 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cellular nanotechnology: making biological interfaces smarter
Paula M.
Mendes
School of Chemical Engineering, University of Birmingham, Birmingham, B15 2TT, UK. E-mail: p.m.mendes@bham.ac.uk; Fax: +44 (0)121 414 5324; Tel: +44 (0)121 414 5343
First published on 7th October 2013
Abstract
Recently, there has been an outburst of research on engineered cell–material interfaces driven by nanotechnology and its tools and techniques. This tutorial review begins by providing a brief introduction to nanostructured materials, followed by an overview of the wealth of nanoscale fabrication and analysis tools available for their development. This background serves as the basis for a discussion of early breakthroughs and recent key developments in the endeavour to develop nanostructured materials as smart interfaces for fundamental cellular studies, tissue engineering and regenerative medicine. The review covers three major aspects of nanostructured interfaces – nanotopographical control, dynamic behaviour and intracellular manipulation and sensing – where efforts are continuously being made to further understand cell function and provide new ways to control cell behaviour. A critical reflection of the current status and future challenges are discussed as a conclusion to the review.
 Paula M. Mendes | Paula M. Mendes received her MSc (1997) and PhD (2002) degrees in Chemical Engineering from the Faculty of Engineering, University of Porto, Portugal. She undertook post-doctoral research firstly (2002–04) in the School of Chemistry, University of Birmingham, UK, and subsequently (2004–06) at the Department of Chemistry and Biochemistry, University of California, Los Angeles, USA. She began her academic career in the School of Chemical Engineering, University of Birmingham in 2006, as an academic fellow, and has been a Professor of Advanced Materials and Nanotechnology and an EPSRC Leadership Fellow since 2013. The research in her group lies in the development of novel methods for controlling the structure and functionality of materials at the molecular and nanometer scales and their application in biology, medicine and energy. |
Key learning points• Nanofabricated structures, recapitulating the length scale of naturally occurring nanotopographic structures, are now being used to elucidate how physical cues can direct cell behaviour and orchestrate complex cellular processes such as stem cell differentiation and tissue organization.• Advances in nanotechnology have unlocked our ability to create stimuli-responsive interfaces for spatially and temporally controlling extracellular physical and biochemical cues. • With nanomaterials being over one hundred-fold smaller than cells, nanomaterial-based platforms can be used for intracellular sensing and delivery at the sub-cellular level. • The field of nanoengineered cell–material interfaces is rapidly evolving, carrying with it the potential for major breakthroughs in fundamental cellular studies and regenerative medicine. |
1. Introduction
While the word ‘cell’ was first introduced by Robert Hooke in 1665 to describe the fundamental units of multi-cellular organisms, it was not until 200 years later when compound achromatic microscopes became available that all plants and animals were discovered to be composed of one or more cells. Throughout its history, cell biology research has been characterized by its great capacity to apply and integrate concepts, principles and tools from other disciplines, notably physics, chemistry, engineering, computer science and mathematical sciences. It was through these collaborations that one of the greatest scientific breakthroughs of recent times – the mapping of the human genome – was accomplished.1 It is important to recognize that while our genome plays a central role in the function of almost all human cells, it is only one piece of the puzzle required to elucidate the molecular basis of cell function. Advances in cellular and molecular technologies – in particular, in imaging – have made it possible to understand a vast range of cellular and molecular mechanisms underlying cell function but many more remain obscure. For instance, the temporal element of cell communication has long been known, but its significance in receptor based signalling is poorly understood.2Cell biology today is on the verge of a nanotechnology-driven research era, one in which the availability of sophisticated new experimental techniques and tools of nanotechnology is set not only to emulate more complex, in vivo like extracellular environments, but also monitor dynamic complex biological processes in real time at the single cell level. Ultimately, the goal is to establish a fully integrated knowledge of how the building blocks of humans – cells – work at the molecular level. It is only by a detailed knowledge of how cells work, independently and together, in healthy and diseased states that one will be able to understand and anticipate the onset and effects of disease and create an appropriate and effective means to prevent and treat disease. The unravelling of cellular and molecular mechanisms that could be used to reprogram or instruct cells would enable unprecedented advances in tissue engineering and regenerative medicine. In this tutorial review, the spotlight is focused on nanostructured materials – materials with one or more dimensions measuring less than 100 nanometers (nm) – which can be used as smart interfaces to further understand and control the complex interplay of events and interactions occurring within living cells.
2. Nanostructured materials
The unprecedented ability to control and characterise materials and structures on the nanometre scale has led to a rapid development of a plethora of nanostructured materials, including nanoparticles, nanowires, nanorods, nanocapsules, nanofibers, nanotubes, nanocomposites, nanostructured surfaces and thin solid films with nanoscale thickness (Fig. 1).3 In many cases, the properties of such nanostructured materials can be very different from those of corresponding bulk materials, and desirable novel electrical, mechanical, chemical, optical, magnetic, thermal, chemical and/or biological properties may be obtained.3 For instance, graphene, which is only one atomic layer thin, is imbued with a unique combination of high electrical conductivity, mechanical flexibility and chemical stability. Among many other properties, nanostructured materials have the increased ability to cross biological barriers which are inaccessible to larger materials or be specifically functionalised to bind to specific biological targets, such as ligands, proteins, antigens or cell types (e.g. tumour cells).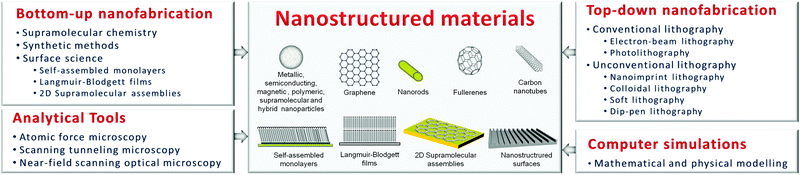 | ||
| Fig. 1 Schematic representation of examples of nanostructured materials, which can be fabricated and controlled by a powerful range of nanotechnology tools and techniques – bottom-up and top-down nanofabrication technologies, new or improved analytical techniques and high-performance computer simulations. | ||
Nanostructured materials are fabricated and controlled by a powerful range of nanotechnology tools and techniques – advanced synthetic methods, supramolecular chemistry, surface science, nanolithography, new or improved analytical techniques and high-performance computer simulations.4 Analytical tools, such as atomic force microscopy (AFM), scanning tunneling microscopy (STM) and near-field scanning optical microscopy, have provided revolutionary improvements in our ability to visualise structures and events all the way down to the molecular and atomic scale. High-performance computer simulations based on advanced mathematical and physical modelling are at present a necessary tool in the development, design, and understanding of nanostructured materials.
Synthetic methods, supramolecular chemistry and surface science are considered bottom-up approaches of nanofabrication since they involve building up nanostructured materials from nano- or sub-nano-scale entities (namely, atoms or molecules), in much the same way that nature does. With advancements in synthetic methods, nanostructured materials can be synthesised with precisely controlled morphologies, sizes and seemingly limitless chemical functional groups. Remarkable progress in the field of supramolecular chemistry has enabled the design of molecular components to interact favorably with each other in such a way that they can self-assemble, through noncovalent interactions, into larger, well-defined entities on the nanoscale with tailored properties.5 The development of synthetic methods and supramolecular chemistry has permitted access to carbon-based nanoparticles such as fullerenes and metallic (e.g. Au, Ag, Pt), semiconducting (e.g. CdSe, CdS, ZnS, GaAs), magnetic (e.g. Fe3O4), polymeric and hybrid (e.g. core–shell) nanoparticles. These bottom-up techniques have also allowed achieving other morphologies such as nanowires, nanorods, nanocapsules, nanofibers and nanotubes. Among the different nanotubes, carbon nanotubes (CNTs), which were discovered in 1991, have received much attention due to their unique physical/mechanical, electronic, chemical, optical and other properties.
