 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Site-specific peptide and protein immobilization on surface plasmon resonance chips via strain-promoted cycloaddition†
Angelique E. M. Wammesa, Marcel J. E. Fischerb, Nico J. de Molb, Mark B. van Eldijka, Floris P. J. T. Rutjesa, Jan C. M. van Hesta and Floris L. van Delft*a
aRadboud University, Institute for Molecules and Materials, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands. E-mail: f.vandelft@science.ru.nl; Fax: +31-2436-53393; Tel: +31-02436-52373
bUtrecht University, Medicinal Chemistry and Chemical Biology, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands
First published on 12th March 2013
Abstract
Surface plasmon resonance (SPR) is a powerful label-free diagnostic tool to study biomolecular interactions. However, one of the drawbacks of SPR is the lack of controlled immobilization of ligands on the sensor surface. We have developed a modular platform for the fast, reagent-free and site-specific immobilization of azide-containing ligands by strain-promoted cycloaddition onto a cyclooctyne-modified SPR sensor surface. The usefulness of the concept was shown in a study with a papain model system, and up to 150 experiments were performed without loss of surface quality. Furthermore, azide-containing green fluorescent protein (GFP) was also effectively immobilized. Taken together, cyclooctyne-modified SPR chips enable smooth and site-selective immobilization of ligands and prove to be more robust than traditionally functionalized systems.
Introduction
Surface plasmon resonance (SPR) has been widely used as an optical tool for real-time and label-free analysis of biomolecular interactions with small quantities of material.1,2 With SPR, the strength and kinetics of biomolecular interactions can be studied and quantified, or new potential binding partners for a particular ligand can be screened and identified. However, although detection of the biomolecular interaction is label-free, the ligand itself requires immobilization on the sensor surface prior to (non-covalent) binding of the analyte. Not surprisingly, controlled covalent attachment of ligands is a critical step, since the amount of ligand immobilized on the surface of the SPR chip is closely related to its performance and, consequently, to the quality of the data retrieved.3 For long-lasting SPR chip activity, the surface has to be packed with optimal density and should be stable under regeneration conditions, an essential step for re-use.There are various ways to immobilize a ligand on the sensor surface. The most commonly used chip surface involves a hydrophilic dextran surface displaying carboxylic end groups that are activated as N-hydroxy succinimide (NHS) ester, thereby enabling ligand immobilization by amide bond formation. As it appears, such carboxy-terminated chips are used in approximately 90% of SPR applications.3,4 Despite its popularity, however, the presence of multiple amines in the ligand, e.g. a peptide or a protein, will hamper site-specific modification, which will affect the binding process.5 In addition, reaction of other nucleophilic groups with activated esters may lead to further uncontrolled immobilization on the surface, while the hydrolytic sensitivity of the activated esters makes them only suitable for immobilization procedures that proceed quickly.
To avoid a coupling approach based on amino groups in the ligand, other immobilization methods have been developed, e.g. based on the streptavidin–biotin interaction.6,7 However, such alternative methods show other disadvantages, such as long-term reversibility or non-specific binding events leading to a change of the affinity profile for the biomolecular interaction.3 It is clear, therefore, that a more generally applicable immobilization strategy is desirable. Ideally, such immobilization is fast, selective, compatible with water and fully orthogonal to biomolecular functionality. Given these conditions, we embarked on the development of a new immobilization strategy employing strain-promoted ‘click’ chemistry between cyclooctynes and azides (Fig. 1).8–16
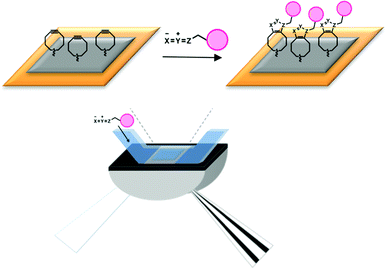 | ||
| Fig. 1 Schematic representation of the SPR detection system and surface functionalization by 1,3-dipolar cycloaddition. | ||
Cyclooctynes are well known to undergo efficient (3 + 2) cycloaddition with a variety of 1,3-dipoles, such as azides (SPAAC),9–12 nitrones (SPANC)13,14 or nitrile oxides (SPANOC).15,16 The incorporation of such a dipole into a protein or a small molecule can be easily achieved by either chemical17 or enzymatic means.18,19 Most recently, extremely fast (4 + 2) cycloaddition of cyclooctynes with tetrazines has also been reported, which further broadens the application of strain-promoted chemistry.20
We here wish to describe the application of cycloaddition chemistry for the modification of SPR chips based on initial functionalization of the chip surface with bicyclo[6.1.0]nonyne (BCN), followed by ligand immobilization via strain-promoted azide–alkyne cycloaddition (SPAAC).12 We show that the BCN-functionalized SPR chips are modular in nature and can be smoothly functionalized in reagent-free fashion with azide-containing compounds. In particular, immobilization of small molecule inhibitors of papain21 allow rapid and quantitative protein detection. The power of the approach is further demonstrated by attachment of a whole protein, i.e. azide-functionalized green fluorescent protein (Anl-GFP).
