 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemical information matters: an e-Research perspective on information and data sharing in the chemical sciences
Colin L. Bird and Jeremy G. Frey*
Chemistry, Faculty of Natural and Environmental Sciences, University of Southampton, University Road, Highfield, Southampton SO17 1BJ, UK. E-mail: J.G.Frey@soton.ac.uk; Tel: +44 (0)23 8059 3209
First published on 20th May 2013
Abstract
Recently, a number of organisations have called for open access to scientific information and especially to the data obtained from publicly funded research, among which the Royal Society report and the European Commission press release are particularly notable. It has long been accepted that building research on the foundations laid by other scientists is both effective and efficient. Regrettably, some disciplines, chemistry being one, have been slow to recognise the value of sharing and have thus been reluctant to curate their data and information in preparation for exchanging it. The very significant increases in both the volume and the complexity of the datasets produced has encouraged the expansion of e-Research, and stimulated the development of methodologies for managing, organising, and analysing “big data”. We review the evolution of cheminformatics, the amalgam of chemistry, computer science, and information technology, and assess the wider e-Science and e-Research perspective. Chemical information does matter, as do matters of communicating data and collaborating with data. For chemistry, unique identifiers, structure representations, and property descriptors are essential to the activities of sharing and exchange. Open science entails the sharing of more than mere facts: for example, the publication of negative outcomes can facilitate better understanding of which synthetic routes to choose, an aspiration of the Dial-a-Molecule Grand Challenge. The protagonists of open notebook science go even further and exchange their thoughts and plans. We consider the concepts of preservation, curation, provenance, discovery, and access in the context of the research lifecycle, and then focus on the role of metadata, particularly the ontologies on which the emerging chemical Semantic Web will depend. Among our conclusions, we present our choice of the “grand challenges” for the preservation and sharing of chemical information.
 Colin L. Bird | Having obtained his BSc and PhD in Chemistry at the University of Southampton, Colin Bird joined IBM UK Laboratories. After contributing to IBM's electrochromic display technology, he transferred to the IBM UK Scientific Centre to develop advanced image and visualisation applications. His work on content-based image retrieval led to a one-year secondment in 1999 back to the University of Southampton. On returning to IBM, he was involved in various aspects of information management, specialising in classification and metadata, and became an information architect. When he left IBM, he resumed his collaboration with Professor Jeremy Frey on e-Research projects, which began in 2000 as an industrial partner for the CombeChem project. |
 Jeremy G. Frey | Jeremy Frey obtained his DPhil on experimental and theoretical aspects of van der Waals complexes, in the PCL, Oxford, followed by a NATO/SERC fellowship at the Lawrence Berkeley Laboratory. In 1984 he took up a lectureship at the University of Southampton, where he is now Professor of Physical Chemistry. His experimental research probes molecular organization in environments from single molecules to liquid interfaces using laser spectroscopy from the IR to soft X-rays. He investigates how e-Science infrastructure can support scientific research with an emphasis on the way appropriate use of laboratory infrastructure can support the intelligent access to scientific data. |
Introduction
Future innovation in chemistry, as indeed in all the physical and life sciences, depends on collaboration and interdisciplinary research. Success in both respects depends strongly on making progress towards adopting open standards and open data in the chemical sciences, with particular emphasis on the exchange and sharing of chemical data.Each year computing facilities and storage become more powerful, almost as a necessity just to keep pace with the expanding volume of data. Corresponding increases in the capabilities of other equipment and technology have amplified the complexity of the data collected and, although storage costs have fallen, the costs associated with data preservation have risen. All of these considerations have contributed to an increased focus on making the best possible use of the data available. It was these same factors that drove the 21st Century e-Science and e-Research programmes and that continue to influence the technologies that comprise and contribute to cheminformatics, the amalgam of chemistry, computer science, and information technology.
Two reviews of the history of cheminformatics have been published recently;1,2 other authors have previously contributed observations on the evolution of the discipline. Guha et al. have also reviewed the field recently, considering the latest trends and projecting how they believe cheminformatics will develop over the next five years. They see the role of cheminformatics as enabling chemists to make better-informed decisions by facilitating chemists to deploy the extensive information resources available to them.3
If cheminformatics has a core principle, it is that structure determines properties, and this principle motivates the search for relationships in data to reveal information and in turn patterns in that information to generate knowledge. This principle underlies the rather poorly defined but useful concept of chemical space.4 Chemists and other scientists seek the principles that convert knowledge to wisdom, which is the top layer of a pyramid built upon data. Unless that data is freely available and willingly shared, progress towards the ultimate goal of wisdom will inevitably be inhibited. Regrettably the majority of chemists have yet to develop the necessary culture of sharing data, information, and knowledge.5
For data exchange and sharing to be effective, researchers must attend to the concepts of preservation, curation, discovery, access, and provenance. They must also subscribe to the principles of openness: open standards, open source, and above all, open data. While these principles are clear, implementing them can have far-reaching consequences on other aspects of the scientific investigation process, for example: open access and publishing economics; reward and academic recognition; and patent protection. The recently published JISC/CNI Workshop report notes that systems for assessment have not changed substantially in the last 15 years. The report also calls for the development of common standards for information sharing.6
In this review we examine the key components for manipulating and integrating chemical structures and data: identifiers, structure representations, and property descriptors. We consider the role of electronic laboratory notebooks, the function of metadata, the importance of capture at source, and the potential of publication at source. We examine the contribution that Semantic Web technologies can make to data exchange and to the assurance of quality and provenance. Preservation and assured discovery are essential for organizations that must provide audit trails and demonstrate due diligence. Although we focus primarily on academic research, we acknowledge the vital significance of data management for commercial organizations that depend on chemistry, for example the pharmaceutical industry: to date, drug discovery has been the foremost application of cheminformatics.
In this review we have concentrated on the information processing aspects that modern computing and software have brought to the chemical sciences, focussing on the management and sharing of chemical information. Accordingly, we regard the various forms of computational chemistry, including the use of High Performance Computing (HPC) in areas such as quantum chemistry and molecular dynamics simulations, as being outside our scope. An alternative differentiation is that computational techniques are applied to individual molecules, whereas cheminformatics brings together the data and information for a set, sometimes a large number, of molecules.3 Such integrative methods depend on collaborative infrastructures that rate sharing highly.
This review therefore reflects progress within the chemical sciences towards realising the six changes listed in the recent Royal Society report concerning open science and intelligent access to supporting data, which called for:
(1) a shift away from a research culture where data is viewed as a private preserve; (2) expanding the criteria used to evaluate research to give credit for useful data communication and novel ways of collaborating; (3) the development of common standards for communicating data; (4) mandating intelligent openness for data relevant to published scientific papers; (5) strengthening the cohort of data scientists needed to manage and support the use of digital data (which will also be crucial to the success of private sector data analysis and the government's Open Data strategy); and (6) the development and use of new software tools to automate and simplify the creation and exploitation of datasets.7
The European Commission support this stance in their Horizon 2020 Framework Programme for Research and Innovation:
Further steps will be taken towards Open Access, to ensure that research results are available to those who need them.†
Overall, access to data is seen as essential for the correct pursuit of science and the self-correcting nature of scientific enquiry; without this access science loses its pre-eminence as a way to investigate the world.
Cheminformatics the discipline
Cheminformatics is commonly defined in terms of the application of information technology and computer science to the chemical sciences.8 The discipline has also evolved to maximise the value obtainable from a wide range of data, seeking to understand that data and to extract information and patterns. Therefore, cheminformatics also embraces the distribution, management, access, and sharing of chemical data, and it is on these aspects that this review focuses. As Frey et al. observed in their introduction to the CombeChem e-Science project: All progress depends on individual scientists building on the results already produced by others,9 a principle long accepted in scientific endeavours. These words reflect those commonly attributed to Newton: “If I have seen a little further it is by standing on the shoulders of Giants”.‡History and development
The full history of cheminformatics is outside the scope of this review, but has very recently been covered in depth by Warr2 and Willett.1 Increases in computing power have led not only to a growth in capability but also to a dramatic expansion of the volume of data produced and, consequentially, a demand for more sophisticated information technology to keep pace with the increased quantities of data. Moreover, as chemistry and biology have evolved, the greater information processing capacity available has stimulated differentiation and specialization within these disciplines, leading to sub-categories within each field.At its most basic, chemometrics applies mathematical and statistical methods to the design of experiments with chemical systems, the analysis of the data obtained, and the understanding of those systems. As such, chemometrics clearly predates cheminformatics. Similarly, biostatistics, the application of statistical methods to biology, came before bioinformatics.
In general terms, chemometrics does not entail knowledge of chemical structure, being concerned mainly with obtaining information from data. The same might be said of biostatistics. Cheminformatics and bioinformatics seek to discern the patterns in the information, to elicit chemical and biological knowledge. Any distinction between these two branches of informatics relies mainly on the size and complexity of the molecules studied. Fig. 1 depicts the relationship between the four disciplines, but without clear divisions, owing to the potential overlaps. The scope for their application remains large, as demonstrated in the recent review of the enumeration of chemical space by Reymond et al.4
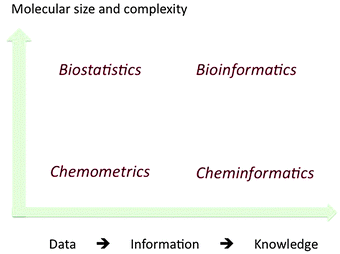 | ||
| Fig. 1 The scope of the information processing sub-disciplines of chemistry and biology. | ||
However, the purpose of this review is not to probe distinctions; its intention is much more to investigate how openness and data sharing can help to overcome any difficulties and conflicts that arise from divergences.
Quoting the second recommendation in the Royal Society report:7
Universities and research institutes should play a major role in supporting an open data culture by: recognising data communication by their researchers as an important criterion for career progression and reward; developing a data strategy and their own capacity to curate their own knowledge resources and support the data needs of researchers; having open data as a default position, and only withholding access when it is optimal for realising a return on public investment.
Cheminformatics is commonly considered to be associated primarily with drug discovery, a field that also relies fundamentally on bioinformatics.10 Indeed, the design and discovery of new drugs is arguably the most significant application of these two disciplines to have benefitted from the increases in the power and range of computational techniques. Establishing efficient links between the worlds of cheminformatics and bioinformatics is essential for good drug design services: in the Collaborative chemistry section we examine how open approaches to data and experiments can contribute to achieving that goal.
Hastings et al. assert that the application of cheminformatics is critically dependent on the data exchange process. The precise description of chemical entities is a key aspect of that process, so they are developing the Chemical Information Ontology (CHEMINF) to facilitate accurate exchange.11 The ontology relies on more general considerations of the physical measurements and general scientific approaches, as well as chemical knowledge. We appraise the attributes of ontologies and other types of vocabulary in the Chemical Semantic Web and chemical ontologies section.
Many cheminformatics tools depend on formal data descriptions that are based on controlled vocabularies. Prominent among such metadata constructs is the Chemical Markup Language (CML) for describing molecular species, first proposed in 1995.12 We examine the scope and role of CML in the Metadata section.