Advances in surface science now permit the fabrication of nanoscale molecular-assembly structures onto a variety of substrate surfaces using techniques such as Langmuir–Blodgett films (LBFs), self-assembled monolayers (SAMs) and two-dimensional supramolecular assemblies.4 Among these three techniques, SAMs are the most widely used for surface functionalisation. SAMs are formed via adsorption of molecules from solution or the gas phase, creating two-dimensional quasi-ordered molecular assemblies on conducting, semiconducting or insulating surfaces with a wide range of terminal functionalities (e.g. carboxylic acid, amine, nitro, hydroxyl and methyl groups).
In contrast to the bottom-up technologies mentioned above, one can fabricate nanostructured materials via etching away bulk material to achieve the required smaller structural architectures. This process type is regarded as the top-down approach and it is generally achieved by nanolithography. The nanolithographic processes could be likened to sculpting a block of stone to obtain the required image. Many lithographic techniques of different complexity, efficiency, areal scale, and cost have been utilised for patterning surfaces with nanometre resolution. These techniques include novel nanolithography techniques such as nanoimprint lithography, colloidal lithography, soft lithography and dip-pen lithography, as well as conventional techniques such as electron-beam lithography and photolithography. Photolithography or electron-beam lithography (EBL) uses UV light or electrons to irradiate a sensitive polymer layer (called resist) to form nanopatterns on the resist that are subsequently transferred to the substrate material, often by etching. While EBL and photolithography are able to generate features with extremely high resolution (e.g. sub-5 nm (ref. 6) and sub-20 nm (ref. 7) for EBL and ArF double exposure immersion photolithography, respectively), these techniques have their limitations in terms of materials that can be patterned and the high costs of investment in equipment.
Novel nanolithography-based techniques have emerged that enable limitations associated with conventional lithography techniques to be overcome. Soft lithography, which includes techniques such as contact printing, replica molding and embossing, is a very useful alternative because it has been developed specifically for making large-scale nanostructures with a low-cost, flexible processing capability. Soft lithography is characterized by the use of a patterned elastomer as the stamp, mold, or mask to generate structures with feature sizes as small as 30 nm.8 The drawback of soft lithography is that it still requires the assistance of conventional lithographic techniques to design the masks or masters. In an attempt to address this challenge, an increasing amount of attention has been paid to the use of two-dimensional arrays of nanospheres as masks for etching or templates for deposition. This technique for sub-10 nm patterning has been termed nanosphere lithography9 or colloidal lithography10 and its application is most suitable under situations in which a periodic arrangement of the nanostructures is required.
These enabling bottom-up and top-down technologies, in addition to advanced analytical techniques and high-performance computer simulations, are driving the integration of nanotechnology in cell biology. In the next three sections, three main areas in which these contributions are being made – nanotopographical control, dynamic behaviour and intracellular manipulation and sensing – are discussed. Present research on nanotechnology-based drug and gene delivery systems, which is focused on achieving targeted delivery of drugs to specific cells or tissues, improved delivery of poorly water-soluble drugs, multiple drug administration, monitoring drug delivery by combining therapeutic agents with imaging biomarkers, and real-time read on the in vivo efficacy of a therapeutic agent, lies beyond the scope of this review. For a detail discussion on the progress made on nanotechnology-based drug and gene delivery systems, refer to reviews by Farokhzad and Langer11 and Kumar and co-workers.12
3. Cell sensing and response to nanotopographic cues
The complexity of cellular processes is astonishing. Cellular processes, such as adhesion, migration, proliferation, morphogenesis, polarity, differentiation and apoptosis, as well as tissue development, functionality and regeneration are influenced by the chemical composition, morphology of the extracellular environment through complex interactions between cells and a myriad of external chemical and physical stimuli. These multiple extracellular cues, including gradients of growth factors, chemokines and cytokines and other secreted proteins from close and distant neighbouring cells, chemical and physical interactions with the extracellular matrix (ECM) and direct cell–cell contacts, are expressed in a spatially and temporally coordinated manner and interpreted and integrated by cells to initiate signalling cascades leading to the activation of specific cellular processes.Studies on the effects of soluble cues have been conducted in well-defined media and assisted by synthetic methods aimed at generating natural and unnatural biologically active molecules. In contrast, elucidating how cells receive and transduce information from the ECM and neighbouring cells has proven to be a much harder task to accomplish. Most of our knowledge on cell responses to insoluble extracellular triggers and the mapping of intracellular signalling pathways is derived primarily from studies in plastic-based cell culture. However, there remain significant fundamental questions surrounding cell–matrix interactions for which nanotechnological advances are essential; how do cells probe and respond to the natural nanotexture of their surroundings? What sensory and regulatory mechanisms are involved?
While the effect of microtopographic structures on cells has long been recognized, it is only relatively recently that the full strength and depth of cellular sensing and reaction to nanotopography has been appreciated. These findings are perhaps not unexpected in view of the nanoscale organization of ECM, which comprises a great variety of molecules, including the collagen family members, elastic fibers, proteoglycans and adhesion molecules. The combination, immobilization and spatial organization of these molecules provide cells with an ECM that ranges in stiffness from soft to rigid and has a complex mixture of pores, protrusions and ridges having nanoscale sizes (5–200 nm)13 that vary considerably in different tissues. This wide spectrum of expression provides a source of great diversity for cell–matrix interactions.
With advancements in nanofabrication techniques, the role of nanotopography on cell behaviour has been examined by using numerous patterns of feature sizes ranging from 5 nm to several hundreds of nanometers, and geometries, including gratings, posts and pits (Fig. 2). The generation of such nanostructured surfaces relies on techniques such as photolithography, e-beam lithography, colloidal lithography, polymer phase separation, to name but a few. By using polymer phase separation – a bottom-up approach to create nanostructured polymers on surfaces, Dalby and co-workers14 were able to investigate the limits of cell sensing. Spontaneous phase separation of polystyrene and poly(4-bromostyrene) allowed the formation of polymer demixed islands with nanoscale vertical dimensions of 13, 35 or 95 nm on glass surfaces. Compared to flat polymer surfaces, human endothelial cells cultured on the nanopatterned surfaces produced an arcuate morphology that is typical of the endothelial phenotype in vivo. Of the nanotopographies, the 13 nm nanoisland elicited the greatest response from the cells, with highly spread cell morphologies containing a well-defined cytoskeleton.