Results and discussion
The first and most important part of the designed SPR experiments is an effective route to obtain a cyclooctyne-functionalized surface. To this end, we selected bicyclononyne (BCN), recently reported by us,12 as the preferred cyclooctyne of choice because of its beneficial balance between accessibility, reactivity and relatively low lipophilicity. Thus, a BCN-functionalized SPR chip was first prepared by attachment of the commercially available amino-derivative of BCN according to the standard coupling protocol of a CM5 chip.5The power of the BCN-based immobilization platform was demonstrated with two model systems, one involving attachment of azide-containing small organic molecules, and one involving a whole protein charged with an azido group. For the small molecule ligand, a peptide-based enzyme inhibitor from an in-house screening program was chosen to study binding interactions with papain, a cysteine protease from papaya fruit.21–25 After preparation of the BCN surface, a reaction with azide-containing ligand 1 was performed, leading to the desired ligand-containing SPR surface (Fig. 2, left). Loading of the SPR surface was corroborated by treatment of each individual chip with a 1 μM papain solution, leading to a clear response (200 m° at equilibrium) by comparison with papain treatment of a reference cell, created by SPAAC reaction of 2-azidoethanol with the cyclooctyne surface. The successful SPR experiment was a first indication of the usefulness of SPAAC for biosensor chip functionalization. As a control, a direct ligand-functionalization of the surface was also executed from the carboxy-terminated chip via the amine immobilization protocol (ESI†), leading to similar response in comparison to the reference chip.
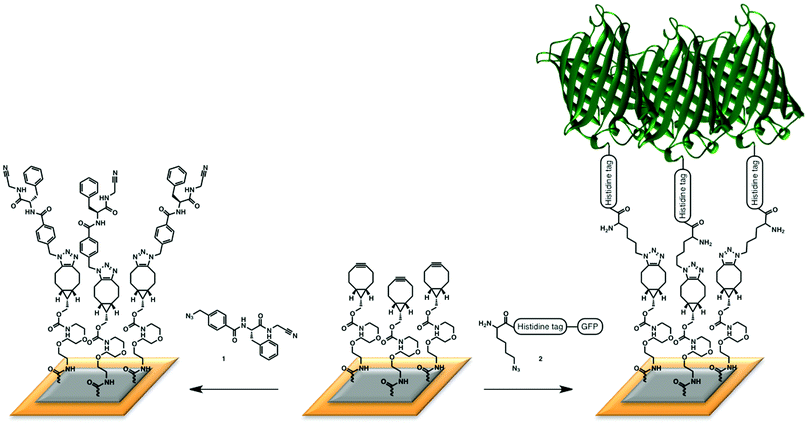 | ||
| Fig. 2 Immobilization of (left) the small nitrile ligand, that was used in a study with papain; (right) azido-containing GFP. | ||
To more closely examine and compare the surfaces obtained by the different immobilization methods, an assay with papain was set up to accurately determine binding constants. Firstly, the binding constant for the interaction at the SPR sensor chip (KC) was determined by addition of different papain concentrations and fitting of the equilibrium signal to a Langmuir binding isotherm (Fig. 3). Next, competition experiments were performed to obtain the binding constant for binding in solution (KS).26 While the binding constant KS is in theory independent from effects at the sensor surface and should be comparable to KC, in practice differences have been observed.3
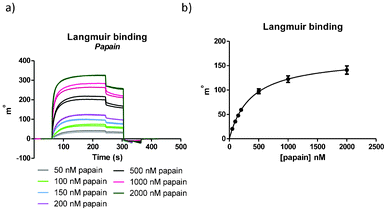 | ||
| Fig. 3 (a) SPR signals for the Langmuir binding experiment (per chip performed in duplicate; in total per experiment two chips were used) and (b) Langmuir binding graph. For ligand 1, KD = 360 ± 30 nM was found. | ||