Although attitudes have changed with the advent of the newer technologies, older approaches are still in evidence. Taylor et al. highlight the difficulties that can result from failures to recognise what is required for data exchange and reuse:
Present chemical data storage methodologies place many restrictions on the use of the stored data. The absence of sufficient high-quality metadata prevents intelligent computer access to the data without human intervention. This creates barriers to the automation of data mining in activities such as quantitative structure–activity relationship modelling.13
Much of the research described in this review is intended to maximise the benefits obtainable from computing technology, beyond those provided by pure computation for computational chemistry. Efficient data management is essential if the techniques deployed downstream are to be effective. For example, the data mining and text mining techniques used in drug discovery rely on unifying data systems that often involve distributed sources in a variety of formats.14
Scope of the discipline
In recent years, one of the main concerns of the drug discovery process has become the understanding of what to do with the large volume of data available: how to integrate and manage it. In his editorial for the first issue of the Journal of Cheminformatics, Wild presents the current status of the discipline and identifies four “grand challenge” areas.15 The first three are application areas:• Overcoming stalled drug discovery;
• Green chemistry & global warming;
• Understanding life from a chemical perspective.
Wild's final challenge is aimed more at extending the field, by exploiting the information in various sources, which is a goal of particular relevance to this review. His fourth area is:
• Enabling the network of the world's chemical and biological information to be accessible and interpretable.
In the Conclusions section of this review, we present our own choice of “grand challenge” areas for open chemical science.
Wild and his team at Indiana University have developed an infrastructure of Web services to provide unified access not only to computational techniques but also to the data to which those techniques can be applied.16 They note that Web services can be used in workflow tools as well as application mashups, both of which enable new functionality by linking together data, applications, and services; in the spirit of openness, they welcome the participation of others in the development of services, mashup APIs,§ and tools.
Such capabilities depend strongly on links and relationships, involving not only informational entities but also people and their activities. Rzepa and Willighagen demonstrated the potential of social graphs based on the Friend-of-a-Friend (FOAF) ontology in a presentation about RDF-metadata enhanced social networking in chemistry, delivered at the 2008 ACS meeting. They illustrated the use of FOAF for relationships other than the purely social, thereby enabling other software to navigate those links.17 Frey and Bird have recently published a comprehensive review of the use of web-based services for drug design and discovery, and refer the reader to that article for further information and relevant references.18
The wider perspective for chemical information and cheminformatics
In the Introduction, we noted how the growth in the capabilities of computer systems had driven the 21st Century e-Science and e-Research programmes. Fig. 2 illustrates the evolution of these programmes and the distributed systems that underpin them. This figure also provides the wider perspective that places chemical information in the context of the distributed data infrastructure that is essential for future collaboration and innovation in science and other e-disciplines.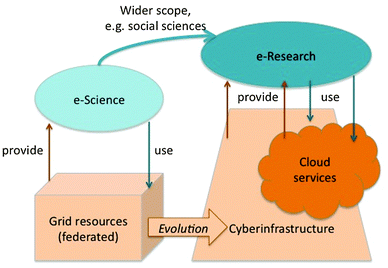 | ||
| Fig. 2 Diagrammatic glossary of e-Science, e-Research, and the underlying infrastructures that support e-disciplines. The initiative that became known as e-Science deployed distributed but federated computing and storage resources to apply advanced information technology to scientific research. The scope of e-Science has widened to include disciplines such as the social sciences, giving rise to the more generic description of e-Research. Moreover, the underlying technologies have also evolved to use service-oriented architectures (SOAs) to access the resources required, through either cloud computing or a more comprehensive cyberinfrastructure or e-infrastructure. | ||
Writing for Science magazine in 2005, Hey and Trefethen set forth the case for an e-infrastructure, as it is known in Europe, or in the USA, a cyberinfrastructure, the goal being to enable collaboration by providing shared resources and data.19 They set out the basic principles and illustrated them with examples from the UK e-Science programme, including CombeChem and Smart Tea, two projects that we describe elsewhere in this review.
The future of cheminformatics and indeed that of e-Research in general will inevitably be influenced by the rate of progress towards openness and data sharing. In this section, we present an overview; we will cover the current issues in more detail in later sections of this review. We have already noted the assertion by Hastings et al. assert that the data exchange process and therefore the precise description of chemical entities is vital to the application of cheminformatics.11 We might add not just to informatics but also to many areas that would not think of themselves as informatics. Many of the tools used in cheminformatics depend on providing formal data descriptions that require controlled vocabularies, thus emphasising the importance of agreed metadata constructs.
Both cheminformatics and bioinformatics depend on data, but it is essential that such data is reliable, and of an assured quality; moreover, that quality must be capable of being assessed. This requirement is particularly pertinent to the drug discovery process, for which the emphasis of cheminformatics has shifted from techniques to the management, curation, and integration of the large amounts of potentially useful data.
As a generic discipline, informatics is manifestly broader than the aspects applied specifically to drug discovery.20 Moreover, informatics has been applied to areas of chemistry with few if any links to drugs or biology. For example, the Master Chemical Mechanism (MCM) website, operated by the University of Leeds, is a resource that facilitates collaboration among, and information sharing by, researchers and other users with an investment in atmospheric chemistry. Of particular interest is the chemistry involved in the tropospheric degradation of volatile organic compounds (VOCs).¶
Similarly, the eMinerals project is another e-Science initiative that addresses environmental problems. The project uses molecular simulations to gain an understanding of the fundamental mechanisms associated with minerals-related issues such the transport of pollutants and the containment of radioactive waste.|| More generally, the interaction between materials science research and cyberinfrastructure was the subject of a 2006 workshop sponsored by the National Science Foundation. The workshop report appraised how the innovations produced by material science have revolutionised computing as well as assessing the influence of cyberinfrastructure on materials development. The report also drew attention to the need to adopt data interchange standards, owing to the importance of data exchange and sharing for materials sciences, as for other scientific areas.21
In his article entitled “e-Science and the Web”, De Roure considers the challenges presented to 21st century science by massively increased data collection rates and automation. He shows how e-Science creates opportunities for global collaboration and more effective sharing of services, data, and software. One manifestation is the emergence of virtual organisations (VOs), in which people and resources came together on a flexible basis to meet specific needs.22
De Roure identifies the Semantic Web and linked data as offering exciting opportunities with regard to recording information for reuse. Taylor et al. demonstrate how Semantic Web technologies can be deployed in the storage and access of molecular structures and properties. Using unique identifiers and relationships, represented as resource description framework (RDF) triples, they create a semantic database with the potential to enrich the exploitation of the data therein.13 The essential point is explicitly to capture and represent the relationships between different components of chemical information as an aide to the automated processing of that information.
The ability to test the provenance of both raw and derived data is becoming of increasing significance. In 2005, Simmhan et al. published a survey of data provenance in e-Science.23 Although the CMCS is the only chemistry project they examined in their survey, they raised several general issues that remain pertinent today, including, but not limited to: rich provenance information can become larger than the data it describes; provenance usability depends on federating descriptive information; coping with missing or deleted data requires further consideration.
Before examining these issues in greater detail, in the Concepts of sharing and exchange section, we consider why the data matters, and the data descriptions that are needed for working collaboratively with chemical data. Science is increasingly global, and collaborations cross cultural and language barriers.
Chemical data matters
Adams, writing early in 2009, made the following observations:**… unlike other scientific, technical and medical fields, chemistry has not evolved a culture of data and knowledge sharing.
Chemistry is a conservative discipline which is nevertheless starting to participate in the semantic web.
Although examples of data and knowledge sharing undoubtedly exist – the Cambridge Structural Database (CSD)†† was established in 1965, and the Protein Data Bank‡‡ in 1971 – Adams' point is a general one: in 2009 chemists were still reluctant to adopt a more open culture.
Historically, scientists – and chemists were no exception – placed greater emphasis on the conclusions they drew from their data than on the data itself. Re-examination, reuse, and repurposing of raw data were notions whose time was yet to come. With advances in cheminformatics has come recognition that scientists must consider the lifecycle of their data.
Enthusiasm for sharing one's data and for exploiting data made available by other researchers will be tempered by systemic pressures to maximise the value obtained from the data before releasing it, a state of affairs that seems likely to continue, at least in the short term. Academic reward and recognition are not the only factors contributing to conservative attitudes towards data sharing. The need to preserve intellectual property rights militates against sharing. Recent patent reforms that will make “first to file” the criterion for determining the right to a patent seem likely to inhibit sharing until the application has been filed.§§
Scientists retain data for a purpose, perhaps several purposes; they do not keep it for its own sake. When considering purposes, we often use the terms data and information interchangeably, but they do not have the same meaning. To take a simple example, a table of numbers remains data until we label the rows and columns: only then do we have information. If we can identify patterns in that information, we might reach a level of understanding that amounts to knowledge.
The data-information-knowledge-wisdom (DIKW) hierarchy is often represented in the form of a pyramid, as shown in Fig. 3. The origins of the hierarchy concept are not entirely clear, but the first mention is commonly attributed to Ackoff.24 His version included understanding as a category, whereas our use of that term as the label for the vertical axis in the diagram recognises that understanding and the ability to predict future behaviours are key features of the scientific method.
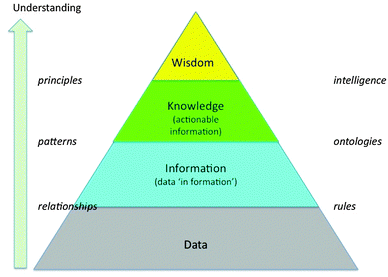 | ||
| Fig. 3 Diagram illustrating how, in particular, information and knowledge derive from raw data through the understanding of relationships and then patterns. | ||
Text box 1: the DIKW modelTo give a non-chemical illustration of the DIKW model, consider a banana:• Our data is the colour of the banana: green, yellow, or going black; • Our information is that the colour of the banana skin is indicative of the state of the fruit; • Our knowledge is that a green banana is unripe, a yellow banana is ripe and sweet, and a black banana is probably rather soft and mushy; • Our wisdom is to buy a yellow banana if we want to eat it now, a green banana if we want to eat it some days later, and to be uncertain about a black banana: if it was yellow when stored in the fridge, it might be fine to eat. |
We also acknowledge that the value of data changes over time: new wisdom supersedes old. Chakravarti explores such issues in the commercial context, making the point that …knowledge changes constantly and has a shelf life.¶¶ Such transience has implications for the data lifecycle, and especially for how we preserve and curate our data.
In this section, we take a broad view of information and data, so we note with interest Keller's recent exposition of the case for understanding ‘information’ as inherently embodied.25 The relevance for this review is that she draws inspiration from the work of Jean-Marie Lehn, the ‘father’ of supramolecular chemistry, which Lehn himself has reviewed in this journal.26 In terms of the DIKW diagram, molecular structures are capable of using their relationships with other molecules to set up self-organising systems. On this basis, Lehn perceives chemistry as an information science. Collaboration is essential to achieve the necessary understanding of how self-organisation processes work. Supramolecular assemblies involve mechanical as well as chemical bonds. In the abstract of his review of the chemistry of the mechanical bond, Stoddart makes the following observation about the importance of the actionable information, the knowledge:
The challenge to make more and more sophisticated compounds is predicated upon our fundamental understanding of the nature of the mechanical bond and how this associated knowledge base can be employed to do complex systems chemistry in very different environments where emergent phenomena become the order of the day.27
Valcarcel and Simonet make similar points in a different context, that of analytical chemistry, arguing the importance of relationships for understanding and managing chemical information.28 They present the data-information-knowledge sequence in an analytical chemistry context and, in their final remarks, urge the need for harmonisation between laboratories. Our response would be that a culture of openness and sharing offers the best way to meet that need.