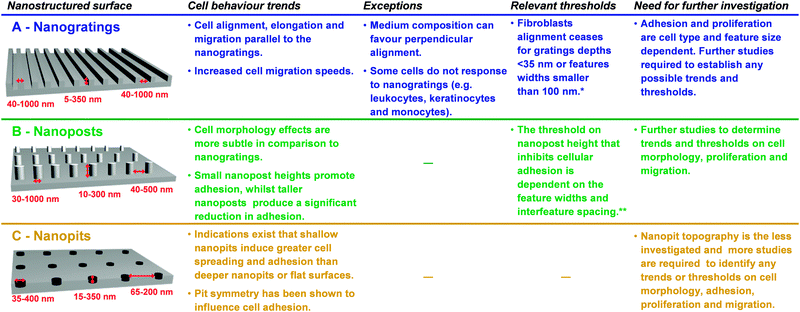 | ||
| Fig. 2 Summary of the main effects of the interaction of mammalian cells with nanoscale topography in the form of (A) nanogratings, (B) nanoposts and (C) nanopits with different feature widths, depths and interfeature spacings.16–18 Progress has been made, but yet there is still a great deal of work to be done in fully understanding the influence of nanotopography on cell behaviour. * The threshold is associated with a particular spacing (ridge-grating ratio) and the cell type investigated. Different threshold numbers are expected depending on the cell type and ridge-grating ratio used.19 ** The threshold is based on several studies with different cell types and features with different aspect ratios and interfeature spacing. | ||
This study14 and others15 provide evidence for the importance of nanoscale topographical cues for both cell function (e.g. cellular morphology, adhesion, motility, proliferation) and gene expression. Whether a nanostructured surface-induced impairment or enhancement of cell adhesion, motility and proliferation is observed depends not only on the cell type, but also on the size, geometry and aspect-ratio of the nanofeatures (Fig. 2).16–19 Nanogratings, which resemble the ECM collagen fibrils morphology in size and shape, are one of the most studied nanoscale geometries and their topology-related alignment and guidance has already proven to be useful, for instance, to construct cardiac tissue that more closely resembles the structural and functional aspects of natural heart muscle tissue.20
The research on stem cell biology has predominantly focused on the use of soluble chemical cues to retain the stem cell phenotype and direct lineage commitment, but now there is mounting evidence that substrate stiffness21 and nanotopography22–24 could potentially be used as versatile tools for the non-invasive manipulation of stem cell proliferation and differentiation. A recent example of nanotopography regulation of stem cell fate includes carbon nanotube network structures on gold surfaces that provide selective adhesion and growth of human neural stem cells.23 Long-term maintenance of mesenchymal stem cell phenotype and multipotency has been also demonstrated on 120 nm square pits symmetrically in a polycaprolactone substrate that was fabricated by e-beam lithography and processed into polycaprolactone by hot embossing.22 Although the exact mechanism for how the nanotopography maintains the stem cells in culture over time remains to be elucidated, this approach brings with it the potential to generate large quantities of stem cells for clinical use and treat various diseases such as arthritis and Alzheimer's.
Current research efforts are also directed at regulating the behaviour of human neural stem cells (NSCs) – a self-renewing and multipotent cell population in the central nervous system, since it can open up significant opportunities in neural regeneration. Graphene, with its unique structural properties, has been shown to be a suitable nanostructured scaffold for promoting NSC adhesion and differentiation into neurons (rather than glial cells) for long-term periods, i.e. one month.24 More recently, a three-dimensional (3D) microporous graphene foam, which was synthesised by chemical vapor deposition using a nickel foam template and coated with the ECM protein laminin, was shown not only to support NSC growth but also sustain a more active proliferation state than that of 2D graphene films.25 Cell proliferation was assessed after 5 days of culture. The 3D graphene-based scaffold that resembles more the in vivo situation was also able to induce NSC differentiation more towards neurons than two types of glial cells – astrocytes and oligodendrocytes (Fig. 3). Furthermore, these 3D scaffolds were also demonstrated to be suited to act as a conductive scaffold to electrically stimulate cells.
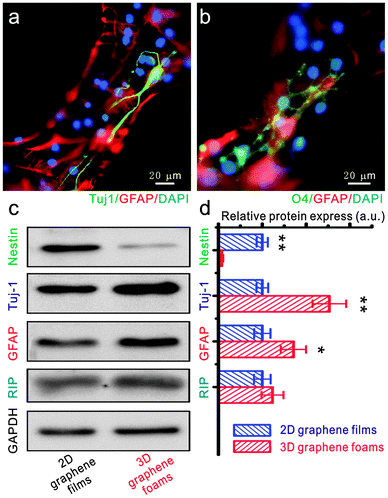 | ||
| Fig. 3 The differentiation of NSCs on a 3D graphene foam scaffold. (a and b) Fluorescence images of differentiated NSCs that were immunostained for Tuj-1 (marker for neurons – green, a), GFAP (marker for astrocytes – red, a and b), O4 (marker for oligodendrocytes – green, b) and DAPI (marker for nuclei – blue, a and b). Western blot analysis of nestin (marker for NSCs), Tuj-1, GFAP and RIP (marker for oligodendrocyte) protein expression of differentiated NSCs on 2D graphene films and 3D graphene foam scaffold. (d) Relative optical densities of nestin, Tuj-1, GFAP and RIP bands shown in (c). Reproduced with permission from ref. 25 (copyright 2012, Nature Publishing Group). | ||
At this stage it is not possible to predict which nanoscale features will be most effective in eliciting a particular cellular response, especially because two critical questions are largely unresolved: which cell sensing mechanisms and intracellular signalling pathways mediate such effects? Is the cellular response a direct result of the surface nanotopography or conveyed through an adsorbed layer of ECM molecules on the nanostructured surfaces, or both? Although some indications exist in the literature26,27 that alteration of cellular function is mainly due to adsorption of ECM molecules, and thus their spatial organization on the nanostructured surfaces, available data are too limited to ascertain the exact origin of the effect of topographical cues on cell function.
A large number of cellular components are expected to be involved in nanotopography sensing and response but to date few have been investigated. Filopodia are cytoskeletal projections that extend from the plasma membrane, mainly from the leading edge, and have a tip diameter of approximately 100 nm. Evidence is growing that filopodia are instrumental in sensing the environment at the nanoscale.28 In addition, heterodimeric transmembrane cell adhesion proteins known as integrins are believed to play a key role in the nanotopographical signal transduction processes.29 Upon binding to extracellular ligands, integrins cluster in the membrane and recruit many different cytoskeletal proteins and signalling proteins, forming new focal adhesions (FAs) and initiating signal transduction pathways required for regulating cell adhesion, morphology and motility.30 A series of studies has been conducted on precisely tailored nanostructured surfaces, with differences in the surface density and spatial arrangement of the integrin binding tripeptide Arg-Gly-Asp (RGD), a sequence present in most of the adhesive ECM proteins (e.g. fibronectin, vitronectin, laminin and collagen) and identified to be the minimum recognition required to support cell adhesion. These studies have provided important insights into how the organization of RGD ligands can influence integrin clustering and FA formation and subsequent adhesion and spreading of cells.29 It has been revealed that a critical lateral interdistance of about 60–70 nm between integrin RGD ligands exists, above which integrin clustering and focal adhesion formation is hindered, limiting cell attachment and spreading.31 It can be speculated that this threshold in spacing might be of physiological relevance since ordered structures occur in the native ECM, such as the 67 nm D-periodicity of collagen fibrils. The mechanism is presently unclear but the stress level at the actin cytoskeleton32 and impairment of integrins crosslinking by cytoskeletal proteins such as talin, a 60 nm elongated actin cross-linking protein,31 have been proposed to explain such spacing-induced instability. To date, efforts have focused primarily on the grafting and presentation of the RGD motif but this peptide represents one of many ligand motifs that interact with cell receptors to trigger cell responses. In particular, carbohydrates are a prime target for investigation at the nanoscale since multivalent carbohydrate–protein interactions are involved in mediating a variety of physiological and pathological processes. As the field of nanotechnology matures the generation of heterogeneous surfaces, comprising multiple ligand motifs patterned into distinct regions at the nanoscale, will enable us to understand how these multiple interactions interact to influence cell behaviour.