The first experiment performed on each chip was the Langmuir binding assay and the equilibrium signals were used to obtain KC. As can been seen in Fig. 3, after 60 s of baseline stabilization, the analyte papain was added to the surface for an association time of three minutes, followed by 60 s dissociation and finally regeneration of the surface with NaOH. Different concentrations of papain were added to the surface in a range from 50 to 2000 nM leading to good, reproducible SPR signals (Fig. 3a). We were pleased to find that similar KC values were determined, irrespective if the route of immobilization of the ligand proceeded via SPAAC or via direct amide coupling (Fig. 3b and ESI†). Finally, binding constants obtained in this way compared well with the binding constant determined with isothermal titration calorimetry (ITC) and a UV assay (ESI†). Competition experiments were performed to observe the influence of the cyclooctyne-derived triazole moiety on the affinity between the ligand and papain. These experiments showed that the influence of the BCN moiety is minimal, which is consistent with results from a UV assay (ESI†). The papain experiments showed that a cyclooctyne-modified surface could be effectively applied for the reagent-free attachment of an azido-containing ligand and that binding to the analyte is warranted. It was established that up to 150 experiments could be performed without detectable loss of quality of the surface (ESI†).
In order to further broaden the scope of ligand immobilization by SPAAC, attachment of an azide-containing protein was next explored. To this end, an engineered green fluorescent protein (GFP), containing a single azide functionality, an N-terminal His-tag and a C-terminal FLAG-tag, was prepared by expression in a methionine auxotrophic bacterial system using azidonorleucine (Anl) as a methionine surrogate (ESI†).27 Having obtained the azide-functionalized GFP, the cyclooctyne surface was prepared using the same protocol as for the papain experiments, only in this case a PBS measuring buffer was used. To this end, a protein solution was prepared in 10 mM formic acid buffer (pH 4) and SPAAC reaction was performed for 20 min to ensure effective attachment (Fig. 4). A Langmuir binding study with different antibodies corroborated the fact that GFP was effectively immobilized. Two different anti-GFP, an anti-His and an anti-FLAG antibody were used (Table 1 and ESI†). For all four antibodies KD values were found in the low nanomolar range. The same control experiment as for the papain experiments was performed, in which GFP without an azide (Met-GFP) was immobilized following the amine immobilization protocol (ESI†). The same KD constants were found as for our new SPAAC strategy. However, one important difference we observed is that GFP immobilized by the EDC/NHS strategy led to a significantly enhanced loss of material (50%) from the surface after 50 measurements, compared to a loss of about 20% for the SPAAC strategy (ESI†). We can thus conclude that the SPAAC strategy leads to a more stable and robust surface for both peptide and protein-based ligands.