Although the R&D sector of the pharmaceutical industry has for many years pressed for data integration as the answer to the volume and complexity of the data available, Slater et al. argue that it is not sufficient to bring together data and information from multiple sources.29 Semantics are necessary to interpret the information and derive knowledge. They propose a knowledge representation scheme that corresponds to the Semantic Web vision of data and resources described for use by humans and machines.30,31
In the Concepts of sharing and exchange section, we examine the potential of Semantic Web tools to integrate data from disparate sources for reuse in data-driven research. The lack of robust, usable tools has held back the wider adoption of semantic web technology, but this paucity is now changing. Ultimately, such tools can exploit machine reasoning to enhance understanding and thus ascend the DIKW pyramid.
Communicating chemical data
It is a general truth that we must be able to identify the entities to which data and information relate, and this truism is especially important in the field of drug discovery, to which the history of cheminformatics is inextricably linked. The search for new drugs, agrochemicals, and even materials has become increasingly data-driven and dependent on the integration of data from multiple sources.It is of equal importance to describe accurately the relationships between chemical and biological entities. Guha et al. argue for a holistic view of the relationships between small molecules and biological systems32 while Kohler, in her review of the three-volume set “Chemical Biology: From Small Molecules to Systems Biology and Drug Design”,33 stresses the importance of integrating chemical and systems biology.34 This fusion leads to the emerging discipline of systems chemistry, reviewed in 2008 by Ludlow and Otto from a complex systems perspective. They restrict themselves to synthetic systems in solution, for example combinatorial chemistry, but also cover other multivariate systems, including models that might contribute to the understanding of biological systems.35 Given the importance of the similar property principle in cheminformatics, it is tempting to draw an analogy with Tobler's first law of geography: Everything is related to everything else, but near things are more related than distant things.36 As noted previously, the concept of chemical space relies on such relationships.
Blomberg et al. discuss a range of initiatives aimed at increasing the interoperability of data and information, notably the formation and objectives of the Open PHACTS consortium, which we cover in the Open chemistry section.37
Identifiers and structure representations
Chemists have for many years communicated using a combination of linear notations and 2D and 3D structure depictions but in the digital era, machine readability of identifiers and structure representations has become a necessity and is fundamental to chemical information discussions. Willett discerns two foundations for modern cheminformatics, one being the efficient searching of databases containing large numbers of structures.1 The other is the modelling of molecular properties, which we cover in the Property descriptors section. Willett's review covers the development of structure and substructure search from the early days of connection tables through to the use of 3D atomic coordinates. Stumpfe and Bajorath have reviewed the principles and practices of similarity searching, in which they note that the concept of molecular similarity can depend on how structures are represented.38More generally, it is unique chemical identifiers that are the keys to communicating chemical structures and data. The systematic chemical names used in publications are capable of being unique but they are not machine-readable, and uniqueness is not always realised in practice. Molecular formulae, while amenable to machine interpretation, are not necessarily unique and are frequently ambiguous. The first attempt to obtain both uniqueness and machine interpretation was the Wiswesser Line Notation (WLN), for which a commonly cited source is Smith and Baker.39 However, toward the end of the 20th century, WLN gave way to the Simplified Molecular-Input Line-Entry System (SMILES).40 There are some limitations with SMILES representations, such as the existence of different SMILES versions of the same compound (the canonicalization issue). With the objective of establishing a unique label, IUPAC introduced the International Chemical Identifier (InChI).|||| Internet search engines have difficulty finding InChI strings, so IUPAC added the InChIKey to improve discovery. The InChIKey is a fixed-length (27 character) hash code representation of the InChI itself. Because hashing is an irreversible procedure, lookup tables are used to obtain the InChI from the InChIKey. With the exception of polymeric molecules, the majority of compounds, including some inorganic and organometallic molecules, can be represented with InChI identifiers. The Journal of Cheminformatics has very recently published an Article collection entitled The IUPAC International Chemical Identifier (InChI) and its influence on the domain of chemical information.***
Looking to the future, IUPAC have a project underway with the objective: To develop a standard machine-readable, indexable and searchable representation of chemical reactions based on the IUPAC International Chemical Identifier (InChI).†††
Systems biologists have developed the MIRIAM (Minimal Information Requested In the Annotation of biochemical Models) guidelines to facilitate the exchange of data and other objects. These guidelines require that “all the components of a model need to be unambiguously identified in a perennial and standard way”. Consequently, data and other objects are identified with MIRIAM URIs.41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ‡‡‡ Although the MIRIAM guidelines are a valuable initiative for systems biology, they are most unlikely to displace the InChI as the chemical identifier of choice.
‡‡‡ Although the MIRIAM guidelines are a valuable initiative for systems biology, they are most unlikely to displace the InChI as the chemical identifier of choice.
Williams notes the importance of the InChI for the Semantic Web in chemistry.42 Taylor et al. highlight the unique nature of the InChI and consider the construction of a URI from an InChIKey.43 Such URIs enable links between chemical properties, data, and publications, or entries in an ELN. Coles et al. have investigated the potential of the InChI for chemical information retrieval.44 Using the InChI strings for a corpus of 104 molecules whose crystal structures were published under the eCrystals/eBank project, they obtained high values for both precision and recall (though the available dataset was very limited and it would be useful to repeat the test now that more structures have been published). Tests with other corpora were similarly encouraging. ChemSpider, the Royal Society of Chemistry structure database, provides powerful and wide-ranging search services, using InChI, SMILES, systematic and trade names, or registry numbers.§§§
The eCrystals archive¶¶¶ illustrates the importance of the InChI and InChiKey for linking both raw and processed data, using these identifiers for linking to the data resulting from a single crystal X-ray structure determination, produced, for example, by the UK National Crystallography Service (NCS).45
CML, the Chemical Markup Language, which we examine in the Metadata section, defines an element, but is not prescriptive about the content. InChI identifiers can be used, but must be specified with the @value attribute.|||||| Murray-Rust and Rzepa, in their article describing the evolution and design of CML, consider the potential of CML to create an extensive, semantically-rich resource of chemical information, with the proviso that the chemical community still needs to agree identifier systems for molecules, reactions, spectra, and other components.46
Property descriptors
It is as important to have unambiguous descriptors for molecular properties as it is for the molecules themselves, particularly if the values are used in computations. Property descriptors, also known as molecular descriptors, can be obtained by experiment or derived from theory or calculations. Experimental measurements include dipole moment, NMR shifts, spectral peaks (electronic and vibrational), solubility, logP, polarizability, and Hammet parameters. Other descriptors provide structural information, for example, defining a characteristic sub-structure. Todeschini and Consonni define molecular descriptors as follows:The molecular descriptor is the final result of a logic and mathematical procedure which transforms chemical information encoded within a symbolic representation of a molecule into a useful number or the result of some standardized experiment.47
The principal use for property and other molecular descriptors is in quantitative structure–activity relation (QSAR) and Quantitative Structure Property Relation (QSPR) models, which use mathematical regression techniques to predict chemical properties and biological activity. Detailed consideration of QSARs is outside the scope of this review, but Warr, for example, provides a overview of the state of QSAR modelling.2 The 1995 review by Katritzky et al. of the use of QSPR for predicting chemical and physical properties gives a clear description of the principles of QSPR,48 while the reference handbook, Statistical Modelling of Molecular Descriptors in QSAR/QSPR, delivers broad and recent coverage of many aspects of models that make use of molecular descriptors.****
Units are an important aspect of property description, especially if the data is intended for semantic processing. Taylor et al. start with the premise that all values must have units associated with them, and then discuss how to describe those units other than with plain text.43 In the same paper, they present a layered model for describing data values in a manner suitable for semantically aware processing. The CombeChem project used RDF to classify chemical descriptors.9 Chepelev and Dumontier have implemented CHESS (Chemical Entity Semantic Specification) for representing chemical entities and their descriptors,49 a key aim for CHESS is to facilitate the integration of data derived from various sources, thereby enabling more effective use of Semantic Web methodologies.
Descriptors can be obtained by means other than experiment and computation. Downing et al. have applied text-mining tools to extract chemical entities and property information from electronic theses.50 Their underlying purpose is to develop the infrastructure to support machine-readable semantic representations of chemical information, which they argue to be the basis for machines to take over much of the routine work of discovering, analyzing, and collating chemical information.
With regard to open access to property information and to the exchange of molecular descriptions, the outlook is encouraging. For molecules with fewer than 1000 atoms and 1000 bonds, the PubChem database provides free download of structure and property information about many millions of compounds and substances.†††† Comprehensiveness can, however, become an issue if some of the information is proprietary. Masek et al. discuss the ‘safe’ exchange of relevant chemical information in the context of property and/or activity data.51 They propose guidelines for exchanging data that is relevant for QSARs and QSPRs without revealing the chemical structure (which might compromise intellectual property rights and/or competitive advantage). However, Filimonov and Poroikov argue that the safe exchange of relevant chemical information without disclosing structures is impossible, which would prevent the pharmaceutical industry from engaging in such information exchange.52
Infrastructures for sharing and exchange
Where Newton stood on the shoulders of giants, today's scientists depend upon the insights of a broad spectrum of other researchers, but to obtain access to that knowledge, an infrastructure for data sharing and analysis is essential. Haak et al., writing for Science magazine, argue for data exchange standards as a first step towards a Web-based infrastructure. They point out that a single database solution is not feasible, but unified standards would enable researchers to access data, tools, and other resources from a distributed network. They stress the importance of identifying interdisciplinary work and propose that:With these underlying exchange standards, the first step is to create a Web-based registry for data, or expand an existing one such as DataCite, to meet the needs of a global, multidisciplinary, multi-stakeholder community. Specific Web-based user interfaces (“wrappers”) can interact with the registry and fulfill discipline-specific requirements, such as metadata and related code needs.
It would be very difficult to gainsay their reasoning, although their subsequent remarks about the provision of metadata appear remarkably optimistic, given the observations elsewhere in this review about the burden of curation:
Providers would document their data with standard metadata, including data elements, sample frame, access levels, terms of use, and any fees, which could vary according to the amount and nature of use (e.g., scholarly, commercial, or algorithm development), and vary across providers…53
As indicated by Wild's first “grand challenge”, drug discovery is the primary purpose for mining chemical, biological, and pharmacological data sources.15 Wild has reviewed the mining techniques that can be applied to such sources and in the same paper examines a future predicated on intelligent, semantic aggregation of information.54 The authors of this review consider that view to be key to progress towards openness and data sharing in the chemical sciences.
Hohman et al. propose a community-based platform to provide traditional drug discovery informatics in conjunction with Web 2.0 tools.55 They summarise the challenge and opportunity as follows:
What is probably required of new collaborative software for biologists and chemists is a combination of capabilities that ensure privacy but allow selective collaborations when intentionally desired. For mainstream applicability, the tool must handle free text and also complex, heterogeneous drug discovery data and molecular structures. Furthermore, this complex data must be presented so that humans can easily draw conclusions and prioritize experiments from the data, procedures and ancillary information.