4. Stimuli-responsive interfaces as in vitro model systems
Surfaces with stimuli-responsive properties are also essential for the realization of smart, highly engineered cell–material interfaces. Surfaces equipped with molecular cues mimicking certain aspects of structure or function of natural ECM offer new opportunities for mechanistic studies of the pathways by which cells sense, integrate and respond to changes in their environments. Aside from being important for fundamental cellular studies, responsive artificial ECM models can also affect cell behaviour, and play an active regulating role in promoting tissue regeneration. Active and switchable interfaces comprising domains that confer tailored responsiveness and biochemical and/or physical cues have sufficiently progressed to emulate important aspects of the ECM. Understanding cell–matrix interactions necessitates the study of the role of local and temporal topographical cues. To this end, substrates have been incorporated with physical cues in the form of thermally responsive polymer surfaces to dynamically control surface topography and dictate the morphology of stem cells (Fig. 4A).33 Thermally activated poly(ε-caprolactone) shaped memory polymers with a transition temperature (Ttrans) near physiological temperature (∼37 °C) were used to fabricate surfaces displaying a dynamic transition from a primary to a secondary surface pattern.33 In order to fabricate such a dynamic surface, the liquid prepolymer was cast into a mold of the primary shape and photocured. The primary shape was subsequently mechanically deformed into a secondary shape at temperatures that exceed Ttrans using a second replica mold. The temporary shape was fixed by cooling down below Ttrans. Recovery of the primary shape was attained by heating the substrate above Ttrans. When human mesenchymal stem cells were cultured on substrates with temporary shapes consisting of 3 μm × 5 μm channel arrays, cells showed marked alignment along the channel direction as previously observed for other nanograting topographies. However, when the substrate was heated at 40 °C, and the surface switched from a 3 μm × 5 μm channel array to a planar surface (primary shape), the cell morphology changed from an aligned to a stellate shape.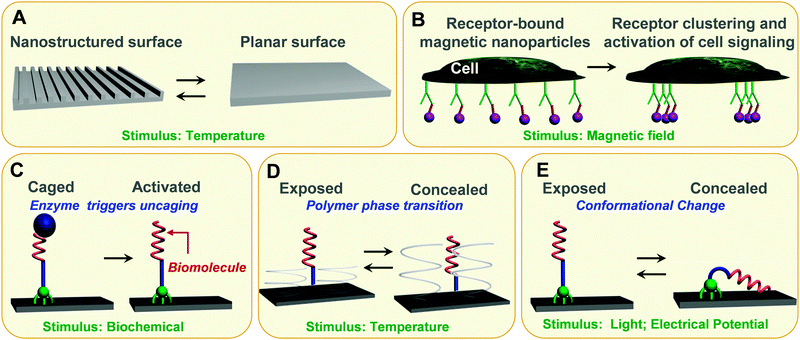 | ||
| Fig. 4 Stimuli-responsive interfaces as in vitro model systems. (A) Shape-memory polymer surfaces, which transitions are induced by temperature, allows for dynamic topographical control of cell behaviour. (B) Superparamagnetic nanoparticles (purple), each linked with a single ligand, bind evenly across the plasma membrane to cell surface receptors. By applying a magnetic field, the nanoparticles aggregate together, driving the receptors into a clustered arrangement and activating cell signalling. (C) Enzyme triggers activation of a surface-tethered RGD peptide, thereby promoting cell adhesion. (D) Specific interactions between cell integrins and immobilised RGD moieties can be non-invasively thermally regulated for cell attachment/detachment using surface-grafted thermoresponsive polymers. (E) Surface-tethered electrical and photoresponsive molecules are able to expose or conceal biomolecules on demand. | ||
Magnetic nanoparticles have been also exploited to manipulate cells and create 3D tissue culture leveraging magnetic levitation.34 In this approach, cells were incubated with a hydrogel composed of filamentous bacteriophages, magnetic iron oxide and gold nanoparticles, which partially enter the cells or bind to cell membranes. By spatially controlling the magnetic field while cells were dividing and growing, the cells could be levitated and the geometry of the cell mass controlled. Magnetic cell levitation was able to induce multicellular clustering of different cell types in co-culture, and perhaps more importantly, it could more closely recapitulate in vivo protein expression. A nanoscale magnetic actuation mechanism have been also employed to accomplish spatiotemporal control of cellular processes such as transmembrane signal transduction.35 Superparamagnetic nanoparticles, coated with monovalent ligands and bound to cell surface IgE–Fc1RI receptor complexes, magnetize when exposed to magnetic field, inducing clustering of the bound receptors that triggers a specific intracellular signalling response (Fig. 4B).
Integration of dynamic biochemical cues on surfaces has been largely exploited as a means to control cell adhesion by tuning the interactions between the integrin transmembrane receptors and an RGD peptide. Successful strategies for in situ temporal modulation of cell adhesion have included direct modification of the chemical structure of the RGD peptide with an enzyme-responsive molecule (Fig. 4C)36 and thermally responsive platforms based on poly(N-isopropyl)acrylamide (PNIPAM) polymer37 (Fig. 4D). PNIPAM is a thermo-responsive polymer that has a low critical solution temperature (LCST) of 32 °C in aqueous solution. Below its LCST, PNIPAM polymer is in an extended, solvent-swelled conformation, but when heated up above the LCST, the polymer undergoes a phase transition to yield a collapsed morphology that excludes solvent. The convenient PNIPAM reversible volume phase transition, in response to a moderate thermal stimulus near physiological temperatures, has found extensive applications in thermo-responsive cell culture dishes. In particular, the possibility to create cell sheets in vitro by using thermo-responsive polymers as substrates for cell growth and release has been established as a reliable, effective tissue reconstruction technology.38 Cell sheets are tissue-like cellular monolayers, which have been developed as a new tool for regenerative medicine and have already been applied in human clinical studies (e.g., cornea reconstruction, the treatment of esophageal ulcerations after endoscopic submucosal dissection). Cell sheets can be harvested intact with the associated extracellular matrix (ECM) by reducing cell culture at temperatures below the LCST and transplanted effectively to the damaged sites of tissues/organs as a regenerative medical treatment.39 Modification of PNIPAM with the cell-adhesive, protein-derived tripeptide RGD sequence, has been shown to significantly enhance cell adhesion and spreading under serum-free conditions at physiological temperature (37 °C).40 Moreover, cells were detached when the temperature was lowered below the LCST (20 °C) as the hydrated polymer chains extended impeding the cell's integrin receptors from accessing the immobilised RGD ligand (Fig. 4D).
Efforts have also been directed toward the development of electro-switchable self-assembled monolayers to regulate biomolecular interactions and cell adhesion (Fig. 4E).41–43 In our previous work,43 electrically controlled switching has been applied to regulate the conformational changes of modified positively charged oligolysine peptides tethered to a gold surface, such that bioactive molecular moieties (biotin) incorporated into the oligolysines could be reversibly exposed (bio-active state) or concealed (bioinactive state) on demand, as a function of surface potential. The dynamics of the switching and the biological properties of the surface were studied by observing the binding events between biotin and fluorescently labeled neutravidin. Fluorescence microscope images and surface plasmon resonance (SPR) spectral data clearly revealed opposite binding behaviors when +0.3 V or −0.4 V were applied to the surface. High fluorescence intensities were observed for an applied positive potential, while minimal fluorescence was detected for an applied negative potential. SPR has further shown that these responsive surfaces can control binding ability to greater than 90%. Following this work, Gooding and coworkers44 have extended the concept of molecular mechanical motions of surface-bound electro-switchable molecules to control cell adhesion (Fig. 5). The two-component SAMs comprised a protein-resistant ethylene glycol chain, which contained a charged moiety at its distal end, and a terminated RGD component on which cellular adhesion receptors, integrins, can bind. Two SAM surfaces were prepared with two different ethylene glycol derivatives, one with a sulfonate (anionic) distal moiety and the other with an ammonium (cationic) distal moiety. If the electrode possessed a potential of the same polarity as the charged moiety, the ethylene glycol molecules project out from the surface and conceal the RGD peptides from the cells, hence resisting cell adhesion. Switching the potential to the opposite polarity causes the ethylene glycol molecules to flip towards the surface, exposing the RGD peptides and allowing cells to adhere.
