 Open Access Article
Open Access ArticleThe mechanistic influence of aligned nanofibers on cell shape, migration and blebbing dynamics of glioma cells†
Puja
Sharma
a,
Kevin
Sheets
a,
Subbiah
Elankumaran
b and
Amrinder Singh
Nain
*ca
aSchool of Biomedical Engineering and Sciences, ICTAS, 325 Stanger Street, Virginia Tech, Blacksburg, Virginia 24061, USA. E-mail: nain@vt.edu
bDepartment of Biomedical Sciences and Pathobiology, VMRCVM, Virginia Tech, Duck Pond Drive 0442, Blacksburg, Virginia 24061, USA
cMechanical Engineering Department, 100S Randolph Hall, Virginia Tech, Blacksburg, Virginia 24061, USA
First published on 3rd June 2013
Abstract
Investigating the mechanistic influence of the tumor microenvironment on cancer cell migration and membrane blebbing is crucial in the understanding and eventual arrest of cancer metastasis. In this study, we investigate the effect of suspended and aligned nanofibers on the glioma cytoskeleton, cell shape, migration and plasma membrane blebbing dynamics using a non-electrospinning fiber-manufacturing platform. Cells attached in repeatable shapes of spindle on single fibers, rectangular on two parallel fibers and polygonal on intersecting fibers. Structural stiffness (N m−1) of aligned and suspended nanofibers (average diameter: 400 nm, length: 4, 6, and 10 mm) was found to significantly alter the migration speed with higher migration on lower stiffness fibers. For cells attached to fibers and exhibiting blebbing, an increase in cellular spread area resulted in both reduced bleb count and bleb size with an overall increase in cell migration speed. Blebs no longer appeared past a critical cellular spread area of approximately 1400 μm2. Our results highlighting the influence of the mechanistic environment on the invasion dynamics of glioma cells add to the understanding of how biophysical components influence glioma cell migration and blebbing dynamics.
Insight, innovation, integrationThe influence of the biophysical microenvironment on tumor cell behavior is well recognized. Glioblastoma cells prefer migrating along nano-micron sized aligned white matter tracts which are challenging to recreate for in vitro studies. Our study integrates a novel non-electrospinning STEP (Spinneret based Tunable Engineered Parameters) technology with time lapse video microscopy to investigate individual glioblastoma migration and blebbing dynamics on suspended and aligned nanofibers. For the first time, we demonstrate that the structural stiffness (N m−1) of the suspended nanofiber influences glioblastoma cell migration with cells migrating faster on lower stiffness nanofibers. In addition, for the first time we show a reversible blebbing and non-blebbing phenomenon where the blebbing dynamics of glioblastoma cells is affected by cell spread area. STEP nanofibers are an improvement over the conventionally-used flat cell culture surfaces as the suspended and aligned fibers better represent the white matter tracts in vivo. This study also warrants that cancer should be viewed as a system that constantly interacts with its biophysical and biochemical microenvironment. |
1. Introduction
Cancer is the second leading cause of death in the US, with an overall loss of $263.8 billion in medical and morbidity costs.1 Glioblastoma multiforme (GBM) originates from the glia or its precursors in the central nervous system.2 Characterized as a grade IV tumor by the World Health Organization, GBM is the most aggressive and also the most common glioma in humans.3 Approximately 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people die of GBM every year.4 It can be described as a complex organ lesion with diverse anatomy, genetic mutations, and genetic variability within the tumor.2,5 Lack of understanding of causative agents, tumor progression, invasiveness and the location of tumors in vital areas of the brain have limited treatment protocols to surgery, chemotherapy, and radiation therapy, making GBM one of the most difficult conditions to treat. Even with extensive therapy, the median survival rate is only 15 months.2,6
000 people die of GBM every year.4 It can be described as a complex organ lesion with diverse anatomy, genetic mutations, and genetic variability within the tumor.2,5 Lack of understanding of causative agents, tumor progression, invasiveness and the location of tumors in vital areas of the brain have limited treatment protocols to surgery, chemotherapy, and radiation therapy, making GBM one of the most difficult conditions to treat. Even with extensive therapy, the median survival rate is only 15 months.2,6
Described as one of the hallmarks of cancer, the ability of tumors to invade surrounding tissues and metastasize to other parts of the body causes 90% of cancer deaths in humans.7 One of the major challenges in treating GBM arises from its highly invasive behavior, which allows tumors to progress despite surgery, chemo and radiation therapy. About 15–25% of the central nervous system volume is occupied by the extracellular space, which contains metabolites, hormones, proteins, and extracellular matrix (ECM) molecules that the glia and neurons produce. It is composed of proteins such as glycosaminoglycans (GAG), hyaluronan (HA), fibronectin, laminin, proteoglycans, and nanofibrous collagen structures.8 Cells typically attach to and utilize their immediate ECM to migrate. Probing the mechanical components of the extracellular environment has shown that it directly affects cellular migration, proliferation, and cytoskeletal organization, thus establishing mechanical stimuli as a necessary factor that influences cellular behavior.9 It is well known that elasticity (N m−2) of the environment affects migration.10 The rigidity of the substrate has also been known to alter the cytoskeletal organization of glioma cells6 with stiffer constructs enhancing integrin expression and progression.11 However, the influence of structural stiffness (N m−1) on cell migration has not been investigated. Also, a recent study has suggested that the local mechanical property of the substrate may have a stronger influence on cell behavior than the bulk mechanical property of the substrate.12 Therefore, it is important to understand the influence of structural stiffness on glioma cell migration to get a better picture of how the biophysical environment influences glioma metastasis.