With regard to controlling access to sensitive information, they describe temporarily restricted data sharing, in which the data exchange can be controlled either by mutual agreement or through a trusted intermediary.
Groom and Allen have recently published a focus article about the Cambridge Structural Database (CSD) and its role in collaborative research.56 In their abstract, they note the use of the CSD for understanding molecular recognition processes such as protein–ligand interactions. Other chemical reaction databases can serve a corresponding role in understanding reactivity, although that goal requires data about unsuccessful procedures. The Dial-a-Molecule Grand Challenge network has recognised this point and encourages researchers to share their negative data and unpublished results.‡‡‡‡ Achieving the aim of making a step change in the ability to deliver molecules quickly and efficiently – in days rather than years – will also depend on databases like ChemSpider§§§§ and the ChemSpider SyntheticPages.
Collaborative chemistry
The earliest advocacy of open collaboration in chemistry was arguably that of Kouzes, Myers, and Wulf, who set the scene in 1996, citing Wulf's 1993 definition of a collaboratory as a:… center without walls, in which the nation’s researchers can perform their research without regard to geographical location-interacting with colleagues, accessing instrumentation, sharing data and computational resource, and accessing information in digital libraries.
They acknowledged the existence of both social and technological barriers to adoption, which we can relate to the issues raised by Borgman, as discussed in the Open chemistry section. However, their conclusion was a confident prediction (since borne out) that collaboratories would be part of our future.57
In the open chemistry context, the first significant initiative was the Collaboratory for the Multi-scale Chemical Sciences (CMCS),58 which Myers et al. describe further in the context of its application to combustion science.59 CMCS specifically supported combustion research, and this work led subsequently to the independent development of a cyberinfrastructure to underpin the sharing not only of data but also of tools and other resources. The goal of this cyberinfrastructure was to facilitate exchanges both within and between the sub-disciplines of combustion research.60
The authors of the CMCS report also commented on the social barriers that could influence the development and exploitation of shared digital resources, noting the differences between disciplines. The sharing of resources is more effective when a culture of open exchange already exists within a discipline, for example with publications. Moreover, they observe that researchers develop their own “one-off ad hoc” tools when shared infrastructures do not provide for their needs.
The goals of the CMCS can be seen in terms of the DIKW diagram in the Chemical data matters section. The system relies strongly on metadata to facilitate the collaborative analysis and reuse of chemical data. We examine the CMCS usage further in the Metadata section. The CMCS team have also designed the Knowledge Environment for Collaborative Science (KnECS) as an open-source informatics toolkit.61 Although KnECS evolved from CMCS, the two projects are now separate: KnECS is a general-purpose toolkit and CMCS is regarded as an extension of KnECS. This paper identifies six areas of challenge for applications developed using KnECS, all of which are relevant in the wider context of this review: data management; integration; reuse; usage; open-source; and collaboration.
The team at Indiana University have developed the Chemical Informatics and Cyberinfrastructure Collaboratory (CICC), one aim being to provide consistent access to integrated cheminformatics resources such that they can be combined in innovative ways to obtain chemical insights.3 Guha et al. present an evaluative overview of the methods and models deployed in the CICC and explain in some detail the web service infrastructure that they have developed, and how it interfaces with their workflow tools. They also describe the cheminformatics education programs that they deliver for training chemists and to ensure the future supply of cheminformatics specialists.
Yu et al. have developed the Collaboratory for MS3D (C-M3SD) as an environment to:
facilitate collaborations among biochemists, biophysicists, structural biologists and mass spectrometrists utilizing chemical cross-linking and covalent labelling techniques to investigate the structure and function of macromolecules and macromolecular complexes.62
The C-M3SD infrastructure, which builds on KnECS, adopts a workflow approach and is implemented as a Web portal. Workflow approaches are increasingly being used, spreading out from bioinformatics and life sciences, via drug discovery, to more mainstream computational chemistry. In chemistry, workflows have the potential to transform locally written scripts into more versatile and interchangeable ways for linking computational chemistry programs and services.
The value of an e-Science infrastructure for multidisciplinary collaborations is illustrated by the electrochemical research conducted in three separate laboratories, two in Australia and one in the UK. All three use a common set of tools and workflows that provide a flexible but integrated platform.63 More specific community efforts include the basis set exchange database proposed by Schuchardt et al.,64 which uses an XML data format, and the AnnoCryst system for collaborative discussion of 3D crystallographic models.65 The latter uses RDF graphs to represent annotations, held in an extended Annotea server: AnnoCryst is undoubtedly a Semantic Web project.
Alsberg and Clare report the use of MediaWiki for managing chemometric projects.66 They believe the complete openness about research projects to be the most important advantage of their wiki approach. However, they also note several shortcomings, some of which are inherent aspects of wikis. However, they also point out the lack of semantic annotation and the outstanding issue with integrating large amounts of structured data: the authors of this review regard these as highly significant deficiencies. Bradley and others have used wikis for a range of collaborative purposes, such as: reports, experiment plans, and open notebook science, although semantic annotation does not feature in the UsefulChem home page.¶¶¶¶ We review open notebook chemistry in the following section.
In 2008, Llinas took collaboration into a new area by posing a challenge to other researchers to predict the solubility of 32 druglike molecules, based on the reported solubility data for 100 other druglike molecules.67 The results were published in the following year.68 For the purposes of this review, the significant points come at the end of that paper. The authors foresee open and objective challenges becoming important for evaluating the capabilities and progress of computational chemistry, but they also note the dependence of any evaluation on data quality. The 2012 JISC/CNI Workshop report captures well the general concern about data quality: “The challenge of assessing the quality and value of datasets, and rewarding their creators appropriately, stalks the future”.6
Open chemistry
In a blog given over to science and Web 2.0, Nielsen posted the following definition:Open science is the idea that scientific knowledge of all kinds should be openly shared as early as is practical in the discovery process.||||||||
Much of the research reported in this review has been directed towards, and informed by, collaboration between scientists in different establishments. Indeed, the Open Access movement is founded upon collaboration, and Web 2.0 technologies are available to facilitate collaboration. Guha et al. formed the Blue Obelisk movement to promote collaboration and interoperability in cheminformatics, with several open source and open data projects.69 They discuss the social aspects of Blue Obelisk and emphasize that the value of interoperability extends beyond the open initiatives.
Martinsen, reporting on the 223rd ACS National Meeting and Exposition, discusses the impact of Web 2.0 technologies on scientific communication.70 He summarises papers about semantic tagging, data sharing, information discovery, and interaction with wikis and podcasts. Martinsen also reports findings about the evolving usage of journal articles now that almost all published scientific content is available on the Web. In concluding, he suggests that, owing to the potentially disruptive nature of such changes, we should monitor their effects on human behaviour and make adjustments for undesired results. In the first issue of the Journal of Cheminformatics, Bachrach made a strong case for data to be published for reuse, as open data.71 Many other advocates of open data in chemistry have echoed his views,72–75 and Losoff, from the perspective of a science librarian, argues that the integration of databases with journal literature and other research-related resources is an important component in furthering scientific progress.76
As noted earlier, there is evidence that open access increases the exposure and reuse of the results of scientific investigations.77 As mentioned previously, Hohman et al. describe temporarily restricted data sharing, in which the data exchange can be controlled either by mutual agreement or through a trusted intermediary, as a means for controlling access to sensitive information.55
Regrettably, the movement towards openness and data sharing in the chemical sciences still faces barriers. With regard to professional incentives, in their survey of research chemists at Cambridge and Imperial College, Downing et al. invited respondents to indicate the factors that would encourage them to share research data in an open access repository.5 Based on the responses, they found a clear reluctance to allow immediate open access to research results, permitting only other group members to see information before publication. They also found a tendency to store data as hard copy, and where data was preserved electronically, for individuals to choose different formats or even to use a range of formats to represent the same type of data. These attitudes to storing data are potentially inhibitory to effective sharing.
When the sharing community extends beyond a single research group, access control issues can arise. The combustion research cyberinfrastructure incorporated provisions for authentication and authorisation before allowing access to the data, tools, or resources available in the shared infrastructure.60 Discussing open access to data, Borgman notes that current practice tends to discourage data sharing.78 She identifies four categories of reasons for not contributing data to repositories, which we summarize as follows:
• Reward systems favour publication rather than data curation;
• Significant effort is required to organize, manage, and curate data; we refer to this elsewhere as the burden of curation;
• Research tends to be competitive, leading to a reluctance to share data until papers have been published and/or data is no longer commercially sensitive;
• Researchers value ownership of their data: it is their intellectual property.
Future systems set up for organised data exchange will have to tackle these issues of provenance, protection, citation, and reward. As reported earlier, the JISC/CNI Workshop report observes: Systems for assessment, reward and recognition have not changed in substance over the last 15 years of ubiquitous web use.
On the positive side, the need for data aggregation and integration in drug discovery has led to the formation of the Linking Open Drug Data (LODD) task force, as described by Samwald et al.79 They note that some of the LODD datasets are not fully open, owing to considerations that the task force is actively exploring, patient confidentiality being one such issue. Blomberg et al., concerned with increasing the interoperability of data and information to address the bottlenecks in small molecule drug discovery, describe the formation and objectives of the Open PHACTS consortium, which aims to develop an open source, open standards, and open access platform as the basis of an Open Pharmacological Space (OPS), adopting a Semantic Web approach to drug discovery.37,80
Other initiatives that promote openness and data sharing include the OpenTox project, which aims to provide semantic services to assist integration of toxicology information with the rest of the drug discovery process.***** The Chem2Bio2RDF repository exploits semantics to facilitate interoperation between chemistry and biology by integrating chemogenomics repositories with other chemical biology resources.81 Steinbeck et al. first presented the Chemistry Development Kit (CDK) in 2003.82 The CDK is an open-source toolkit for cheminformatics and bioinformatics, written in Java. Kuhn et al. have combined the CDK with the Taverna workflow engine to create an open workflow environment for cheminformatics.83
Open access to reports and details of experiments can be taken to an extreme with Open Notebook Science. Three scientists who are notable proponents of this practice, Jean-Claude Bradley, Cameron Neylon, and Matthew Todd, have used open notebooks very successfully and with fruitful results. Bradley is a leading exponent of open science: he provides all the experimental results from his work on anti-malarial compounds online.††††† Neylon and Todd have also made some of their laboratory notebooks available in electronic laboratory notebooks that are open to public access, for example “Cameron's LaBLog”.‡‡‡‡‡ Todd has coordinated a whole research project in public view as Project Lab Books on the ourexperiment.org site, for example the Pictet–Spengler route to Praziquantel.§§§§§
In concluding this section, we note that several of the observations made in The Independent Climate Change E-mails Review, published in July 2010, are pertinent to promoting a culture of collaboration and the willing sharing of information.84 The review team note the need for research data, methods, and other information to be publicly accessible; for scientific debate to become more open; and for:
all scientists to learn to communicate their work in ways that the public can access and understand; and to be open in providing the information that will enable the debate, wherever it occurs, to be conducted objectively.