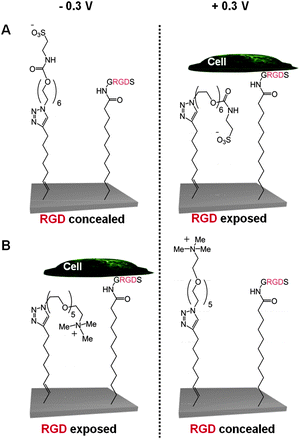 | ||
| Fig. 5 Schematic representation of two electro-switchable surfaces that are able to control cell adhesion under an electrical potential.44 In one case (A), the surface is constituted by a RGD and EG6-sulfonate mixed SAM that promotes cell adhesion under a positive potential of +0.3 V, while in the other case (B) the surface comprises a RGD and EG5-ammonium mixed SAM that prevents cell adhesion at the same positive potential of +0.3 V. | ||
Apart from SAMs, electrically-responsive polymer surfaces have also been investigated for eliciting a particular cellular response and have shown to improve bone implants45 and enhance nerve regeneration.46 For instance, biodegradable conductive composites made of polypyrrole (PPy) (2.5%) and chitosan (97.5%) have been prepared in order to electrically stimulate Schwann cells, which are essential for nerve regeneration.46 This conductive PPy–chitosan composite was shown to support cell adhesion, spreading, and proliferation in the presence or absence of electrical stimulation. However, electrical stimulation significantly enhanced the proliferation of Schwann cells and the expression and secretion of the nerve growth factor (NGF) and brain derived neurotrophic factor (BDNF) when compared with control cells without electrical stimulation. The enhanced cell secretion of NGF and BDNF is expected to promote nerve regeneration by enhancing survival and outgrowth of damaged nerves.
Molecular approaches that are similar to those already discussed can be used to engineer cell–material interfaces that respond to light. Studies have been focused on developing reversible systems based on photoisomerisable moieties such as azobenzene moieties. The azobenzene isomerises upon illumination with UV light (λ = 300–400 nm) from the stable trans form to the cis state, while reverse isomerisation can be triggered by irradiation with visible light (λ = 425–500 nm). Isomerisation of azobenzene is accompanied by an appreciable shape change as the trans isomer adopts a more linear conformation than the cis isomer. When azobenzene moieties were immobilised on surfaces and derivatised with an RGD-containing peptide, this change in molecular conformation enabled the ability to modulate cell adhesion.47 The photoswitchable RGD peptide-SAM supported cell adhesion in the trans-azobenzene configuration, while in the cis form a few cells adhered to the surface. This investigation suggests that small changes in the conformation/orientation of the peptide by the azobenzene isomerisation can modulate the availability and potency of the active sites for cell surface receptors.
Another means by which azobenzene SAMs have been able to control cell adhesion was through the use of host–guest complexation. It is well known that the host–guest interaction between azobenzene and α-cyclodextrin (CD) or β-CD can be controlled through the photoisomerisation of azobenzene. Gong and coworkers48 have taken this unique feature and presented an α-cyclodextrin (α-CD)-terminated silane SAM which allowed to assemble with azobenzene-glycine-arginine-glycine-aspartate-serine (azo-GRGDS) via host–guest interactions for controlling cell adhesion (Fig. 6). Cells were shown to adhere to the α-CD azo-GRGDS complex SAM, which upon UV irradiation led to the release of the azo-GRGDS and cells.
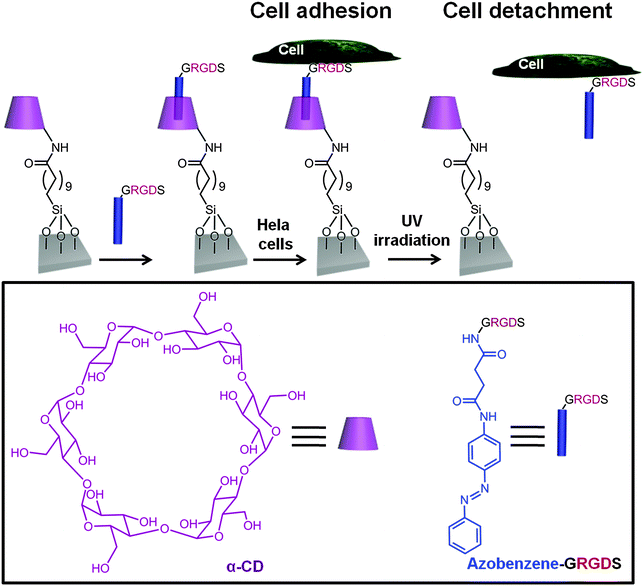 | ||
| Fig. 6 Schematic representation of a α-CD terminated-silane SAM which can form an inclusion complex via host–guest recognition with an azobenzene-GRGDS peptide.48 Hela cells were cultured on the substrate when the azobenzene was in its trans conformation. Upon UV irradiation at 365 nm, the trans-azobenzene underwent isomerization to the cis isomer and both the azo-GRGDS and the cells were detached from the substrate. | ||
Integrin-mediated adhesion regulates a variety of signalling pathways, including those that control cell migration, cell cycle and cell differentiation,49 and although approaches to switching cell adhesion on or off selectively have been demonstrated, their potential has still to be harnessed effectively to provide new insights into the complex signalling mechanism underlying cell adhesion. Quantitative experimental research on cell cue-signal–response relationship is expected to be most fruitful. Within the living body, the adhesive interactions between integrin receptors and ECM proteins are dynamic in nature, which is a result of constant molecular binding and unbinding events. Despite reversibility being a basic and crucial feature of the cell adhesion process, reversible control over integrin binding to artificial interfaces is still a challenging endeavour with limited success. The next advances in stimuli-responsive surfaces are likely to emerge from the design of surfaces conferring superior reversibility characteristics and expanding to other ligands other than RGD. Controlling the activity of larger biomolecules, such as proteins, on surfaces would go a long way toward resolving critical cellular mechanisms that, when defective, lead to significant and diverse diseases such as cancer, cardiovascular disease and immunological and metabolic disorders.