The normal brain ECM differs significantly from that of patients with GBM. Glioma cells modify their microenvironment as they migrate by rearranging the normal brain ECM (approximate modulus of 500 Pa) to create an ECM that is more rigid to facilitate proliferation.6,13 In particular, glioma cells have been known to migrate via normal brain parenchyma, collect below the pial margin, border around blood vessels and neurons and directionally migrate along highly aligned white matter tracts.2,14,14b,15 Studies on dimensions of white matter tracts show considerable variations suggesting that they can even be characterized into small and larger tracts.16 Consequently, the range of diameters of these aligned anatomical white matter tracts has been shown to be anywhere from less than 500 nm to 7 μm.15–17 Therefore, suspended, aligned and multilayer nanofibers of varying diameters and lengths serve as an excellent platform to study individual glioblastoma cell behavior in vitro. In this study, a constant starting diameter of 400 nm is selected to resemble glioma migration in smaller white matter tracts. There are two reasons for starting at a lower diameter: (a) in future, we aim to extend the study by progressively exploring glioma migration in fibers with larger diameters to understand mechanisms by which fiber curvature influences migration, and (b) the STEP technique allows high spinnability of polystyrene in the 400–900 nm fiber diameter range.
In addition to metastasizing and migrating to secondary locations, the ability of cancer cells to resist apoptosis is also described as a hallmark of cancer.7a A phenomenon that interlinks cytoskeleton organization, migration, and apoptosis is blebbing. Observed in the 1900s, blebs were described as blisters that are now characterized as short lived (<1 minute) circular extensions (about 2–15 μm in diameter) that expand off the cytoplasm and retract to the initial site of origin.18 While blebs have been associated with apoptosis, migration, cytokinesis, cell spreading, virus infection, cellular protection against injury and migration, it is still a largely unknown phenomenon whose complete functions are yet to be recognized.18,19 The dynamics of blebbing in cancerous cells requires more attention as it has been associated with invasiveness, ability to escape apoptosis, and motility.19a–c In 1970, Blaser et al. demonstrated how blebbing was used for cellular motility in zebra fish germ cells.20 Similarly, Babiychuk et al. showed that human embryonic kidney cells utilize blebs to trap detrimental constituents of the cytosol to enhance survival of injured cells.19b Blebbing has also been associated with changes in nuclear shapes, mitotic disturbances causing genetic instability, and multidrug resistance in tumor cells.21 A study by Caspani et al. has shown that glioma cells exhibit reversible blebbing and non-blebbing phenotypes both in vivo and in vitro, and that the blebs do not contribute significantly in glioma migration.14b This makes the study of blebs a crucial component in the understanding of glioma migration as blebs are being increasingly associated with tumor cell behaviors.
Using suspended and highly aligned nanofibers in single and multiple layers to represent the aligned physiological pathways used by glioma for migration can help us better understand how individual cells are influenced by changes in their microenvironment.15 In this study, the migratory behavior in response to single nanofiber structural stiffness (N m−1), associated cell shape, and blebbing dynamics of malignant glioma cells are investigated.22
2. Materials and methods
2.1 Cell culture
Passage 18 DBTRG-05MG (Denver Brain Tumor Research Group-05MG) cell line was purchased from ATCC (American Type Culture Collection, Manassas, VA). The cells were maintained in RPMI-1640 media (ATCC), supplemented with 10% FBS (HyClone, Canada), 1% penicillin/streptomycin (HyClone, Logan, UT), additional 30 mg L−1L-proline, 35 mg L−1L-cystine, 3.57 g L−1HEPES (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid), 15 mg L−1hypoxanthine, 1 mg L−1adenosine triphosphate, 10 mg L−1adenine, and 1 mg L−1thymidine (Sigma Aldrich, St. Louis, MO) as recommended by ATCC. Both T75 and T25 cell culture flasks (Corning Inc., Corning, NY) were used to culture the cells at 5% CO2 and 37 °C in the incubator (Thermo Scientific, Barrington, IL). DBTRG-05MG passage numbers 19–30 were used for data collection. Media were renewed 2 times a week. In order to suspend the cells, media were removed, and the cells were rinsed with PBS twice. They were then dispersed with 0.25% trypsin (HyClone, Logan, UT) for one minute and the cells were resuspended in fresh medium.2.2 STEP fibrous substrate
Substrates of suspended, parallel, and intersecting polystyrene (PS) nanofibers (lengths: 4, 6 and 10 mm; diameters: approximately 400 nm) were manufactured using the STEP platform.22b,23 The diameters of the fibers were confirmed using scanning electron micrographs. Square Thermanox® plastic cover slips (NUNC Brand Products, Rochester, NY) were used as frames for the substrates. Subsequently, they were cut out (4, 6, and 10 mm) to form hollow substrates upon which suspended fibers were deposited. The substrates were mounted on glass bottom 6-well plates (MatTek Corp., Ashland, MA), sterilized with 70% ethanol for 5 minutes, rinsed with PBS twice and coated with fibronectin (2 μg mL−1) overnight before seeding the cells. The structural stiffness (N m−1) in the middle of suspended fiber (750 μm on either sides from the center of the fiber span length) was calculated for 4, 6 and 10 mm polystyrene fibers by Atomic Force Microscopy (AFM, Veeco BioScope II, Plainview, NY; using tip less cantilevers of stiffness 0.2 N m−1 from AppNano, Santa Clara, CA). A three-point bending test23a,24 was used to determine fiber structural stiffness within 750 μm on either side from the center of the fiber, and the average values were calculated.2.3 Cell seeding
The cells were resuspended to obtain a concentration of 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL. While 30 μL of this suspension was placed on the 4 × 4 and 6 × 6 mm2 fibronectin coated substrates, 60 μL of the suspension was placed on 10 × 10 mm2 fibronectin coated substrates. The seeded substrates were placed and maintained at 5% CO2 and 37 °C in the incubator until the cells attached onto the nanofibers (2–6 hours). About 1 hour after seeding, 2 mL of media was added to each well. Once cells attached, time-lapse video micrographs of the substrates were taken continuously for 10 hours (every 10 or 15 minutes) using a Zeiss microscope with incubating capacity (Zeiss AxioObserver Z1, Jena, Germany).