Several government reports have endorsed the review team's call for greater openness. Most recently the Royal Society made its first recommendation that:
Scientists should communicate the data they collect and the models they create, to allow free and open access, and in ways that are intelligible, assessable and usable for other specialists in the same or linked fields wherever they are in the world. Where data justify it, scientists should make them available in an appropriate data repository. Where possible, communication with a wider public audience should be made a priority, and particularly so in areas where openness is in the public interest.7
Concepts of sharing and exchange
Historically, before the widespread use of digital storage, a researcher wishing to make use of data or information generated by another worker might need to go through the stages of: confirming the existence of the data; locating the source; accessing that source to retrieve the data (probably in a non-digital form); and validating the data for correctness and relevance.Few if any scientists would deny the need to preserve data throughout the research lifecycle: chemists have long acknowledged the importance of capture and preservation during the earlier stages of that lifecycle, but have paid less attention to curation, which is now seen as vital for discovery and then access of data. The concepts of preservation, curation, provenance, discovery, and access are embedded within the research lifecycle, as illustrated in Fig. 4.
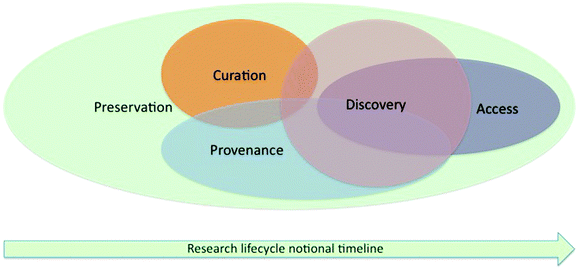 | ||
| Fig. 4 The concepts of preservation, curation, provenance, discovery, access in the context of the research lifecycle. | ||
For the ideal scientific methodological enquiry, effective data management is required from planning onwards. Properly curated data, with its provenance recorded from the outset can be analysed in conjunction with subsequent results to determine what factors are significant. Frey highlights the importance of recording at source, and argues for designing curation into experiments.85 Opportunities for the reuse of chemical data will be severely limited if that data is not preserved and curated in a form that enables ready access by human and computer proxies. In many cases, reuse depends on the extent to which the researcher builds in metadata capture at source: we consider this point in the Capturing metadata section.
Boyack et al., using computational scientometrics, studied the structure and evolution of chemistry research over a 30-year time frame to understand the growth of collective knowledge over time, concluding that the influence of Biochemistry and Bioengineering is increasing, notably in Chemistry.86 They computed knowledge exchange via citation linkages, thus illustrating the importance of linking for discovery and access. We believe that these findings demonstrate the growing need for flexible and effective mechanisms for knowledge flow and data exchange, especially at an interdisciplinary level. This requisite applies to closely related disciplines as much as when addressing the grand challenges of global environmental change or the societal challenges of aging populations. The numerous articles published by Boyack about science mapping, accuracy, and metrics illustrate the increasing interdisciplinarity of science.¶¶¶¶¶
At the beginning of its lifecycle, we cannot know what we will require of our data, so we must endeavour to capture everything relevant to that data. Although this is a challenging requirement, it becomes more tractable when we consider the components of “everything relevant”: the science that prompted the experiment; the thinking that went into the design; the origins of the materials used; the equipment that recorded the data, with the recording conditions; and the disposition of the data itself, the first step towards its preservation. In the remainder of this review we consider how these considerations relate to openness and data sharing.
Preservation
It is clear that preservation continues throughout the research lifecycle, and all the other lifecycle phases depend upon the quality of the preservation.When researchers began to use computers for recording their observations, whether measurements they had made themselves or those taken directly from equipment, the preferred medium was the flat file. The format ranged from a sequence of alphanumeric values through name-value pairs to proprietary data formats. Such data files could be copied, carried around (on so-called ‘floppy disks’), and stored in (again) so-called ‘safe places’. Data was thereby preserved, with a degree of permanence, and the researcher could say that the data was ‘protected’.
Although deceptively simple, the flat file is an unsatisfactory medium for good curation: the difficulties with finding, classifying, searching, and accessing such data, especially after the researcher has moved on, can be significant. Moreover, if the files contain only alphanumeric data, with no descriptive metadata, discovery and interpretation are likely to be compromised. Evidence of such practices emerged from the survey conducted by Downing et al., in which they found a tendency to store data in hard copy form or, when preserving data electronically, to use a range of formats.5
… our survey has shown that chemists still do not sufficiently appreciate the value of preservation of data in digital form. Moreover, few have had experience of the processes involved. Many were unaware that institutions have digital repositories and that there is little active archival of primary publications or data.
In the early days of the UK e-Science programme, Frey et al. argued that Data Grids enable e-dissemination of full experimental records, which they describe as publication at source.87 They also pointed out the potential advantages of Grid-based knowledge management for long-term preservation.
Subsequently, Frey compared the use of flat files, attractive for their simplicity, with database management systems (DBMSs), valued for their integrity and scalability.88 Reese suggests that relational databases are appropriate for data that changes frequently and for which maintaining integrity is important. He argues that data that does not change is best preserved in flat files, in tabular form wherever possible, and also proposes that, as well as the raw data, the archive should also contain a codebook that records how the data is entered and the descriptive metadata.89 Agrafiotis et al. describe a drug discovery informatics platform based on federated DBMSs.14 However, Frey, in part from considering the requirements for long-running experiments, argues for an intermediate approach, as described later in this section.
Kuhn et al. argued for more rigorous digital preservation of spectral data, noting that much valuable information is lost with traditional methods.90 They propose an information architecture that supports the capture, preservation, and dissemination of spectral data, based on a full formal specification in CML, on the basis that spectroscopic data was often stored in proprietary formats. However, their case has achieved no traction against the established JCAMP-DX format,|||||||||| which IUPAC supports for the electronic publication of spectroscopic data.
The requirements for secure preservation and ready access led Borgman to argue for a public rather than an institutional response:
Achieving a critical mass of datasets in public repositories, with links to and from publisher databases, is the most promising solution to maintaining and sustaining the scholarly record in digital form.78
The UK Research Councils now mandate that all grant proposals include a data management plan that includes provisions for data access by other researchers: the DCC present a useful summary of these requirements.****** The USA funding agencies have similar provisions, following on from requirements that publications should now include data. Furthermore, the UK HEFCE Joint Information Systems Committee (JISC now reorganized as a separate entity “Jisc”) funded a two-phase study to understand the costs of data preservation and develop a cost model for preserving research data in UK universities.91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ††††††
††††††
For all the above work it is clear that public and institutional repositories are only part of the response to the requirements of openness and data sharing. At least as significant, and possibly more so, are the networks of connections and the services that enable those connections to be exploited.
The Semantic Web provides such connection technologies, which Frey believes can cover the middle ground between the uncontrolled flat files and the rigid relational database.88 Semantic Web technologies employ URI-style links to data and metadata, thus enabling data analysis to connect with the appropriate version of the data, rather than relying on moving the content itself, a point to which we return in the Provenance section.
To illustrate URI-style links, Fig. 5 states that there is a substance, ethyl acetate, which has two identifiers, one a SMILES representation, the other an InChI. There is also an NMR spectrum for ethyl acetate, which can be accessed at http://example.org/NMR/012345.
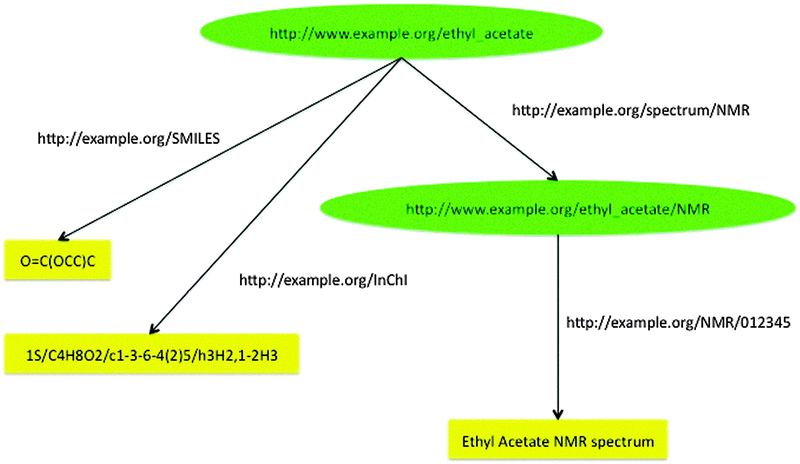 | ||
| Fig. 5 RDF graph linking to two identifiers for ethyl acetate and to its NMR spectrum. | ||
Curation
According to the Digital Curation Centre (DCC): Digital curation involves maintaining, preserving and adding value to digital research data throughout its lifecycle. This definition is followed by a list of the steps that comprise the digital curation lifecycle.‡‡‡‡‡‡ Although in this review we consider curation from a research lifecycle perspective, the DCC's comprehensive view serves to emphasise the importance of curation for data sharing and reuse. The principles and policies that provide a foundation for the stewardship of research data can be obtained the web publications of the DCC and the Research Information Network.§§§§§§ In their curation policy report, the DCC makes the following observation:Curation policies related to specific research projects are on the increase. The majority of the main UK research funders expect applicants to consider creation and management of their research outputs at the proposal stage in order to submit a data management and sharing plan.
A report commissioned by the JISC acknowledged the difficulties of the typical researcher in meeting the burden of curation, to ensure that shared data could be understood and applied appropriately. The report called for investment in training people for data curation and preservation work, thereby supporting metadata capture at the time of data acquisition and ingestion.91
In the Preservation section, we referred to the study, funded by the JISC, of the costs of data preservation: the Phase 2 report makes the following very telling point:
Even if the data is in no imminent danger of disappearing, engaging in curation activities early in the digital life cycle may be a less expensive strategy than postponing them until the future. The costs of preserving uncurated data long after it was originally created can be quite costly; retrospective metadata creation, for example, is often extremely expensive.¶¶¶¶¶¶ [pages 57–58]
The cost of bit-perfect preservation is very high. While such quality might be required for a musical performance, it is arguable that science does not require the same standard for its digital records. However, the long-term retention of physical artefacts such as samples might be necessary, at least until they can be reproduced reliably.
Throughout this review, we aim to show that basic curation is only part of the story: the creators and owners of data must ensure ready availability, preferably with open access. Despite acknowledging the value of curation, researchers can feel discouraged by the effort required to curate their data properly and also, for several reasons, feel inhibited about providing open access to their data, as discussed in the Open chemistry section.78 However, designing curation into experiments and capturing metadata at source can mitigate the burden of curation.85
We expect curation to be concerned in reality with a package, of which the data itself – the raw data – is but one component. Once captured, raw data should not change. However, the data might move to a different storage location, albeit undesirably; its metadata is likely to evolve, and we can expect the records of analysis and other usage to expand. The Collaboratory for the Multi-scale Chemical Sciences (CMCS) acknowledges the potential evolution of metadata: Metadata can also be added at any time during the lifetime of the data, that is, not only at file creation.58 However, while true, this statement blurs the principle of curation that as much metadata as possible should be created as – or even before – data is recorded. Metadata can still be added when the data is processed further: it is then viewed as annotation from the perspective of the data and context metadata from the perspective of the process involved.