5. 3D nanostructured surfaces for intracellular manipulation and sensing
Another emerging area of great interest is the interfacing of three-dimensional (3D) nanostructured surfaces, based on vertically oriented nanomaterials on a solid support, with living cells for bidirectional flow of information. Achieving direct interconnection of cells to an external platform through integrated nanomaterials holds great opportunities to stimulate and analyse cellular processes, occurring inside cells, across membranes, and between neighbouring cells all with high spatial and temporal resolution. Developments in microscopy techniques and fluorescent probe technology have enabled the imaging of dynamic processes such as protein translocation and protein–protein interactions with single molecule sensitivity, leading to profound insights into the movement of proteins and their interactions in living cells.50 However, in order to explain the intricate processes underlying cellular function a greater number of high spatial and temporal resolution tools are necessary to that can not only monitor a broad range of chemical and physical properties but can also modulate them at well-defined sub-cellular locations (e.g. cytoplasm, nucleus, mitochondria).With nanomaterials having dimensions down to a few nanometers, 3D nanostructured surfaces are unparalleled as high spatial resolution tools for cell biology. So far, high-aspect-ratio nanomaterials, such as carbon and silicon dioxide nanotubes, and metallic and semiconductor nanowires and nanorods have been investigated to interface with cells. These nanomaterials offer a high surface area to ensure appropriate cell–nanomaterial communication but are much smaller than the typical cell dimensions enabling minimal invasiveness. Surface-modified vertical silicon nanowires have been shown to penetrate mammalian cells without affecting cell viability51 and deliver biomolecules such as proteins and DNA plasmids into living cells.52 Other examples include cell viability studies using vertical arrays of indium-arsenide (InAs) nanowires53 and development of intracellular pH sensors based on high density zinc oxide (ZnO) nanorods.54
Intracellular electrical coupling has been also achieved with vertical nanowire electrode arrays, and the first steps have been taken to perform multiplexed intracellular electrical stimulation and recording. Park and co-workers55 used standard silicon nanofabrication technology to fabricate vertical nanowire electrode arrays, comprising 16 stimulation/recording pads that could be individually addressed (Fig. 7A). Each pad contained a 3 × 3 array of 9 silicon nanowires with a diameter of 150 nm and height of 3 μm that were spaced 3 μm apart. The nanowires were made of a highly doped silicon core encapsulated by an insulating silicon dioxide shell, and capped by a conductive titanium and gold tip. The conductive silicon core and metal tip offered electrical access to the interior of the cell, while the insulating shell prevented current leakage through the nanowire sidewalls. When rat cortical neurons were cultured on top of the silicon nanowire platform, some nanowires were shown to penetrate through the cell membrane whereas other nanowires only entered the cells when a short high voltage pulse was used (a process named electroporation). The nanostructured array platform was shown to be able to record and stimulate neuronal activity and map multiple individual synaptic connections.
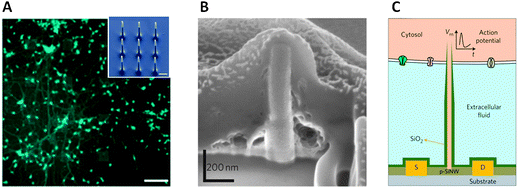 | ||
| Fig. 7 (A) Rat cortical neurons cultured on top of a device consisting of 16 stimulation/recording pads for parallel multi-site interrogation of neuronal circuits. Scale bar, 120 μm. Inset: SEM micrograph of the nine silicon nanowires that constitute each pad. Scale bar, 1 μm. Reproduced with permission from ref. 55 (copyright 2012, Nature Publishing Group). (B) SEM micrograph illustrating that a Pt nanopillar electrode is engulfed tightly by the cell following electroporation. Focused ion beam (FIB) milling was employed to expose the interface cross-section. Reproduced with permission from ref. 56 (copyright 2012, Nature Publishing Group). (C) Schematic diagram showing a branched intracellular nanotube FET device, in which a hollow silica nanotube (green) penetrates the cell membrane, bringing the cytosol (pink) into contact with a p-type silicon nanowire (p-SiNW). Reproduced with permission from ref. 57 (copyright 2011, Nature Publishing Group). | ||
Cui and co-workers56 also built vertical nanowire electrode arrays of similar-sized nanowires but made of platinum to record both the extracellular and intracellular action potentials in cardiac muscle cells (cardiomyocytes) derived from mice. Like in the nanowire platform used by the Park and co-workers, membrane electroporation was an effective strategy for getting the nanowires into the cell's interior (Fig. 7B). The multiplexing capability of the nanoarray system was demonstrated by performing simultaneous recordings from multiple cells over four days. The potential of the technology in drug screening was also demonstrated by detecting subtle changes in the action potentials induced by drugs that target ion channels in cells.
Instead of vertical nanowire electrode arrays, Lieber and co-workers57 used the high-resolution electrical recording capabilities of field-effect transistors (FETs) to measure intracellular action potentials from cultured cardiomyocyte cells. By employing bottom-up and top-down nanofabrication approaches, a branched intracellular nanotube FET device was developed that combined a silicon nanowire FET detector with an electrically insulating silicon dioxide nanotube on top of it (Fig. 7C). The silicon dioxide nanotube had an outer diameter that tapered from 150 nm at its base to 55 nm at its tip. In order to penetrate the cell membrane, the nanotubes were modified with phospholipids. Once inside the cells, the intracellular fluid (the cytosol) entered the hollow nanotubes and came into contact with the FET, enabling intracellular electrical recordings to be collected. The use of multiple BIT-FETs enabled multiplexed recording from both single cardiomyocyte cells and networks of these cells. The devised nanoscale probes based either on FETs or vertical nanowire electrode arrays for intracellular recording and stimulation represent an important progress in the ongoing efforts for developing non-invasive electrophysiological techniques with multiplexing capabilities.
Efforts have also been directed towards the development of electrochemical probing platforms based on carbon nanotubes,58 which are expected to be highly beneficial for monitoring and determining the precise location of redox activities occurring within the cell (Fig. 8). Electrochemical detection inside the cells offers several advantages over conventional fluorescence measurements. It includes less expensive components with simple sample preparation steps, and of utmost importance, the capability of measuring quantities down to the zeptomole level – allowing detection of trace levels of electroactive species – in complex turbid environments. The electrochemical sensing platform was fabricated using indium tin oxide (ITO), which is the underlying conducting surface. The surface was modified with an amino-terminated thin film via electrochemical grafting that allowed for the self-assembly of highly conductive vertically aligned carbon nanotubes (VACNTs). The CNTs, which were shown to be approximately 60 nm in height, were further modified with DNA. The vertically aligned CNTs were shown to enter immune cells (macrophage cell line) naturally without application of any external force and sense the intracellular presence of a redox active moiety, methylene blue (MB).
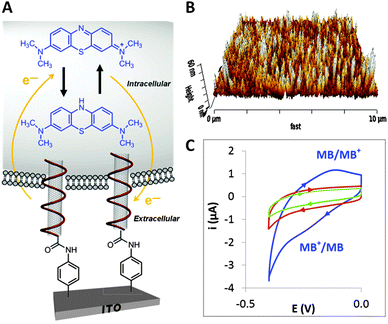 | ||
| Fig. 8 (A) DNA-wrapped CNTs vertically anchored to an ITO surface that are naturally taken up by a mouse macrophage cell and used for electrochemical probing the intracellular environment. (B) AFM image of the CNT modified ITO surface. (C) Cyclic voltammograms of methylene blue (MB) stained cells on DNA-wrapped CNTs vertically anchored to an ITO surface (blue curve) and MB stained cells on CNTs vertically anchored to an ITO surface (green curve). Cyclic voltammogram of cells not stained on DNA-wrapped CNTs vertically anchored to an ITO surface (red curve). The control experiments (green and red curves) did not display any redox peaks, providing strong evidences that the voltammetric peaks observed on the DNA-wrapped CNTs vertically anchored to an ITO surface (blue curve) are due to the reduction and oxidation of MB within the cells. Reproduced with permission from ref. 58 (copyright 2013, American Chemical Society). | ||
Another distinct approach has been developed for intracellular electrochemical measurements using platinum (Pt)/Pt black nanoelectrodes.59 The fabrication of the platinized nanoelectrodes relied on the electrodeposition of Pt black on an etched Pt nanoelectrode under AFM control. The Pt/Pt black nanosensor was introduced into a macrophage inducing a mechanical stimulation and triggering an oxidative response. This response was monitored intracellularly with the Pt–Pt black nanosensor, showing changes in reactive oxygen and nitrogen species (ROS and RNS) in the cytoplasm upon stimulation. These measurements in single cells have offered insight into the intracellular processes of oxidative stress.