000 cells per mL. While 30 μL of this suspension was placed on the 4 × 4 and 6 × 6 mm2 fibronectin coated substrates, 60 μL of the suspension was placed on 10 × 10 mm2 fibronectin coated substrates. The seeded substrates were placed and maintained at 5% CO2 and 37 °C in the incubator until the cells attached onto the nanofibers (2–6 hours). About 1 hour after seeding, 2 mL of media was added to each well. Once cells attached, time-lapse video micrographs of the substrates were taken continuously for 10 hours (every 10 or 15 minutes) using a Zeiss microscope with incubating capacity (Zeiss AxioObserver Z1, Jena, Germany).
2.4 Cell tracking/blebbing analysis
Time lapse videos were performed at 10× or 20× magnification and were analyzed using AxioVision software (Zeiss, Germany). Approximately 30 cells were sampled per substrate. Cell displacements were measured every hour from the center of the cell for the entire time-lapse period (10 hours). The highest displacement in the entire 10 hour period was recorded. Cells that migrated less than 10 μm h−1 (about 30%, N = 157) were considered non-moving cells and were not included in the migration study. Cellular migration was recorded at the middle of the span length of suspended nanofibers (about 750 μm on either side from the center of the nanofiber).Time-lapse video micrographs obtained at 20× were used to analyze the blebbing dynamics of the DBTRG-05MG cells. Using AxioVision software, cell spread area, bleb count, and bleb sizes were measured. Bleb size and count were compared as a function of cell spread area or cell size.
2.5 Cell cytoskeleton staining
DBTRG-05MG cells on flat and suspended nanofibers were stained for F-actin stress fibers, focal adhesions and the nucleus. The cells were fixed using 4% paraformaldehyde in phosphate buffered saline (PBS) solution for 15 minutes, and washed two times with PBS. The cells were exposed to permeabilization solution (PBS + 0.1% Triton-X 100 solution) for 15 minutes, PBS washed twice, soaked in anti-goat blocking buffer (Invitrogen, Grand Island, NY) for 30 minutes. Diluted primary paxillin (pY31, Invitrogen, Grand Island, NY) was used (PBS with 1% Bovine Serum Albumin and Triton-X 100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution ratio), and the substrates were refrigerated (2–8 °C) for 1 hour. After washing it with PBS three times, the secondary stain (Goat, anti-rabbit, Alexa Fluor 488 or 647, Grand Island, NY) and Phalloidin (Santa Cruz Biotechnology, Santa Cruz, CA) were used at 1
100 dilution ratio), and the substrates were refrigerated (2–8 °C) for 1 hour. After washing it with PBS three times, the secondary stain (Goat, anti-rabbit, Alexa Fluor 488 or 647, Grand Island, NY) and Phalloidin (Santa Cruz Biotechnology, Santa Cruz, CA) were used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilutions. The substrates were placed in room temperature for 1 hour, away from light. After washing with PBS three times, DAPI (4′,6-diamidimo-2-phenylindole) was used to stain the nucleus for 5 minutes. The substrates were then rinsed with PBS two times, and observed using a Zeiss® microscope.
200 dilutions. The substrates were placed in room temperature for 1 hour, away from light. After washing with PBS three times, DAPI (4′,6-diamidimo-2-phenylindole) was used to stain the nucleus for 5 minutes. The substrates were then rinsed with PBS two times, and observed using a Zeiss® microscope.
2.6 Statistical analysis
JMP software was used to analyze the data for statistical significance. ANOVA and Student's t-tests were used to test for significant differences between variables when required.3. Results
Our previously reported non-electrospinning pseudo dry-spinning method called the STEP technique is able to deposit highly aligned polymeric micro/nanofibers in single and multiple layers. The fiber networks have uniform diameters and can be manufactured at user defined geometrical spacing (Fig. S1, ESI†).22 Specifically, high aspect ratio (length/diameter) fibers can be precisely deposited with control of fiber dimensions (diameter: sub 100 nm-micron, length: mm-cm, parallelism: ≤2.5 degrees, spacing: sub-micron to microns) in single and multiple layers. In this study, both highly aligned parallel single suspended (SS) and orthogonally arranged double suspended (SD) fibers were deposited on hollow square plastic frames, and used for migration and blebbing dynamics study (Fig. S1, ESI†). For this study, fiber diameter was kept constant at ∼400 nm.3.1 DBTRG-05MG cells on flat and STEP fibers
DBTRG-05MG cells mainly adopted 3 major morphologies on STEP fibers: spindle shape on single fibers, rectangular shape on parallel fibers and polygonal shape at orthogonal fiber intersections (Fig. 1). This observation was consistent with those observed by Sheets et al. where mouse myoblasts also adopted three major morphologies when cultured on STEP fibers.25 In spindle shaped cells (Fig. 1(i) and (v)), the nucleus was predominately located at the cell center. The stress fibers were around the nucleus connecting the poles of the spindle, and focal adhesions were concentrated near the poles of the spindles with occasional expressions along the cell-fiber interface. Cells between two parallel nanofibers formed (Fig. 1(ii) and (vi)) rectangular cell structures with nucleus at the center and stress fibers mostly around the perimeter. Focal adhesions were mostly concentrated in the corners of the rectangular morphologies, with some along the cell–nanofiber interface. On orthogonal fibers, cells formed polygonal (kite like) structures (Fig. 1(iii) and (vii)) with the nucleus at the center. Focal adhesions were pronounced in the corners of the cell with occasional expressions along the cell–nanofiber interface.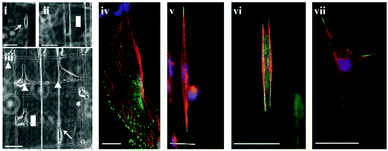 | ||
| Fig. 1 Optical images of cellular morphologies on SS: (i) spindle (indicated by an arrow), (ii) rectangular (indicated by a rectangle), and on SD nanofibers (iii) circular (indicated by *), spindle (indicated by an arrow), rectangular (indicated by a rectangle) and polygonal (indicated by triangles), immunostained images of DBTRG-05MG cells (iv) on flat, (v) spindle shaped on a single nanofiber, (vi) rectangular between two parallel nanofibers, and (vii) polygonal on an intersection of two orthogonal nanofibers. Stains: blue (DAPI, nucleus), red (Phalloidin, F-actin), and green (Alexa Fluor®, focal adhesion paxillin). Scale bar = 50 μm. | ||
On flat control substrates, however, DBTRG-05MG cells mostly showed spread configurations with pronounced focal adhesions around the circumference of the cells (Fig. 1(iv)). Stress fibers were visibly seen to be connecting the focal adhesions forming netlike structures of the cytoskeleton. Cells were observed to migrate at about 30 μm per hour on average on flat substrates, and were found to have no obvious directional bias.