The SPECTRa project aims to provide its embargo repository with a package that comprises comprehensive descriptive information as well as the CML-based data packages.5 Recognising the need, as an aspect of curation, for the formal representation of chemical reactions, Holliday et al. have extended CML by adding the CMLReact vocabulary.92
Frey, describing the CombeChem project, outlines the capture of synthetic methods as a series of linked steps, for which the materials, and processes and the way they are linked are clearly stated and explicitly recorded in RDF; not just as simple free text.85 In the general Semantic Web context, the capture of relationships as RDF triples, which constitute the metadata, is essential to the process of curation. An earlier paper about this e-Science project makes the following assertion, which manifestly states the case for semantically aware curation:
The CombeChem e-Science project has demonstrated the advantages of using Semantic Web technology, in particular RDF and triplestores, to describe and link diverse and complex chemical information, covering the whole process of the generation of chemical knowledge from inception in the synthetic chemistry laboratory, through analysis of the materials made which generates physical measurements, computations based on this data to develop interpretations, and the subsequent dissemination of the knowledge gained.9
Provenance
The provenance of a digital object is a record of the processes in that object's lifecycle, and the scientific method depends on the reproducibility of those processes.93 The importance of reliable provenance in a culture of openness and sharing is self-evident. We treat provenance as a distinct concept, although the capture and preservation of provenance information can be seen as aspects of curation. The provenance is, of course, an important part of the metadata!Capturing provenance has recently been recognized as an important challenge for informatics, but there is very little understanding of the full needs of researchers, let alone solutions that go beyond static identifiers or tracking data through a predictable lifecycle. Research data needs its own form of version-control, tracking changes in a way that is linked to the metadata description and that should move with the data.7
Borkum et al., describing the oreChem project, point out the importance of the relationship between the level of trust in reported results and the provenance, or pedigree, of the data from which those results were derived.94 In considering metadata quality assessment, Margaritopoulos et al. list provenance as one of the indicators of metadata quality, later refining their view to suggest that the provenance reflects the probability of having quality metadata.95
Data curation presents additional challenges to commercial organisations, especially those with a statutory obligation to maintain audit trails of their investigations and of the data produced thereby. Such organisations tend to deploy either proprietary Laboratory Information Management Systems (LIMS) or relational database technology, for which the associated database management system (DBMS) provides assured integrity, reliability, and availability. While curation solutions based on a LIMS or a DBMS do provide the required assurance, they are comparatively inflexible and require dedicated maintenance.
In his keynote review, Frey examines the potential for Semantic Web technologies in the laboratory context, making several observations about the importance of provenance.88 He refers particularly to the development of Provenance Explorer at the University of Queensland.96 Semantic Web technologies employ URI-style links to data and metadata thus enabling data analysis to connect with the appropriate version of the data, rather than relying on moving the content itself. When we preserve data with version control metadata, we freeze its provenance; accessing that data with a read-only connection ensures the integrity of that provenance. Another researcher wishing to use that data and its metadata has assured knowledge about how, when, why, and for what purpose, the data was generated.
This practice is most in evidence with web applications, with which we have moved away from keeping our own versions of documents and datasets to obtaining copies from a server, working with them, and then returning them to the server. The server administrator handles all the maintenance concerns, particularly backup. This reasoning is a manifestation of connectivism.|||||||||||| In the context of experimental records, the connectivist would argue strongly for the documents and data that comprise a compound record to be linked, or included by reference, rather than creating a compound document by copying. Inclusion by reference is also known as transclusion, which in today's networked world is a very realistic approach to building content, although transclusion does raise new issues for provenance, notably version control and link maintenance. In essence, when we retrieve information by following a link, unless we have an assured provenance trail we might question whether what we acquire is what the originator of the document intended us to obtain.
Murray-Rust and Rzepa present the alternative view.97 They prefer to aggregate the components by copying them into a datument at the time of publication, thereby ensuring the continued integrity of the compound record, or datument. Although other authors cite this paper, there is no evidence of general adoption of the datument concept. Moreover, compound documents (datuments) are by their nature static, whereas a record comprising links has the innate capacity to enlarge its sphere of influence: Nodes always compete for connections because links represent survival in an interconnected world.98 [page 106] It is perhaps worth noting that Murray-Rust and Rzepa themselves admitted that their article was an expansion of a short “slightly tongue-in-cheek” presentation.
Discovery
For this section of the review, we distinguish the discovery of discrete pieces of chemical information from the mining of multiple or aggregate data sets to identify patterns, which we cover in the Mining chemical data (Text Mining) section.Researchers in all scientific disciplines now rely predominantly on electronic methods when searching for and accessing scientific information. With regard to studies of information-seeking behaviour, Davis described chemists as an ideal group to study, owing to their heavy use of journal literature.99 Although his paper is not specifically about linking, Davis brings out the importance of links to the process of information discovery by chemists.
In 2007, Brown published an analysis of eight American Chemical Society journals to elicit the use by chemists of Web-based information resources.100 She found that chemists were not taking full advantage of Web-based resources, despite the growth in their number and availability. Nevertheless, she made the following observation:
Even though chemists do not incorporate large numbers of freely available Web-based resources into their publications, an increasingly important component of a chemist's information behaviour for the direct support of his or her research is unfettered bench-top access via the Web.
We can confidently predict that any survey of the use of Web-based resources conducted in 2012 would show a significant increase in usage. For example, scientists now use social networks such as Twitter to assist their colleagues to discover information of value to their work.101
In 2009, findings similar to those of Brown emerged in the study of information use and exchange in the life sciences that was conducted jointly by the Research Information Network and the British Library.102 In the ‘Information lifecycle’ section of their report, the authors note a limited awareness and usage of the information services available. They attributed the little usage of social networking tools to the lack of a critical mass for the users of such services. Although the researchers in their study were in favour of data sharing, they did experience competing pressures: it is perhaps not surprising that they correspond with those identified earlier in this review.78
A recent study sponsored jointly by the British Library and JISC explored the information-seeking behaviour of doctoral students from ‘Generation Y’ (born between 1982 and 1994). Amongst the findings was a reliance on text-based and secondary material, rather than primary sources. The bar chart depicting the types of resource used, analysed by discipline, shows that of students in the physical sciences, a little over 5% accessed raw data. Moreover, few science students exploited large datasets. The majority of doctoral students work alone, rather than in collaborating teams, and tend to share their research outputs only with their immediate colleagues, leading the study team to observe:
Despite their evident reluctance to share their research outputs wider than their immediate work colleagues, overall the doctoral students endorsed in principle the benefits of greater openness and sharing in research.
The study also shows a general lack of understanding of open access.103 We note that this deficiency might well be influenced by the attitudes of students' supervisors.
Although most studies of information-seeking behaviour concentrate on literature, the focus of this review is the management, access, and sharing of chemical data. Moreover, the increasing held view is that when data is the product of publicly funded research, it becomes a public asset and should be available for verification and reuse.******* If the owner of data has deposited it for use by other researchers, how might potential users discover the existence of and then locate that data?
While search is undoubtedly the primary tool of discovery, Zhou et al. have analysed text and chemical search tools and concluded that the addition of a chemical intelligence layer would improve search precision.105 Their solution is Entity-Canonicalization Keyword Indexing (ECKI), in which chemically intelligent filters convert source data to text and replace synonyms with a proxy, the canonical keyword (CK) representation, prior to an indexing stage.
Subscriptions, notifications, and alerts offer a ‘push-based’ alternative to the traditional approach whereby researchers ‘pull’ information from various sources. Murray-Rust and Rzepa have proposed RSS as the basis for a service to alert the chemical community to new information relevant to specified interests.106
The Collaboratory for the Multi-scale Chemical Sciences (CMCS) relies on metadata for data discovery:58Metadata is at the heart of this data-centric infrastructure, enabling the discovery of data across scales and preserving the data provenance or pedigree. In some cases the data, although available, is incomplete, necessitating a recovery stage. Banfi and Patiny explore this need in the context of NMR spectra and put forward a tool that enables the assignment of a structure by drawing lines between atoms and automatically characterized signals.107
Jiao and Wild have used natural language processing and text-mining methods to extract protein and chemical interactions from journal article abstracts.108 Their work relates specifically to Cytochrome P450 (CYP), but they believe that their method can be used to extract other chemical and biological information. Mining a range of heterogeneous data sources also presents implementation challenges. For example, Miled et al. have addressed the efficient design of a database for drug candidates:109 their goal is to optimise data access performance. They consider alternative schema designs and explore parallel processing approaches.
On the other hand, data sets generated by high-throughput screening (HTS), while homogeneous, can be very large. Yan et al. have developed a data mining approach based on an algorithm called ontology-based pattern identification (OPI) for extracting relevant knowledge from such data sets.110 Their OPI method identifies subgroups of structurally similar compounds and ranks them with a probability score: the compound with the highest score is used as the representative of the subgroup. High-throughput techniques and the analysis of the data they produce is a huge area in its own right, which has generated a significant literature of statistical issues. We consider this area to be outside the scope of this review.
Singh et al. note that mining databases to identify compounds that can lead eventually to a novel drug can be complemented by examining the textual context in the literature.111 They propose a methodology called Text Influenced Molecular Indexing (TIMI), which combines their own Latent Semantic Structure Indexing (LaSSI) technique with Latent Semantic Indexing (LSI).
An important aspect of drug discovery is the identification of substructures that affect physicochemical and biological properties. Two groups have reported graph-based chemical representation methods for substructure mining.112,113 Having extracted substructures, they enable data mining techniques to infer relationships between molecular structure and chemical or biological properties, from which quantitative structure–activity relationships (QSARs) can be derived.
In 2006, Bruce et al. conducted a comparison of QSAR classifiers, concluding that the support vector machine was the best of those studied.114 Gedeck et al. studied the quality of QSAR predictions, using a range of data sets and QSAR models.115 Other studies have been published that relate to specific data sets and data mining algorithms. One interesting application of structure–activity modelling is predictive toxicology. Richard has reviewed this area and reached an optimistic conclusion based on the integrated deployment of cheminformatics – QSAR modelling and data mining – with bioactivity profiling.116
In the context of this review, the importance of obtaining the maximum value from the available data was one of the factors encouraging the foundation of the Open PHACTS consortium. This association was in part set up to address the dependence of modern drug discovery on the availability, processing and mining of high quality data. Later in the same article, the authors issue the following invitation: Any partner with an interest and an ability to contribute data, software or expertise is principally considered as an ‘associated partner’.37
Access
This concept is broad, ranging from providing access to obtaining access. Providers might wish to control not only the data and information that they make available, but also to whom they provide that material. They might also wish to constrain the purposes for which the material is used and the extent to which it can be reused. Discovery of resources might be only the first step; obtaining access to those resources might require additional involvement. In this section we examine the methods that chemists can and should use to make data and information available.Publication by definition provides open access, but scientific progress depends on access to the data on which publications are founded. In a recent press release, the European Commission stated its intentions regarding access to scientific information produced in Europe:‡‡‡‡‡‡‡
As a first step, the Commission will make open access to scientific publications a general principle of Horizon 2020, the EU's Research & Innovation funding programme for 2014–2020.
The Commission will also start experimenting with open access to the data collected during publicly funded research (e.g. the numerical results of experiments), taking into account legitimate concerns related to the fundee's commercial interests or to privacy.
