Gehan and T. D. Sharkey, Plant Physiol., 2005, 138, 2280–2291 CrossRef CAS PubMed.
- C. S. Pittendrigh and S. Daan, J. Comp. Physiol., 1976, 106, 291–331 CrossRef.
- A. Pokhilko, S. K. Hodge, K. Stratford, K. Knox, K. D. Edwards, A. W. Thomson, T. Mizuno and A. J. Millar, Mol. Syst. Biol., 2010, 6, 416 CrossRef PubMed.
- M. F. Covington, S. Panda, X. L. Liu, C. A. Strayer, D. R. Wagner and S. A. Kay, Plant Cell, 2001, 13, 1305–1315 CAS.
- C. Sugden, P. G. Donaghy, N. G. Halford and D. G. Hardie, Plant Physiol., 1999, 120, 257–274 CrossRef CAS PubMed.
- A. Kulma, D. Villadsen, D. G. Campbell, S. E. Meek, J. E. Harthill, T. H. Nielsen and C. MacKintosh, Plant J., 2004, 37, 654–667 CrossRef CAS.
- S. L. Harmer and S. A. Kay, Plant Cell, 2005, 17, 1926–1940 CrossRef CAS PubMed.
- W. Huang, P. Perez-Garcia, A. Pokhilko, A. J. Millar, I. Antoshechkin, J. L. Riechmann and P. Mas, Science, 2012, 336, 75–79 CrossRef CAS PubMed.
- T. Legnaioli, J. Cuevas and P. Mas, EMBO J., 2009, 28, 3745–3757 CrossRef CAS PubMed.
- T. Imaizumi, H. G. Tran, T. E. Swartz, W. R. Briggs and S. A. Kay, Nature, 2003, 426, 302–306 CrossRef CAS PubMed.
- T. P. Michael and C. R. McClung, Plant Physiol., 2003, 132, 629–639 CrossRef CAS PubMed.
- K. Baerenfaller, C. Massonnet, S. Walsh, S. Baginsky, P. Bühlmann, L. Hennig, M. Hirsch-Hoffmann, K. A. Howell, S. Kahlau, A. Radziejwoski, D. Russenberger, D. Rutishauser, I. Small, D. Stekhoven, R. Sulpice, J. Svozil, N. Wuyts, M. Stitt, P. Hilson, C. Granier and W. Gruissem, Mol. Syst. Biol., 2012, 8, 606 CrossRef CAS PubMed.
- M. Stitt, Curr. Opin. Biotechnol., 2013, 24, 229 CrossRef CAS PubMed.
- M. Piques, W. X. Schulze, M. Höhne, B. Usadel, Y. Gibon, J. Rohwer and M. Stitt, Mol. Syst. Biol., 2009, 5, 314 CrossRef PubMed.
- M. Thelander, T. Olsson and H. Ronne, EMBO J., 2004, 23, 1900–1910 CrossRef CAS PubMed.
- E. Baena-Gonzalez, F. Rolland, J. M. Thevelein and J. Sheen, Nature, 2007, 448, 938–942 CrossRef CAS PubMed.
- M. Stitt, J. Lunn and B. Usadel, Plant J., 2010, 61, 1067–1091 CrossRef CAS PubMed.
- N. Hädrich, J. H. Hendriks, O. Kötting, S. Arrivault, R. Feil, S. C. Zeeman, Y. Gibon, W. X. Schulze, M. Stitt and J. E. Lunn, Plant J., 2012, 70, 231–242 CrossRef PubMed.
- S. Fragoso, L. Espindola, J. Paez-Valencia, A. Gamboa, Y. Camacho, E. Martinez-Barajas and P. Coello, Plant Physiol., 2009, 149, 1906–1916 CrossRef CAS PubMed.
- A. McBride and D. G. Hardie, Acta Physiol., 2009, 196, 99–113 CrossRef CAS PubMed.
- O. Kartal and O. Ebenhöh, FEBS Lett., 2013, 587, 2882–2890 CrossRef CAS PubMed.
- A. Scialdone, S. T. Mugford, D. Feike, A. Skeffington, P. Borrill, A. Graf, A. M. Smith and M. Howard, eLife, 2013, 2, e00669 Search PubMed.
- C. Nunes, L. F. Primavesi, M. K. Patel, E. Martinez-Barajas, S. J. Powers, R. Sagar, P. S. Fevereiro, B. G. Davis and M. J. Paul, Plant Physiol. Biochem., 2013, 63, 89–98 CrossRef CAS PubMed.
- E. MacRae and J. E. Lunn, in Control of primary metabolism in plants, ed. W. C. Plaxton and M. T. McManus, Blackwell Publishing Ltd., Oxford, 2006, vol. 22 Search PubMed.
- S. Comparot-Moss, O. Kötting, M. Stettler, C. Edner, A. Graf, S. E. Weise, S. Streb, W. L. Lue, D. MacLean, S. Mahlow, G. Ritte, M. Steup, J. Chen, S. C. Zeeman and A. M. Smith, Plant Physiol., 2010, 152, 685–697 CrossRef CAS PubMed.
- J. M. Cross, M. von Korff, T. Altmann, L. Bartzetko, R. Sulpice, Y. Gibon, N. Palacios and M. Stitt, Plant Physiol., 2006, 142, 1574–1588 CrossRef CAS PubMed.
- M. Lohse, A. Nunes-Nesi, P. Krüger, A. Nagel, J. Hannemann, F. M. Giorgi, L. Childs, S. Osorio, D. Walther, J. Selbig, N. Sreenivasulu, M. Stitt, A. R. Fernie and B. Usadel, Plant Physiol., 2010, 153, 642–651 CrossRef CAS PubMed.
- H. Winter, D. G. Robinson and H. W. Heldt, Planta, 1996, 193, 530–535 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3mb70459a |
| This journal is © The Royal Society of Chemistry 2014 |