3.2 DBTRG-05MG migration on STEP nanofibers
Cell migration has been described as a multi-step process involving polarization of the cell, extension of protrusion at the leading edge, contraction of the actomyosin complex, proteolytic degradation of ECM and retraction of the trailing edge.26 Using the STEP platform, the polarization, extension of protrusions at the leading edge, and retraction of the trailing edge of a single cell on a single nanofiber were captured using time lapse images (Fig. 2). Cells polarized, extended their leading edge, and retracted their trailing edge like a ‘sling shot’ on single nanofibers.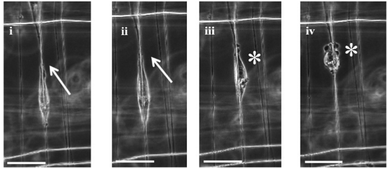 | ||
| Fig. 2 Time lapse of a single DBTRG-05MG migrating along a single nanofiber at 10 minute intervals. (i) and (ii) show the extension of leading edges (marked by arrows), and (iii) and (iv) show retraction of the trailing edge. Cell does not bleb when spread (i) and (ii), and starts blebbing (shown by *) once the spread area reduces, (iii) and (iv). Scale bar = 50 μm. | ||
As tumor cells start remodeling their environment through increased cross linking, the associated increase in ECM stiffening has been observed to affect focal adhesion expression and metastasis.11,27 Prior studies have observed that tumor environments are stiffer than normal ECM, and the stiffness of ECM influences cancer cells.11,13b,27,28 Therefore, we investigated the difference in migration speeds of DBTRG-05MG on SD (2 layers of orthogonally arranged fibers, ESI,† Fig. S1) versus SS (a single parallel layer of fibers, ESI,† Fig. S1) and the role of structural stiffness within SS. It was observed that glioma cells spread more and migrated significantly slower on double suspended (SD) fibers when compared to cells on parallel single suspended (SS) fibers and on flat fibers (Fig. 3(i)).
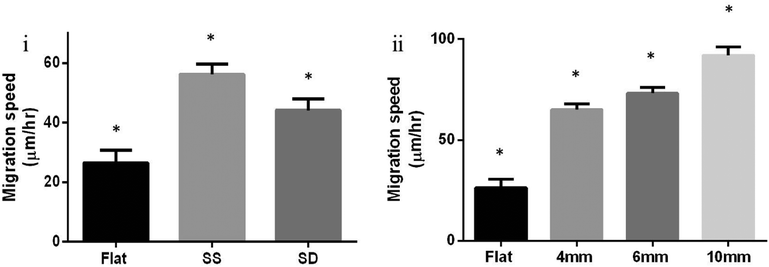 | ||
| Fig. 3 (i) DBTRG-05MG migration on flat (N = 14), single suspended (SS, parallel, N = 56), and double suspended (SD, orthogonal, N = 62) nanofibers ±SE of length 6 mm. A statistical difference was observed between migration on flat, SS, and SD nanofibers (Student's t-test, p = 0.0004 for SS-flat, p = 0.0294 for SD-flat, and p = 0.0171 for SS-SD), (ii) cell migration of single DBTRG-05MG cells on single suspended (SS, parallel) fibers ±SE. Cells migrated significantly faster on fibers of lengths 10 mm (N = 60), 6 mm (N = 101) and 4 mm (N = 120) compared to flat (N = 14). Statistically significant differences were observed between all lengths (Student's t-test, p < 0.0001 for 10 mm-flat, 10–4 mm, 6 mm-flat, 4 mm-flat, p = 0.0001 for 10–6 mm, and p = 0.0439 for 6–4 mm). The average structural stiffness within 750 μm on either side from the center of the 4, 6, and 10 mm nanofibers was experimentally determined to be 3.4, 1.5, and 0.75 mN m−1 respectively. * shows statistical significance. | ||
Furthermore, single suspended nanofibers of lengths 4, 6 and 10 mm were used to study DBTRG-05MG migration behavior on single fibers. Migration data were generated from cells attached at the middle of the span lengths (about 750 μm on either side from the center of the nanofiber). Nanofiber length significantly increased cell migration and cells migrated the fastest on 10 mm nanofibers among the three lengths tested (Fig. 3(ii)). The cells migrated significantly faster on all three lengths tested when compared to flat substrates. Compared to currently reported values in the literature for glioma migration (0.4 to 100 μm h−1),29 in this study, cells were observed to migrate at the higher end of this range. However, glioma migration speeds from 13–196 μm h−1 were recorded on STEP fibers.