At the end of the Open chemistry section, we quote the first recommendation in the Royal Society report about open data for open science, which strongly reinforces the European Commission view.7 The JISC/CNI Workshop report points out the tension between openness and maximising impact, given the role of publication in the research lifecycle.6
The case for sharing chemical data is reinforced, albeit in a different context, by Botstein's conclusion that three articles from the early years of the publication ‘Molecular Biology of the Cell’ had a remarkably high number of citations because the articles included gene expression data. It was the data that attracted the continuing interest.117
Casher and Rzepa propose improvements to electronic publishing with the use of their SemanticEye model.118 Their application enables unique identifiers, such as a Document Object Identifier (DOI), to be embedded in Adobe XMP (extensible Metadata Platform), thereby enhancing navigation between articles from multiple sources. However, a search of the literature since 2006 suggests that SemanticEye has not received the interest that the authors hoped for.
Semantic assertions also enable the authentication and validation of chemical information. Gkoutos et al. explored the potential for such processing for entire documents, or individual components, against a digital signature in an X.509 certificate.119
In the context of electronic theses, Sefton et al. propose the ICE-Theorem infrastructure for eScholarship, with tools for authoring, managing and disseminating semantically-rich thesis documents fully integrated with supporting data.120
One of Borgman's four reasons for the reluctance of researchers to contribute data to repositories is a reluctance to share data until papers have been published and/or data is no longer commercially sensitive.78 To address this issue, Downing et al. propose an embargo mechanism, whereby new material would be held in a “closed archive” until the owner allowed the release of the material, commonly after publication or when no longer commercially sensitive.5 This extends the role of embargo and leaves the power with the author, but does have the potential to suffer from abuse and neglect. The data deposition and release policies of the Protein Data Bank include provisions for data to be held for a period of time before public release.§§§§§§§
The Pistoia Alliance came into being in 2009, following an earlier meeting in Pistoia, Italy, with a mission to facilitate collaboration and innovation at the precompetitive stage of life sciences research. Data management and sharing issues are of particular interest, leading, amongst other topics of mutual interest, to collaborative consideration of the requirements for ELNs.¶¶¶¶¶¶¶ Recently, a new group, the Allotrope Foundation was announced, with a remit to promote an open information framework for analytical laboratories. The framework will comprise data exchange standards, metadata repositories, and open source libraries.||||||||||||||
Metadata
If increased openness and data sharing are to lead in turn to real progress in the chemical sciences, metadata will play a crucial role. Capturing metadata is a vital part of preservation, curation, and provenance. Discovery and access depend on exploiting metadata, as does the use of provenance information. The majority of the publications cited in this review stress the importance of metadata, but few are willing to attempt a rigorous definition of the term. The commonly accepted definition, that metadata is “data about data”, soon runs into difficulty. Pancerella et al. assert: such a definition is very dependent on one's perspective.58 They go on to provide a definition, the final sentences of which are telling for this review:… because metadata must be understood and manipulated, it must be formatted in a way that exposes its meaning in machine-comprehensible form.
In this review, we adopt a less strict interpretation: we consider an item of information to be metadata if it enables us to make deduction(s) (or be more certain about the deductions that are made) about the data with which it is associated.
Metadata formats
In an open world that encourages collaboration and data exchange, proprietary metadata formats are highly unlikely to survive as more than curiosities. Currently, the dominant storage format for metadata is XML (eXtensible Markup Language). For many cheminformatics practitioners, the tools of the Semantic Web offer considerable potential for the advancement of chemistry, and indeed of all science.11,22,88,121,122 RDF (the Resource Description Framework) is the foundation of Semantic Web technologies deployed for data description, providing URI-style links to data and metadata. RDF has several representation formats, one of which is RDF/XML.CML (Chemical Markup Language) is an XML vocabulary developed specifically for describing chemical information.12 Since its introduction a number of extensions have emerged, including a revision as a form compliant with XML Schema121 and, recently PML (Polymer Markup Language).123 Two recent publications by Murray-Rust et al. provide a comprehensive account of the design, evolution, and semantics of CML.124,125 While it has taken some time, CML has now been adopted as a medium for exchanging structural data, often being the format that underpins other, more highly visible, molecular structure file formats. However, CML has not achieved the widespread traction for which its originators must have hoped.
Chemical Semantic Web and chemical ontologies
The Semantic Web will almost inevitably play an increasing part in achieving progress towards openness and data sharing in the chemical sciences. In concluding their survey of semantic e-Science applications, Chen et al. make the following assertion:126As semantic technology has been gaining momentum in various e-science areas, it is important to offer semantic-based methodologies, tools, middleware to facilitate scientific knowledge modeling, logical-based hypothesis checking, semantic data integration and application composition, integrated knowledge discovery and data analyzing for different e-science applications.
A primary purpose of chemical metadata is to categorise or classify the data to which the metadata relates. The act of classification requires the target categories to be organised with a controlled vocabulary, typically a taxonomy or ontology that in effect models how the community understands the information space.
The various forms of controlled vocabulary are described more fully elsewhere, but Fig. 6 provides a schematic comparison of the principal embodiments. A term list is a vocabulary selected and controlled by some form of authority: although simple, term lists are still useful. By organising the terms into a taxonomy, which is typically hierarchical, we add both structure and the descriptive power that derives from the relationships between the terms. Users create a folksonomy by attaching descriptive tags, or labels, to items of information, and use the labels provided by other members of the community, thus generating a vocabulary with a social aspect. An ontology is the most complex and powerful form of controlled vocabulary, mainly because it organises relationships with a network representation and can include social aspects, for example, by using the FOAF ontology mentioned earlier.
 | ||
| Fig. 6 A comparison of the attributes of the principal forms of controlled vocabulary. | ||
Frey notes that the use of a controlled vocabulary ensures that everyone uses the same terms, but these terms have to be agreed and workable. However, he also points out that ontology construction can involve considerable effort.88 The development of ontologies for components of the chemical information space is only part of the problem: common agreement is necessary to enable individual ontologies to be used collectively. Mechanisms such as XML namespaces do exist for integrating ontologies, but are predicated on a shared understanding of the meaning of the terms used. A search of the web for “chemical OR chemistry ontology” reveals an active field, although few developments have achieved significant prominence. Adams, writing early in 2009, ended a brief overview of existing attempts to describe various aspects of chemistry as ontologies by noting the lack of a community effort to formalise chemical concepts.******** Currently, ChEBI (Chemical Entities of Biological Interest) is the most established ontology in chemistry,†††††††† while ChemAxiom seems likely to come into prominence as an ontological framework.127 ChemAxiom is a set of interoperable ontologies that describe both chemical concepts and chemical data.
Molecular structure is the principal basis for representing chemical compounds, so the ability to draw and edit structures on a computer screen is naturally of considerable importance. Sankar et al. have developed the ChemEd tool, written in Java, which enables users to select structural fragments from a fragment ontology.128 The authors list the other commercially and freely available editors but believe that the semantic richness of ChemEd illustrates the potential for developments that add meaning to basic constructs. Ontologies are also beginning to be adopted in chemical engineering, for example in chemical batch process management.129
That the evolution of the Chemical Semantic Web (CSW) requires coordinated activity directed towards the development and dissemination of chemical ontologies is apparent from the article about the CSW vision.‡‡‡‡‡‡‡‡ In discussing new research directions and possible focus areas, Bhat makes the following observation:
At present chemical Semantic Web is in its infancy and a global participation is needed by many data providers to make it to grow. Common vocabularies, general ontologies, efficient and scalable technologies for generating and searching ontologies, and ‘use-case’ based methods to define RDFs for compounds are just a few stumbling blocks for the growth of CSW.
Although we consider general ontologies and common vocabularies to be essential for progress in cheminformatics, a comprehensive survey of the ontologies relevant to the chemical sciences is beyond the scope of this review. The authors believe there is a role here for international organisations here, for example IUPAC in collaboration with the other scientific Unions (ICSU) building on the success of the InChI.
Capturing metadata
If the original researcher has curated the data effectively, discovery, access, and reuse should be reasonably straightforward: other researchers can realize the value of data that has been made openly accessible. Irrespective of how the chemical information space is represented, it is important to automate as much metadata capture as is possible, to relieve the burden of curation. Basic metadata will be acquired together with data produced by measuring equipment, but insofar as is feasible, other information should be captured from the context. Frey notes that the capture of semantic relationships can lead to tension between freedom and control, in that controlled vocabularies inhibit the free text annotation with which researchers might be more comfortable.88Hawizy argues that text mining can be used to process reports and publications extract chemical knowledge and express it in RDF.§§§§§§§§ Park et al. propose a combination of text mining and machine vision for automating the annotation of entries in the chemical database.130 Their purpose is to automate the extraction of information such as synthetic method, chemical and physical properties, and biological activity data, and integrate that information with the chemical structure database. Previously, Gkoutos et al. had presented a machine vision approach to extracting chemical metadata from raster images.131 Text mining techniques have advanced since Postma et al. reported their system for the semiautomatic extraction of information from abstracts describing analytical methods.132 They describe the stages of their text analysis in some detail but conclude that their information extraction task is still a time consuming one.
Under the auspices of the CombeChem project, Frey et al. adopted a Human Computer Interaction (HCI) approach to designing an information system for capturing the data and metadata recorded by chemists during an experiment.133 The Smart Tea project developed an ontology to model the Materials and Processes comprising the experiment, as one part of a system to support the experimental process from planning through to publication (at source). Given the importance of data capture at source, remote access to scientific instruments and sensors is essential. Bramley et al. describe a Common Instrument Middleware Architecture (CIMA) for integrating instruments into a Grid computing environment.134 McMullen and Huffman describe the CIMA Crystallography portal for managing the workflows within individual laboratories, supporting remote operation, and enable cooperative working between laboratories in the federation.135 They handle authentication and authorization issues by using public key infrastructure (PKI) certificates.
Electronic laboratory notebooks (ELNs)
The authors of this review are also preparing a paper about record keeping in the digital era, which will cover the nature and history of the scientific record and also include a survey of ELNs. A prominent theme of that paper will be role of ELNs in fostering openness and data sharing in all sciences.The purpose of ELNs can be stated very simply as capturing, preserving, sharing, and exploiting the information that matters. Prior to the use of computers, the details of experiments and the results obtained were recorded by hand in laboratory notebooks. The information so captured was almost exclusively for the benefit of the individual scientist, with little or no thought given to the reuse or repurposing of that data by other researchers.