Although important progress has been made in the fabrication and use of 3D nanostructures for biology, current research is mainly at proof-of-concept stage and has yet to make its way to tackle real biological questions. The work in this area is still largely focused on the development of nanostructured-based platforms that can stimulate and measure molecular and biochemical events intracellularly without deleterious effects to the cell. The next challenge will be to develop nanotools that can induce experimental perturbations on well-defined and sub-cellular locations and examine the effects in a spatial and temporal manner. At the same time, it will be also important to expand the nanotool set available for manipulation and monitoring and integrate these nanotools into comprehensive, real-time platforms with the sensitivity to perturb and monitor diverse intracellular processes simultaneously (Fig. 9). This represents a very challenging undertaking, particularly due to the necessity of including multiple functional nanomaterials onto the same platform that need to be taken up by cells. Furthermore, perhaps the most challenging aspect is that these tools must not impair the normal functioning of the cell.
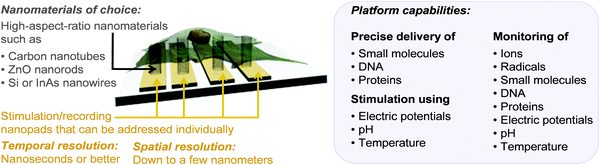 | ||
| Fig. 9 Schematic representation of the vision for 3D nanostructured surface platforms containing nanopads with vertically oriented nanomaterials that can be addressed individually to stimulate and record processes inside cells, across membranes and between neighbouring cells. | ||
6. Concluding remarks and perspectives
Nanotechnology is allowing the creation of in vitro models, which capture more of the relevant complexity that entails the native ECM, leading to a better understanding of the parameters governing cell behaviour. These insights begin to lay a foundation for constructing materials to direct cell fate, either without or synergistically with the use of chemical cues, in applications such as regenerative medicine and tissue engineering.Emerging nanofabrication techniques are also allowing the investigation of cell biology on the smallest of scales and promise insight into cellular interactions beyond the reach of current analysis. Although current 3D nanostructured surfaces provide the foundation for engineering a nanotool interface with single cells, both the integration of multiple measurement nanotools and analysis of complex data remain to be accomplished in a self-contained platform. It is anticipated that, ultimately, such capabilities will take us closer to the ultimate goal of mapping out and understanding at all levels the structure and function of a living cell.
The tutorial review presented here shows clearly that there remains a large potential for further progress in this field. There is no doubt that future research will progress in the direction of more detailed analysis of cell function and precise manipulation of cell response with expanding sets of complex nanoscale tools. Cell biologists and tissue engineers need to adopt and capitalise on such advances to build a better understanding of normal cell behaviour and disease mechanisms, and what pathways and targets might be best exploited for clinical interventions and drug design as part of developing personalised medicine.
Acknowledgements
P.M.M. is grateful to Dr Simon Johnston and Professor Liam Grover for their comments on the manuscript. P.M.M. would like to thank the Leverhulme Trust (F/00 094/BD) and Wellcome Trust for funding (WT091285MA).References
- F. S. Collins, M. Morgan and A. Patrinos, Science, 2003, 300, 286–290 CrossRef CAS PubMed.
- B. N. Kholodenko, Nat. Rev. Mol. Cell Biol., 2006, 7, 165–176 CrossRef CAS PubMed.
- The Chemistry of Nanomaterials: Synthesis, Properties and Applications, ed. C. N. R. Rao, A. Müller and A. K. Cheetham, Wiley-VCH Verlag GmbH, Germany, 2004 Search PubMed.
- P. Iqbal, J. A. Preece and P. M. Mendes, in Supramolecular Chemistry: From Molecules to Nanomaterials, ed. J. W. Steed and P. A. Gale, John Wiley & Sons Ltd, Chichester, UK, 2012, vol. 8, pp. 3589–3602 Search PubMed.
- B. H. Northrop, A. B. Braunschweig, P. M. Mendes, W. R. Dichtel and J. F. Stoddart, Handbook of Nanoscience, Engineering, and Technology, CRC Press, 2007 Search PubMed.
- M. Isaacson and A. Muray, J. Vac. Sci. Technol., 1981, 19, 1117–1120 CrossRef CAS.
- M. Al-Amri, Z. Liao and M. S. Zubairy, in Advances in Atomic, Molecular, and Optical Physics, ed. E. Arimondo, P. R. Berman and C. C. Lin, 2012, vol. 61, pp. 409–466 Search PubMed.
- Y. N. Xia and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 551–575 CrossRef.
- J. C. Hulteen and R. P. Vanduyne, J. Vac. Sci. Technol., A, 1995, 13, 1553–1558 Search PubMed.
- S. M. Yang, S. G. Jang, D. G. Choi, S. Kim and H. K. Yu, Small, 2006, 2, 458–475 CrossRef CAS PubMed.
- O. C. Farokhzad and R. Langer, ACS Nano, 2009, 3, 16–20 CrossRef CAS PubMed.
- V. R. Devadasu, V. Bhardwaj and M. Kumar, Chem. Rev., 2013, 113, 1686–1735 CrossRef CAS PubMed.
- N. W. Karuri, S. Liliensiek, A. I. Teixeira, G. Abrams, S. Campbell, P. F. Nealey and C. J. Murphy, J. Cell Sci., 2004, 117, 3153–3164 CrossRef CAS PubMed.
- M. J. Dalby, M. O. Riehle, H. Johnstone, S. Affrossman and A. S. G. Curtis, Biomaterials, 2002, 23, 2945–2954 CrossRef CAS.
- C. Y. Tay, S. A. Irvine, F. Y. C. Boey, L. P. Tan and S. Venkatraman, Small, 2011, 7, 1361–1378 CrossRef CAS PubMed.
- M. J. P. Biggs, R. G. Richards and M. J. Dalby, Nanomed.: Nanotechnol., Biol. Med., 2010, 6, 619–633 CrossRef CAS PubMed.
- C. J. Bettinger, R. Langer and J. T. Borenstein, Angew. Chem., Int. Ed., 2009, 48, 5406–5415 CrossRef CAS PubMed.
- M. J. Dalby, Int. J. Nanomed., 2007, 2, 373–381 CAS.
- W. A. Loesberg, J. te Riet, F. van Delft, P. Schon, C. G. Figdor, S. Speller, J. van Loon, X. F. Walboomers and J. A. Jansen, Biomaterials, 2007, 28, 3944–3951 CrossRef CAS PubMed.
- D.-H. Kim, E. A. Lipke, P. Kim, R. Cheong, S. Thompson, M. Delannoy, K.-Y. Suh, L. Tung and A. Levchenko, Proc. Natl. Acad. Sci. U. S. A., 2009, 107, 565–570 CrossRef PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. C. Oreffo and M. J. Dalby, Nat. Mater., 2011, 10, 637–644 CrossRef CAS PubMed.