3.3 Blebbing dynamics
DBTRG-05MG cells demonstrated relatively continuous blebbing on flat surfaces (Fig. 4(I)) and exhibited an interesting reversible blebbing–non-blebbing phenomenon on suspended fibers (Fig. 4(II–IV), and ESI,† Movie S1). Separation of the actin cortex from the plasma membrane is considered a hallmark for blebbing.19a This distinction was observed when DBTRG-05MG cells were stained for actin. Fig. 4III demonstrates that the actin cytoskeleton of the bleb is distinct and separate from the actin cortex. As blebbing has been associated with migration, resistance to cell death and multi drug resistance,19b,21c it is important to study the blebbing behavior of glioma cells as they migrate. In particular, we investigated the relationship of blebbing as a function of cell migration and spreading on our STEP fibers. Not all the cells demonstrated plasma membrane blebbing (percentage of blebbing cells: 65% (N = 349) on flat, and 36% (N = 248) on STEP fibers), and those cells that exhibited this phenomenon were considered for the blebbing dynamics study. We observed that blebs on a single cell were more in number (count) and larger in size when cells were in smaller, spherical morphologies. As cells spread along the STEP nanofibers (both SS and SD) and increased in spread area, both bleb size and their occurrence count decreased (Fig. 4(II–IV) and 5, and Movie S1, ESI†). Furthermore, it was observed that blebbing cells had significantly lower migration than cells that were not blebbing (Fig. 6). Both bleb size and count showed a decrease in blebbing as the cells spread in area, and a near elimination of blebbing at a cell spread area of about 1400 μm2 and beyond. However, as highly spread cells retracted to smaller areas, re-occurrence of blebbing was observed (Fig 4(IV), and ESI,† Movie S1). This reversible blebbing–non-blebbing phenomenon based on cell spreading, to the best of authors' knowledge, has not been reported before. Characterizing the occurrence of blebbing with respect to glioma cell migration can add to our understanding of cell migration specific apoptotic and drug resistance behaviors. | ||
| Fig. 4 DBTRG-05MG cell blebbing dynamics. (I) Continuous plasma membrane blebbing of DBTRG-05MG on flat, (II) while the spread cell has no blebs, the circular cell (shown with an arrow) is blebbing on suspended fiber networks, (III) actin (Phalloidin) immunofluorescent stain of a blebbing DBTRG-05MG. The bleb actin cytoskeleton (white arrow) of the bleb is separated from the actin cortex (blue arrow). Scale bars are 20 μm, 50 μm and 20 μm for (I), (II), and (III) respectively, (IV) time lapse images at 20 minute intervals of DBTRG-05MG cell blebbing dynamics on STEP fibers (shown using arrow). Blebbing decreases or disappears as cell spreads along the nanofibers (b, c, f, i, j, and l), and increases or reappears as cells reduce their spreading area (a, d, e, g, h, and k). Scale bar is 50 μm. Respective cell spread areas are shown on top right corners. Video included in the ESI† (Movie S1). | ||
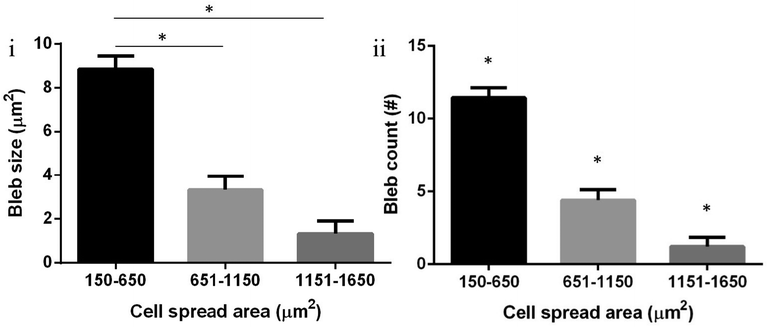 | ||
| Fig. 5 Increasing cell spread area inversely influenced both bleb occurrence and size in single DBTRG-05MG cells. (i) Bleb size for cells with spread area 150–650 μm2 (N = 109) was significantly higher than those for areas 651–1150 μm2 (N = 80), and 1151–1650 μm2 (N = 36) (Student's t-test, both p < 0.01). Bleb size for a cell spread area of 651–1150 μm2 was almost significantly higher than those for areas 1151–1650 μm2 (Student's t-test, p = 0.05). (ii) Similarly, bleb count for cell spread area 150–650 μm2 was significantly higher those for areas 651–1150 μm2 and 651–1150 μm2 (Student's t-test, p < 0.01 for 150–650 μm2 to 651–1150 μm2 and 150–650 μm2 to 1151–1650 μm2, p = 0.02 for 651–1150 μm2 to 1151–1650 μm2). * Shows statistical significance. | ||
 | ||
| Fig. 6 Blebbing DBTRG-05MG cells migrate slowly. DBTRG-05MG cell migration was significantly slower for blebbing cells (N = 31) compared to those that were not blebbing (N = 30) (ANOVA, p = 0.002). * shows statistical significance. | ||
4. Discussion and conclusion
Glioma is an integral system that nurtures itself by conditioning its microenvironment and vice versa. In order to understand the comprehensive migratory and invasive behavior of glioma cells, a thorough understanding of cell–microenvironment interaction is necessary. It is thus essential for in vitro platforms to recapture the in vivo glioma migration environment. Current methods of simulating the ECM environment involve both 2-D and 3-D substrates. As glioma metastasis is known to be favoured by aligned physiological structures such as white matter tracts, suspended and aligned fibers like the STEP fibers serve as an improvement over the conventionally used 2D and 3D platforms to study glioma migration. While studies have reported the material stiffness (N m−2) of white matter tracts in the range of 2.5–10 kPa,30 reference to their structural stiffness is not available. Using the STEP platform, the structural stiffness of the fibers was altered by the choice of fiber length. As structural stiffness decreased with fiber length, cell migration increased. This effect of structural stiffness on cell migration is a new finding and we hope that this will encourage the community to investigate this in detail.Cytoskeletal staining and time-lapse images of DBTRG-05MG suggested that cellular shape and migration are