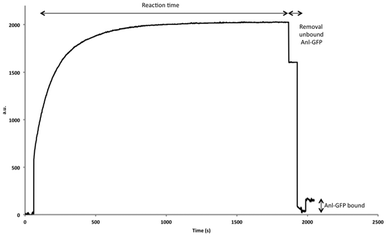 | ||
| Fig. 4 SPR signal of the immobilization of Anl-GFP. After stabilization of the surface (first 60 s), a GFP solution was added to the surface and allowed to react for 20 min. After cleaning of the surface a large increase of mass was observed, indicating the successful immobilization of GFP, which was also confirmed with a Langmuir binding experiment using different antibodies (Table 1). | ||
| Antibody | KD (nM) |
|---|---|
| Anti-His | 32.2 ± 4.8 |
| Anti-GFP (1) | 61.7 ± 8.5 |
| Anti-GFP (2) | 39.7 ± 3.7 |
| Anti-FLAG | 33.9 ± 6.2 |
Conclusions
We have successfully demonstrated that functionalization of SPR sensor surfaces via strain-promoted cycloaddition leads to a versatile, modular system for the efficient and reagent-free attachment of ligands. Using papain as a model system, a synthetic azidopeptide ligand was effectively immobilized to the SPR chip, leading to a highly robust platform that could be applied for the repeated determination of affinity for papain by standard protocols. A protein containing azide functionality was immobilized in a similar fashion and effectively detected and quantified by binding of a range of antibodies. These experiments demonstrate the value of SPAAC-based strategies for the immobilization of all types of azido-containing ligands, including those that do not contain an amine for attachment via amide bond formation or, more commonly, feature multiple amino functions like in the case of proteins and the majority of peptides. In such cases, it must be noted that several strategies were recently developed for the selective introduction of an azide into a protein, either chemically28 or by enzymatic, metabolic or genetic means.19 In this respect, the methodology presented here also compares favorably to a comparable procedure recently reported, based on the strain-promoted reaction of trans-cyclooctenes (TCO) with tetrazines,29 because the site-specific introduction of either of these components into peptides or proteins is much more limited or impossible. Although the latter cycloaddition in principle is much faster than the cyclooctyne–azide reaction, we have found the reaction rate for immobilization not to be a limiting factor, while our system has the distinct advantage of easier accessibility and stability of both BCN (commercially available or readily prepared) and azide. Moreover, we recently demonstrated that (4 + 2) cycloaddition between BCN and tetrazines takes place with near similar reaction kinetics as TCO–tetrazine cycloaddition,20 thereby further enhancing the potential usefulness of BCN-modified SPR surfaces.In conclusion, the BCN-modified SPR chips described here are simple and stable modular systems that may find broad applications for reagent-free immobilization of azide-containing ligands. In addition, it is noted that strain-promoted cycloadditions with other 1,3-dipoles is also feasible, such as nitrones13 and nitrile oxides,15 while the recently reported cycloaddition of BCN to tetrazines by (4 + 2) cycloaddition may further extend the applicability of BCN-modified SPR chips for the controlled immobilization of peptides, proteins or other (bio)molecular structures.
Materials and methods
General
All chemicals, enzymes and the anti-His antibody were purchased from Sigma-Aldrich, except BCN–POE3–NH2 and BCN–PEG3–NH–lissamine rhodamine B conjugate (SynAffix BV, The Netherlands). All other antibodies were purchased from Santa Cruz Biotechnology (GFP(1) = SC-5385; GFP(2) = SC-9996; anti-FLAG = SC-807). Aqueous solutions were prepared with pure water (18.2 MΩ cm resistivity; Milli-Q station, Millipore). For SPR experiments, sensor chip CM5 (research grade) and an amino coupling kit composed of EDC, NHS and ethanolamine hydrochloride (pH 8.5) were purchased from GE Healthcare.Protein analyte
For daily activation of papain, a 10 μM solution was prepared in a 0.1 M sodium phosphate buffer (pH 6.5), containing 2.5 mM EDTA and 15 mM DTT. The solution was incubated at 25 °C for 1 h and the activated enzyme was kept on ice. In this way stability was ensured for 6–8 h. Further dilutions were made in a 0.1 M sodium phosphate buffer (pH 6.5), containing 2.5 mM EDTA, 150 mM NaCl and 0.005% Tween-20 (running buffer A).Monoclonal anti-polyhistidine was diluted in a PBS buffer (pH 7.0) containing 0.005% Tween-20 (running buffer B). Working dilution samples were stored at room temperature for a maximum of 12 h.