- S. Y. Park, D. S. Choi, H. J. Jin, J. Park, K. E. Byun, K. B. Lee and S. Hong, ACS Nano, 2011, 5, 4704–4711 CrossRef CAS PubMed.
- S. Y. Park, J. Park, S. H. Sim, M. G. Sung, K. S. Kim, B. H. Hong and S. Hong, Adv. Mater., 2011, 23, H263–H267 CrossRef CAS PubMed.
- N. Li, Q. Zhang, S. Gao, Q. Song, R. Huang, L. Wang, L. Liu, J. Dai, M. Tang and G. Cheng, Sci. Rep., 2013, 3, 1604 CAS.
- C. J. Wilson, R. E. Clegg, D. I. Leavesley and M. J. Pearcy, Tissue Eng., 2005, 11, 1–18 CrossRef CAS PubMed.
- J. Y. Lim, J. C. Hansen, C. A. Siedlecki, J. Runt and H. J. Donahue, J. R. Soc. Interface, 2005, 2, 97–108 CrossRef CAS PubMed.
- K.-J. Jang, M. S. Kim, D. Feltrin, N. L. Jeon, K.-Y. Suh and O. Pertz, PLoS One, 2010, 5, e15966 CAS.
- B. Geiger, J. P. Spatz and A. D. Bershadsky, Nat. Rev. Mol. Cell Biol., 2009, 10, 21–33 CrossRef CAS PubMed.
- R. O. Hynes, Cell, 1992, 69, 11–25 CrossRef CAS.
- M. Arnold, E. A. Cavalcanti-Adam, R. Glass, J. Blummel, W. Eck, M. Kantlehner, H. Kessler and J. P. Spatz, ChemPhysChem, 2004, 5, 383–388 CrossRef CAS PubMed.
- A. G. F. de Beer, E. A. Cavalcanti-Adam, G. Majer, M. Lopez-Garcia, H. Kessler and J. P. Spatz, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2010, 81, 051914 CrossRef.
- D. M. Le, K. Kulangara, A. F. Adler, K. W. Leong and V. S. Ashby, Adv. Mater., 2011, 23, 3278–3283 CrossRef CAS PubMed.
- G. R. Souza, J. R. Molina, R. M. Raphael, M. G. Ozawa, D. J. Stark, C. S. Levin, L. F. Bronk, J. S. Ananta, J. Mandelin, M.-M. Georgescu, J. A. Bankson, J. G. Gelovani, T. C. Killian, W. Arap and R. Pasqualini, Nat. Nanotechnol., 2010, 5, 291–296 CrossRef CAS PubMed.
- R. J. Mannix, S. Kumar, F. Cassiola, M. Montoya-Zavala, E. Feinstein, M. Prentiss and D. E. Ingber, Nat. Nanotechnol., 2008, 3, 36–40 CrossRef CAS PubMed.
- S. J. Todd, D. Farrar, J. E. Gough and R. V. Ulijn, Soft Matter, 2007, 3, 547–550 RSC.
- M. Ebara, M. Yamato, T. Aoyagi, A. Kikuchi, K. Sakai and T. Okano, Biomacromolecules, 2004, 5, 505–510 CrossRef CAS PubMed.
- J. Yang, M. Yamato, C. Kohno, A. Nishimoto, H. Sekine, F. Fukai and T. Okano, Biomaterials, 2005, 26, 6415–6422 CrossRef CAS PubMed.
- Y. Akiyama, A. Kikuchi, M. Yamato and T. Okano, Langmuir, 2004, 20, 5506–5511 CrossRef CAS.
- M. Ebara, M. Yamato, T. Aoyagi, A. Kikuchi, K. Sakai and T. Okano, Adv. Mater., 2008, 20, 3034–3038 CrossRef CAS.
- X. Y. Jiang, R. Ferrigno, M. Mrksich and G. M. Whitesides, J. Am. Chem. Soc., 2003, 125, 2366–2367 CrossRef CAS PubMed.
- W. S. Yeo, M. N. Yousaf and M. Mrksich, J. Am. Chem. Soc., 2003, 125, 14994–14995 CrossRef CAS PubMed.
- C. L. Yeung, P. Iqbal, M. Allan, M. Lashkor, J. A. Preece and P. M. Mendes, Adv. Funct. Mater., 2010, 20, 2657–2663 CrossRef CAS.
- C. C. A. Ng, A. Magenau, S. H. Ngalim, S. Ciampi, M. Chockalingham, J. B. Harper, K. Gaus and J. J. Gooding, Angew. Chem., Int. Ed., 2012, 51, 1–6 CrossRef PubMed.
- S. Sirivisoot, R. Pareta and T. J. Webster, Nanotechnology, 2011, 22, 1–15 CrossRef PubMed.
- J. H. Huang, X. Y. Hu, L. Lu, Z. Ye, Q. Y. Zhang and Z. J. Luo, J. Biomed. Mater. Res., Part A, 2010, 93, 164–174 Search PubMed.
- D. B. Liu, Y. Y. Xie, H. W. Shao and X. Y. Jiang, Angew. Chem., Int. Ed., 2009, 48, 4406–4408 CrossRef CAS PubMed.
- Y. H. Gong, C. Li, J. Yang, H. Y. Wang, R. X. Zhuo and X. Z. Zhang, Macromolecules, 2011, 44, 7499–7502 CrossRef CAS.
- K. R. Legate, S. A. Wickstroem and R. Faessler, Genes Dev., 2009, 23, 397–418 CrossRef CAS PubMed.
- D. G. Spiller, C. D. Wood, D. A. Rand and M. R. H. White, Nature, 2010, 465, 736–745 CrossRef CAS PubMed.
- W. Kim, J. K. Ng, M. E. Kunitake, B. R. Conklin and P. Yang, J. Am. Chem. Soc., 2007, 129, 7228–7229 CrossRef CAS PubMed.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D.-R. Ahn, M.-H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner, T. S. Gujral, G. MacBeath, E. G. Yang and H. Park, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS PubMed.
- T. Berthing, S. Bonde, C. B. Sørensen, P. Utko, J. Nygård and K. L. Martinez, Small, 2011, 7, 640–647 CrossRef CAS PubMed.
- M. Willander and S. Al-Hilli, in Micro and Nano Technologies in Bioanalysis: Methods and Protocols, ed. J. W. F. R. S. Lee, 2009, vol. 544, pp. 187–200 Search PubMed.
- J. T. Robinson, M. Jorgolli, A. K. Shalek, M.-H. Yoon, R. S. Gertner and H. Park, Nat. Nanotechnol., 2012, 7, 180–184 CrossRef CAS PubMed.
- C. Xie, Z. Lin, L. Hanson, Y. Cui and B. Cui, Nat. Nanotechnol., 2012, 7, 185–190 CrossRef CAS PubMed.
- X. Duan, R. Gao, P. Xie, T. Cohen-Karni, Q. Qing, H. S. Choe, B. Tian, X. Jiang and C. M. Lieber, Nat. Nanotechnol., 2012, 7, 174–179 CrossRef CAS.
- F. J. Rawson, C. L. Yeung, S. K. Jackson and P. M. Mendes, Nano Lett., 2013, 13, 1–8 CrossRef CAS PubMed.
- Y. X. Wang, J. M. Noel, J. Velmurugan, W. Nogala, M. V. Mirkin, C. Lu, M. G. Collignon, F. Lemaitre and C. Amatore, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11534–11539 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2013 |