different on conventionally used flat and suspended STEP fibers. Caspani et al. had observed that actin fibers align along the white matter tracts in migratory glioma cells in vivo, similar to what we observed when DBTRG-05MG aligned along the STEP fibers.14b DBTRG-05MG cells acquired 3 major morphologies while interacting with the nanofibers, and most focal adhesions were observed in the poles of the cells (Fig. 3). Although the relationship between focal adhesion and cell migration is not understood well, focal adhesion size has been associated with cell migration.31 We observed that the DBTRG-05MG cells expressed different focal adhesion patterns on STEP fibers when compared to cells on flat substrates which could suggest why they migrated at different speeds on flat substrates and on the fibers. However, this relationship needs further investigation.
Blebbing and its influence in cancer requires more investigation as the study shows that cell spreading can significantly reduce blebbing. One of the causes of blebbing is explained as the rupturing of this actin cortex, and the propulsion of cytoplasmic contents out of the ruptured site.19a As the cell re-establishes its ruptured actin, blebs retract back to the initial site of propulsion. Actin structures in spherical and spread cells could possibly explain this inverse relation of blebbing and cell spreading. In spherical cells, actin is mostly present in the circumference as actin cortex32 which acts as a single barrier. Once this barrier ruptures, cytoplasmic contents freely expel out from the ruptured region. In spread cells, however, actin stress fibers form net like structures33 and organize along the perimeter, thus acting as a series of multiple barriers that can possibly decrease the likelihood of bleb formation.
This phenomenon is particularly interesting as blebbing is characteristic of amoeboid migration in cancerous cells.19c Described as one of the modes of cancer migration, amoeboid migration is adopted by cancerous cells in their circular configurations which allow the cells to squeeze through pores in the ECM and migrate without degrading the ECM.26 Linear regression analysis suggests that the DBTRG-05MG cells form little or no blebs when they are spread beyond an area of about 1400 μm2. It was also observed that blebbing decreased cellular migration. This suggests that amoeboid mode of migration is slower, and is least likely to occur when the cell spreads beyond this threshold. Blebs have also been associated with resistance to cell lysis, drug resistance, and cell survival.34 Therefore, for better prognosis of cancer, a better understanding of migration and blebbing dynamics is necessary.
Cancer is a system that is influenced by both the biophysical and the biochemical aspects of the tumor microenvironment. Studying cancer cell behavior in environments that resemble glioma migration pathways is therefore critical. This study utilizes a non-electrospinning platform to study cytoskeletal, migration and blebbing dynamics of glioblastoma cells and suggests that glioma cells behave differently on conventionally used flat and suspended STEP nanofiber platforms. Hence, it can be expected that the presented glioma cell behavior might bear a closer resemblance to in vivo glioma migration.
While biochemical components may be equally important, we show for the first time that changes in structural stiffness and organization of individual fibers in the microenvironment can significantly alter cell behavior. The change in migratory and blebbing behavior as a function of cellular spreading could also have clinical implications associated with metastasis and resistance against anticancer drugs. Cellular spreading has been related to reduced blebbing. However, the nature of reversible blebbing and non-blebbing behavior of glioma cells warrants further investigation. The unique strategy of investigating cancer cell behavior and dynamics in a fibrous environment can also be coupled with studies of biochemical cues to advance our current understanding of cancer. Our future works will focus on determining the effect of fiber diameter, curvature and tension on migration dynamics and vulnerability of cells to anticancer agents or lysis under spread configuration.
Acknowledgements
The authors would like to acknowledge the financial support from the Jeffress Memorial Trust, and Andrea and Bill Waide Research funds. They also acknowledge the Institute for Critical Technology and Applied Sciences, Virginia Tech, and Ji Wang for taking detailed SEM images of the STEP nanofibers, and AFM measurements to calculate the structural stiffness of individual nanofibers.References
- A. C. Society, Cancer Facts and Figures 2010, Atlanta, GA, 2010 Search PubMed.
- E. C. Holland, Glioblastoma multiforme: the terminator, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6242–6244 CrossRef CAS.
- D. Louis, H. Ohgaki, O. Wiestler, W. Cavenee, P. Burger, A. Jouvet, B. Scheithauer and P. Kleihues, The 2007 WHO Classification of Tumours of the Central Nervous System, Acta Neuropathol., 2007, 114, 97–109 CrossRef.
- C. B. T. R. o. t. U. States CBTRUS 2002: Statistical Report: Primary Brain Tumors- in the United States, 1995–1999: Central Brain Tumor Registry of the United States, 2002; 2002.
- M. Nakada, D. Kita, T. Watanabe, Y. Hayashi, L. Teng, I. V. Pyko and J.-I. Hamada, Aberrant Signaling Pathways in Glioma, Cancer, 2011, 3, 3242–3278 CrossRef CAS.