Ligands
Detailed procedures for synthesis of the ligands are described in the ESI.†Surface plasmon resonance
SPR measurements were performed on a double channel cuvette-based Autolab Esprit SPR instrument (Metrohm Autolab, Utrecht, The Netherlands) equipped with a CM5 sensor chip (GE Healthcare). All samples and buffers were degassed prior to use.Functionalization by amide coupling: standard EDC/NHS coupling chemistry was performed. For the study with papain, nitrile ligand 7 was immobilized in the sample cell and in the reference cell hydroxyl ligand 8 was immobilized. For the ligand immobilization, a 0.1 mg mL−1 solution in 10 mM maleic acid (pH 6.0) was allowed to react with the activated ester surface for 10 min. For the study with Met-GFP, running buffer B was used. For the immobilization of Met-GFP a solution of 0.1 mg mL−1 in 10 mM formic acid buffer (pH 4.0) was used. After ligand immobilization, 1 M ethanolamine was added for 10 min to the sensor surface to block remaining free ester groups, in both sample and reference cells.
Functionalization by SPAAC: BCN-functionalized surface was prepared with BCN–POE3–NH2 using the same EDC/NHS protocol as described above. The second step involved the immobilization of the azide-containing ligands. For the study with papain, ligand 1 was immobilized in the sample cell, using a solution of 0.1 mg mL−1 ligand in running buffer A for 15 min and remaining free cyclooctynes were blocked with 2-azidoethanol (0.1 mg mL−1). For the reference cell, 2-azidoethanol (0.1 mg mL−1) in running buffer A was used. For the study with Anl-GFP, running buffer B was used. For the immobilization of Anl-GFP a solution of 0.1 mg mL−1 in 10 mM formic acid buffer (pH 4.0) was used. Remaining free cyclooctynes were blocked with 2-azidoethanol (0.1 mg mL−1). The reference cell was treated as described above with 2-azidoethanol.
The net SPR signal is the difference in SPR angle between sample and reference cells, expressed in m°. SPR experiments were performed in duplicate at 25 °C in running buffer. Regeneration of the chips was achieved with a fresh 25 mM NaOH solution using a double 30 s injection.
Acknowledgements
This work is financed by the INTERREG IV A Germany–Netherlands program through EU funding from the European Regional Development Fund, the MWEBWV Ministry of North-Rhine Westphalia, the Dutch Ministry of Economic Affairs, Province of Gelderland and the project management Euregio Rhein-Waal.Plasmids coding for L13N, Y260L, H301L MetRS (NLL-MetRS) and GFP were a kind gift by D. A. Tirrell.
References
- D. G. Myszka, J. Mol. Recognit., 1999, 12, 390–408 CrossRef CAS.
- J. M. McDonnell, Curr. Opin. Chem. Biol., 2001, 5, 572–577 CrossRef CAS.
- R. B. M. Schasfoort and A. J. Tudos, Handbook of surface plasmon resonance, RSC Pub., Cambridge, UK, 2008 Search PubMed.
- R. L. Rich and D. G. Myszka, J. Mol. Recognit., 2008, 21, 355–400 CrossRef CAS.
- B. Johnsson, S. Lofas and G. Lindquist, Anal. Biochem., 1991, 198, 268–277 CrossRef CAS.
- B. Renberg, I. Shiroyama, T. Engfeldt, P. A. Nygren and A. E. Karlstrom, Anal. Biochem., 2005, 341, 334–343 CrossRef CAS.
- P. Nilsson, B. Persson, M. Uhlen and P. A. Nygren, Anal. Biochem., 1995, 224, 400–408 CrossRef CAS.
- M. F. Debets, S. S. van Berkel, J. Dommerholt, A. T. Dirks, F. P. Rutjes and F. L. van Delft, Acc. Chem. Res., 2011, 44, 805–815 CrossRef CAS.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS.