- T. A. Ulrich, E. M. de Juan Pardo and S. Kumar, The Mechanical Rigidity of the Extracellular Matrix Regulates the Structure, Motility, and Proliferation of Glioma Cells, Cancer Res., 2009, 69, 4167–4174 CrossRef CAS.
- (a) D. Hanahan and R. A. Weinberg, Hallmarks of Cancer: The Next Generation, Cell, 2011, 144, 646–674 CrossRef CAS; (b) P. Mehlen and A. Puisieux, Metastasis: a question of life or death, Nat. Rev. Cancer, 2006, 6, 449–458 CrossRef CAS.
- E. Sykova, in The Neuronal Environment Brain Homeostasis in Health and Disease, ed. W. Walz, Humana Press Inc., Totowa, New Jersey, 2002, p. 57 Search PubMed.
- (a) T. Yeung, P. C. Georges, L. A. Flanagan, B. Marg, M. Ortiz, M. Funaki, N. Zahir, W. Ming, V. Weaver and P. A. Janmey, Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion, Cell Motil. Cytoskeleton, 2005, 60, 24–34 CrossRef; (b) V. Swaminathan, K. Mythreye, E. T. O'Brien, A. Berchuck, G. C. Blobe and R. Superfine, Mechanical Stiffness Grades Metastatic Potential in Patient Tumor Cells and in Cancer Cell Lines, Cancer Res., 2011, 71, 5075–5080 CrossRef CAS; (c) J. Schrader, T. T. Gordon-Walker, R. L. Aucott, M. van Deemter, A. Quaas, S. Walsh, D. Benten, S. J. Forbes, R. G. Wells and J. P. Iredale, Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells, Hepatology, 2011, 53, 1192–1205 CrossRef CAS.
- C.-M. Lo, H.-B. Wang, M. Dembo and Y.-l. Wang, Cell Movement Is Guided by the Rigidity of the Substrate, Biophys. J., 2000, 79, 144–152 CrossRef CAS.
- M. R. Ng and J. S. Brugge, A Stiff Blow from the Stroma: Collagen Crosslinking Drives Tumor Progression, Cancer Cell, 2009, 16, 455–457 CrossRef CAS.
- S. P. Carey, C. M. Kraning-Rush, R. M. Williams and C. A. Reinhart-King, Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture, Biomaterials, 2012, 33, 4157–4165 CrossRef CAS.
- (a) R. M. Wiranowska and M. V. Rojiani, in Glioma-Exploring Its Biology and Practical Relevance, ed. G. Anirban, InTech, 2011 Search PubMed; (b) P. Schedin and P. J. Keely, Mammary Gland ECM Remodeling, Stiffness, and Mechanosignaling in Normal Development and Tumor Progression, Cold Spring Harbor Perspect. Biol., 2011 DOI:10.1101/cshperspect.a003228.
- (a) J. Johnson, M. O. Nowicki, C. H. Lee, E. A. Chiocca, M. S. Viapiano, S. E. Lawler and J. J. Lannutti, Quantitative analysis of complex glioma cell migration on electrospun polycaprolactone using time-lapse microscopy, Tissue Eng., Part C, 2009, 15, 531–540 CrossRef CAS; (b) E. M. Caspani, D. Echevarria, K. Rottner and J. V. Small, Live imaging of glioblastoma cells in brain tissue shows requirement of actin bundles for migration, Neuron Glia Biol., 2006, 2, 105–114 CrossRef.
- P. A. Agudelo-Garcia, J. K. De Jesus, S. P. Williams, M. O. Nowicki, E. A. Chiocca, S. Liyanarachchi, P. K. Li, J. J. Lannutti, J. K. Johnson, S. E. Lawler and M. S. Viapiano, Glioma cell migration on three-dimensional nanofiber scaffolds is regulated by substrate topography and abolished by inhibition of STAT3 signaling, Neoplasia, 2011, 13, 831–840 CAS.
- M. Makino, K. Mimatsu, H. Saito, N. Konishi and Y. Hashizume, Morphometric study of myelinated fibers in human cervical spinal cord white matter, Spine, 1996, 21, 1010–1016 CrossRef CAS.
- Y. Benninger, H. Colognato, T. Thurnherr, R. J. Franklin, D. P. Leone, S. Atanasoski, K. A. Nave, C. Ffrench-Constant, U. Suter and J. B. Relvas, Beta1-integrin signaling mediates premyelinating oligodendrocyte survival but is not required for CNS myelination and remyelination, J. Neurosci., 2006, 26, 7665–7673 CrossRef CAS.
- G. T. Charras, A short history of blebbing, J. Microsc., 2008, 231, 466–478 CrossRef CAS.
- (a) G. Charras and E. Paluch, Blebs lead the way: how to migrate without lamellipodia, Nat. Rev. Mol. Cell Biol., 2008, 9, 730–736 CrossRef CAS; (b) E. B. Babiychuk, K. Monastyrskaya, S. Potez and A. Draeger, Blebbing confers resistance against cell lysis, Cell Death Differ., 2011, 18, 80–89 CrossRef CAS; (c) O. T. Fackler and R. Grosse, Cell motility through plasma membrane blebbing, J. Cell Biol., 2008, 181, 879–884 CrossRef CAS; (d) K. F. Shea, C. M. Wells, A. P. Garner and G. E. Jones, ROCK1 and LIMK2 Interact in Spread but Not Blebbing Cancer Cells, PLoS One, 2008, 3, e3398 Search PubMed.