- M. F. Debets, S. S. van Berkel, S. Schoffelen, F. P. J. T. Rutjes, J. C. M. van Hest and F. L. van Delft, Chem. Commun., 2010, 46, 97–99 RSC.
- M. F. Debets, C. W. J. van der Doelen, F. P. J. T. Rutjes and F. L. van Delft, ChemBioChem, 2010, 11, 1168–1184 CrossRef CAS.
- J. Dommerholt, S. Schmidt, R. Temming, L. J. Hendriks, F. P. Rutjes, J. C. van Hest, D. J. Lefeber, P. Friedl and F. L. van Delft, Angew. Chem., Int. Ed., 2010, 49, 9422–9425 CrossRef CAS.
- X. Ning, R. P. Temming, J. Dommerholt, J. Guo, D. B. Ania, M. F. Debets, M. A. Wolfert, G. J. Boons and F. L. van Delft, Angew. Chem., Int. Ed., 2010, 49, 3065–3068 CrossRef CAS.
- C. S. McKay, J. Moran and J. P. Pezacki, Chem. Commun., 2010, 46, 931–933 RSC.
- B. C. Sanders, F. Friscourt, P. A. Ledin, N. E. Mbua, S. Arumugam, J. Guo, T. J. Boltje, V. V. Popik and G. J. Boons, J. Am. Chem. Soc., 2011, 133, 949–957 CrossRef CAS.
- A. M. Jawalekar, E. Reubsaet, F. P. Rutjes and F. L. van Delft, Chem. Commun., 2011, 47, 3198–3200 RSC.
- S. Schoffelen, M. B. van Eldijk, B. Rooijakkers, R. Raijmakers, A. J. R. Heck and J. C. M. van Hest, Chem. Sci., 2011, 2, 701–705 RSC.
- S. Schoffelen, M. H. Lambermon, M. B. van Eldijk and J. C. M. van Hest, Bioconjugate Chem., 2008, 19, 1127–1131 CrossRef CAS.
- J. C. van Hest and F. L. van Delft, ChemBioChem, 2011, 12, 1309–1312 CrossRef CAS.
- W. X. Chen, D. Z. Wang, C. F. Dai, D. Hamelberg and B. H. Wang, Chem. Commun., 2012, 48, 1736–1738 RSC.
- R. Löser, K. Schilling, E. Dimmig and M. Gutschow, J. Med. Chem., 2005, 48, 7688–7707 CrossRef.
- R. E. Babine and S. L. Bender, Chem. Rev., 1997, 97, 1359–1472 CrossRef CAS.
- H. H. Otto and T. Schirmeister, Chem. Rev., 1997, 97, 133–171 CrossRef CAS.
- S. A. Thompson, P. R. Andrews and R. P. Hanzlik, J. Med. Chem., 1986, 29, 104–111 CrossRef CAS.
- E. Dufour, A. C. Storer and R. Menard, Biochemistry, 1995, 34, 9136–9143 CrossRef CAS.
- N. J. de Mol, M. B. Gillies and M. J. Fischer, Bioorg. Med. Chem., 2002, 10, 1477–1482 CrossRef CAS.
- J. A. Johnson, Y. Y. Lu, J. A. Van Deventer and D. A. Tirrell, Curr. Opin. Chem. Biol., 2010, 14, 774–780 CrossRef CAS.
- S. Brase, C. Gil, K. Knepper and V. Zimmermann, Angew. Chem., Int. Ed., 2005, 44, 5188–5240 CrossRef CAS.
- C. Tassa, M. Liong, S. Hilderbrand, J. E. Sandler, T. Reiner, E. J. Keliher, R. Weissleder and S. Y. Shaw, Lab Chip, 2012, 12, 3103–3110 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis of nitrile ligands, ITC and UV experiments, expression and purification of Anl-GFP. See DOI: 10.1039/c3lc41338a |
| This journal is © The Royal Society of Chemistry 2013 |