- (a) A. Ridley, Blebbing: motility research moves n a new direction, Nat. Rev. Mol. Cell Biol., 2009, 10, 164 CrossRef CAS; (b) H. Blaser, M. Reichman-Fried, I. Castanon, K. Dumstrei, F. L. Marlow, K. Kawakami, L. Solnica-Krezel, C.-P. Heisenberg and E. Raz, Migration of Zebrafish Primordial Germ Cells: A Role for Myosin Contraction and Cytoplasmic Flow, Dev. Cell, 2006, 11, 613–627 CrossRef CAS.
- (a) J. Gong, R. Jaiswal, J. M. Mathys, V. Combes, G. E. R. Grau and M. Bebawy, Microparticles and their emerging role in cancer multidrug resistance, Cancer Treat. Rev., 2012, 38, 226–234 CrossRef CAS; (b) D. Gisselsson, J. Björk, M. Höglund, F. Mertens, P. Dal Cin, M. Åkerman and N. Mandahl, Abnormal Nuclear Shape n Solid Tumors Reflects Mitotic Instability, Am. J. Pathol., 2001, 158, 199–206 CrossRef CAS; (c) D. Di Vizio, J. Kim, M. H. Hager, M. Morello, W. Yang, C. J. Lafargue, L. D. True, M. A. Rubin, R. M. Adam, R. Beroukhim, F. Demichelis and M. R. Freeman, Oncosome Formation in Prostate Cancer: Association with a Region of Frequent Chromosomal Deletion in Metastatic Disease, Cancer Res., 2009, 69, 5601–5609 CrossRef CAS.
- (a) A. S. Nain, J. A. Phillippi, M. Sitti, J. MacKrell, P. G. Campbell and C. Amon, Control of Cell Behavior by Aligned Micro/Nanofibrous Biomaterial Scaffolds Fabricated by Spinneret-Based Tunable Engineered Parameters (STEP) Technique, Small, 2008, 4, 1153–1159 CrossRef CAS; (b) A. S. Nain, M. Sitti, A. Jacobson, T. Kowalewski and C. Amon, Dry Spinning Based Spinneret Based Tunable Engineered Parameters (STEP) Technique for Controlled and Aligned Deposition of Polymeric Nanofibers, Macromol. Rapid Commun., 2009, 30, 1406–1412 CrossRef CAS.
- (a) A. S. Nain, C. Amon and M. Sitti, Proximal Probes Based Nanorobotic Drawing of Polymer Micro/Nanofibers, IEEE Trans. Nanotechnol., 2006, 5, 499–510 CrossRef; (b) J. Wang and A. S. Nain, Polymeric nanofibers: isodiametric design space and methodology for depositing aligned nanofiber arrays in single and multiple layers, Polym. J., 2013, 1–6, DOI:10.1038/pj.2013.1.
- (a) E. P. Tan and C. T. Lim, Effects of annealing on the structural and mechanical properties of electrospun polymeric nanofibres, Nanotechnology, 2006, 17, 2649–2654 CrossRef CAS; (b) M.-K. W. Tze-jer Chuang, Nanomechanics of Materials and Structures, Springer, 2006, p. 323 Search PubMed.
- K. Sheets, S. Wunsch, C. Ng and A. S. Nain, Shape-dependent cell migration and focal adhesion organization on suspended and aligned nanofiber scaffolds, Acta Biomater., 2013, 9, 7169–7177 CrossRef CAS.
- A. Pathak and S. Kumar, Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness, Integr. Biol., 2011, 3, 267–278 RSC.
- K. R. Levental, H. Yu, L. Kass, J. N. Lakins, M. Egeblad, J. T. Erler, S. F. T. Fong, K. Csiszar, A. Giaccia, W. Weninger, M. Yamauchi, D. L. Gasser and V. M. Weaver, Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling, Cell, 2009, 139, 891–906 CrossRef CAS.
- E. L. Baker, R. T. Bonnecaze and M. H. Zaman, Extracellular Matrix Stiffness and Architecture Govern Intracellular Rheology in Cancer, Biophys. J., 2009, 97, 1013–1021 CrossRef CAS.
- (a) A. Giese, M. A. Loo, M. D. Rief, N. Tran and M. E. Berens, Substrates for astrocytoma invasion, Neurosurgery, 1995, 37, 294–301 CrossRef CAS ; discussion 301–302; (b) W. McDonough, N. Tran, A. Giese, S. A. Norman and M. E. Berens, Altered gene expression in human astrocytoma cells selected for migration: I. Thromboxane synthase, J. Neuropathol. Exp. Neurol., 1998, 57, 449–455 CrossRef CAS.
- A. Romano, M. Scheel, S. Hirsch, J. Braun and I. Sack, In vivo waveguide elastography of white matter tracts in the human brain, Magn. Reson. Med., 2012, 68, 1410–1422 CrossRef.
- D. H. Kim and D. Wirtz, Focal adhesion size uniquely predicts cell migration, FASEB J., 2013, 27, 1351–1361 CrossRef CAS.
- G. T. Charras, C.-K. Hu, M. Coughlin and T. J. Mitchison, Reassembly of contractile actin cortex in cell blebs, J. Cell Biol., 2006, 175, 477–490 CrossRef CAS.
- S. J. Tumminia, K. P. Mitton, J. Arora, P. Zelenka, D. L. Epstein and P. Russell, Mechanical stretch alters the actin cytoskeletal network and signal transduction in human trabecular meshwork cells, Invest. Ophthalmol. Visual Sci., 1998, 39, 1361–1371 CAS.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Geometric control of cell life and death, Science, 1997, 276, 1425–1428 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3ib40073e |
| This journal is © The Royal Society of Chemistry 2013 |