One of the many benefits of the Electronic Lab Notebook (ELN) is the support for collaboration. Data and other records can be examined, validated, analysed, and reused: ELNs permit more informed discussion of experiments and outcomes. In 2005, Taylor reviewed ELNs in chemistry and biology. He predicted that eventually, all R&D scientists would use ELNs to preserve the records of their research. He notes that the incorporation of data is an essential part of the R&D workflow, but (in 2005) none of the ELN vendors were achieving that effectively; Taylor anticipated that this situation would change.13 Coles and Frey stress the importance of machine-processable links between data and publications.¶¶¶¶¶¶¶¶ Samson describes how ELNs and Laboratory information management systems (LIMS) are being adopted more widely, having originated within chemistry.||||||||||||||||
Conclusions
Chemistry is a long tail science, involving global collaborations of heterogeneous, relatively small scale, research groupings, which can now be more effectively combined by the efficient exchange of chemical information and techniques. As the collaborations grow to involve people more and more distantly acquainted (if at all), and the scale of data exchanges required grows, reliable provenance and clarity of the data become increasingly necessary.In this context of defining and exchanging chemical information, we suggest a number of areas that will be increasingly important in the next 10–20 years and form the Grand Challenges for chemical information in the digital/Web era. Several of these challenges are general to science and engineering, obliging us to cope with the following considerations:
• The increasing ability to create large amounts of data using automated processes;
• The global scale of collaborative efforts;
• The demands, even if unrealistic, to solve problems faster;
• The ever increasing breadth of knowledge required to deal with interdisciplinary research, even within problems specific to the chemical sciences: “the physical chemical biology of engineered systems in the environment”, and how we educate and support 21st century polymaths;
• The need for reproducible science: observational, experimental and computational.
The solutions proposed for many, if not all, of these problems involve the use of computers and computational techniques. However, if this digital revolution approach is to be an effective solution, we require that the information generated by experiments, observations, and calculations can be, and actually is, presented in both a human and computer readable manner. In the past, communicating information across language and cultural barriers was a non-trivial problem, solved in part by the use of one language (English) as the dominant means of formal communication, and internationally agreed nomenclature and terminology. This solution now needs to be extended (but not replaced) to deal with a very different type of “intelligence”, although the necessary work on the descriptions of chemical entities, processes, information and knowledge, via an ontology, internationally agreed, is in its infancy.
From our review it should be clear that context and the provenance of results are essential for the proper validation of chemical data and information. With increasingly complex, automated, interdisciplinary, and delocalised research efforts, recording of the context, the environment, and the details of experiments becomes even more essential. The Dial-a-Molecule community has initiated an effort to ensure better recording of chemical reaction data, such as temperature, concentration, and purity profiles, and equally importantly, to provide a route to disseminate details of reactions that have not worked!
One way forward to improve the reproducibility and transparency of chemical research is to adopt some of the processes and practices of the engineering disciplines. The significant overhead in terms of time and effort that this would traditionally have imposed on a project has meant that previously this has not been a practical approach. However, the increasing use of automation, and the rapidly falling costs of computers, sensors, and other technologies open the possibility of achieving the main aims of process and project engineering controls on a smaller laboratory scale. We believe that a major aim of the developments in chemical information collection and handling should adopt this engineering quality management style.
Very recently, Science Exchange Inc., PLoS ONE, figshare, and Mendeley have launched the Reproducibility Initiative. Their aim is to issue certificates of reproducibility for studies that have been validated by being independently reproduced.********* The journal Organic Syntheses already has each synthetic procedure and the associated characterisation data checked by a member of the Board of Editors.†††††††††
In terms of interdisciplinary research work, the Grand Challenges proposed by Wild are very relevant, with his fourth area being a particularly important aim for future development: Enabling the network of the world's chemical and biological information to be accessible and interpretable. We would, however, aspire to extend the range of this idea. On one side we have ever increasing physical information, which can be advantageously imported into chemistry, and in parallel with the life sciences applications of chemistry we have materials science. Further along the complexity scale, bioinformatics is extending biological science to the medical, environmental, epidemiological, and ecological information spaces. We are on the threshold of realising a truly end-to-end objective with the following characteristics:
• Increasing closeness of the links between computational and laboratory experiments;
• Increasing ability to cope with the variability and uncertainty that comes with the consideration of complex, real-world, chemical environments;
• Increasing automation of chemical experiments with more accurate control and recording of conditions;
• Increasing interaction of basic chemistry with longer term objectives related to global environmental issues (Food, Water, Climate, Population).
In our opinion, the issues that require community action are centred on chemical data. We believe that it is essential to increase the amount of chemical data available for open access, while ensuring that new mechanisms for validating the data are provided. The community should use this data to develop more efficient links between the worlds of cheminformatics and those of materials, environmental informatics, bioinformatics, and medical informatics. The goal is to create ever better chemical design services.
The momentum that is now behind open access to scientific information promises to enhance the sharing and collaboration that will underpin the innovations needed to meet the Grand Challenges of 21st century science and technology: we look forward with cautious optimism.
Acknowledgements
The authors would like to acknowledge the support of the Smart Research Framework (JISC) project, EPSRC funded CombeChem e-Science (GR/R67729/01, EP/C008863/1) and the e-Research South Consortium (EP/F05811X/1).References
- P. Willett, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1, 46–56 CrossRef CAS.
- W. A. Warr, in Chemoinformatics and Computational Chemical Biology. Series: Methods in Molecular Biology, ed. J. Bajorath, Springer, 2011, vol. 672, pp. 1–37 Search PubMed.
- R. Guha, G. D. Wiggins, D. J. Wild, M.-H. Baik, M. E. Pierce and G. C. Fox, In Silico Biol., 2011, 11, 41–60 CAS.
- J.-L. Reymond, L. Ruddigkeit, L. Blum and R. van Deursen, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 717–733 CrossRef CAS.
- J. Downing, P. Murray-Rust, A. P. Tonge, P. Morgan, H. S. Rzepa, F. Cotterill, N. Day and M. J. Harvey, J. Chem. Inf. Model., 2008, 48, 1571–1581 CrossRef CAS.
- A. Swan, Transforming opportunities in scholarly discourse, Jisc, 2012 Search PubMed.
- G. Boulton, P. Campbell, B. Collins, P. Elias, W. Hall, G. Laurie, O. O'Neill, M. Rawlins, J. Thornton, P. Vallance, M. Walport, Science as an open enterprise, 2012.
- S. Kim, J. Chem. Inf. Model., 2006, 46, 938 CrossRef CAS.
- J. Frey, D. De Roure, K. Taylor, J. Essex, H. Mills, E. Zaluska, L. Moreau and I. Foster, in Provenance and Annotation of Data, ed. L. Moreau and I. Foster, Springer, Berlin, Germany, 2006, vol. 4145, pp. 270–277 Search PubMed.
- N. Sukumar, M. Krein and C. M. Breneman, Curr. Opin. Drug Discovery Dev., 2008, 11, 311–319 CAS.
- J. Hastings, L. Chepelev, E. Willighagen, N. Adams, C. Steinbeck and M. Dumontier, PLoS One, 2011, 6, e25513 CAS.
- P. Murray-Rust, J. Chem. Inf. Model., 1999, 39, 928–942 CrossRef CAS.
- K. T. Taylor, Curr. Opin. Drug Discovery Dev., 2006, 9, 348–353 CAS.
- D. K. Agrafiotis, S. Alex, H. Dai, A. Derkinderen, M. Farnum, P. Gates, S. Izrailev, E. P. Jaeger, P. Konstant, A. Leung, V. S. Lobanov, P. Marichal, D. Martin, D. N. Rassokhin, M. Shemanarev, A. Skalkin, J. Stong, T. Tabruyn, M. Vermeiren, J. Wan, X. Y. Xu and X. Yao, J. Chem. Inf. Model., 2007, 47, 1999–2014 CrossRef CAS.
- D. J. Wild, J. Cheminf., 2009, 1, 1 Search PubMed.
- X. Dong, K. E. Gilbert, R. Guha, R. Heiland, J. Kim, M. E. Pierce, G. C. Fox and D. J. Wild, J. Chem. Inf. Model., 2007, 47, 1303–1307 CrossRef CAS.
- H. S. Rzepa and E. L. Willighagen, The 235th ACS National Meeting, American Chemical Society, New Orleans, LA, 2008.
- J. G. Frey and C. L. Bird, Expert Opin. Drug Discovery, 2011, 6, 885–895 CrossRef.
- T. Hey and A. E. Trefethen, Science, 2005, 308, 817–821 CrossRef CAS.
- S. He, Electron. Libr., 2003, 21, 117–122 CrossRef.
- S. J. L. Billinge, K. Rajan and S. B. Sinnott, From Cyberinfrastructure to Cyberdiscovery in Materials Science: Enhancing outcomes in materials research, Arlington, Virginia, 2006 Search PubMed.
- D. De Roure, Computer, 2010, 43, 90–93 CrossRef.
- Y. L. Simmhan, B. Plale and D. Gannon, Chimera, 2005, 34, 31–36 Search PubMed.
- R. L. Ackoff, J. Appl. Syst. Anal., 1989, 16, 3–9 Search PubMed.
- E. F. Keller, Stud. Hist. Philos. Sci., C: Stud. Hist. Philos. Biol. Biomed. Sci., 2011, 42, 174–179 Search PubMed.
- J.-M. Lehn, Chem. Soc. Rev., 2007, 36, 151–160 RSC.
- J. F. Stoddart, Chem. Soc. Rev., 2009, 38, 1802–1820 RSC.
- M. Valcárcel and B. M. Simonet, TrAC, Trends Anal. Chem., 2008, 27, 490–495 CrossRef.
- T. Slater, C. Bouton and E. S. Huang, Drug Discovery Today, 2008, 13, 584–589 CrossRef CAS.
- T. Berners-Lee, J. Hendler and O. Lassila, Sci. Am., 2001, 284(5), 34–43 CrossRef.
- L. Feigenbaum, I. Herman, T. Hongsermeier, E. Neumann and S. Stephens, Sci. Am., 2007, 297, 90–97 CrossRef.
- R. Guha, K. Gilbert, G. Fox, M. Pierce, D. Wild and H. Yuan, Curr. Comput.-Aided Drug Des., 2010, 6, 50–67 CrossRef CAS.
- Chemical Biology: From Small Molecules to Systems Biology and Drug Design, ed. S. L. Schreiber, T. M. Kapoor and G. Weiss, Wiley Online Library, 2008, vol. 1–3 Search PubMed.
- J. J. Kohler, Nat. Chem. Biol., 2007, 3, 528–529 CrossRef CAS.
- R. F. Ludlow and S. Otto, Chem. Soc. Rev., 2008, 37, 101–108 RSC.
- W. R. Tobler, Econ. Geogr., 1970, 46, 234–240 CrossRef.
- N. Blomberg, G. F. Ecker, R. Kidd, B. Mons and B. Williams-Jones, EFMC Yearbook, 2011, pp. 39–43 Search PubMed.
- D. Stumpfe and J. Bajorath, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1, 260–282 CrossRef CAS.
- The Wiswesser Line-Formula Chemical Notation (WLN), ed. E. G. Smith and P. A. Baker, Chemical Information Management, Inc., Cherry Hill, NJ, 1975 Search PubMed.
- D. Weininger, J. Chem. Inf. Model., 1988, 28, 31–36 CrossRef CAS.
- C. Laibe and N. Le Novère, BMC Syst. Biol., 2007, 1, 58 CrossRef.
- A. J. Williams, Drug Discovery Today, 2008, 13, 502–506 CrossRef CAS.
- K. R. Taylor, R. J. Gledhill, J. W. Essex, J. G. Frey, S. W. Harris and D. C. De Roure, J. Chem. Inf. Model., 2006, 46, 939–952 CrossRef CAS.
- S. J. Coles, N. E. Day, P. Murray-Rust, H. S. Rzepa and Y. Zhang, Org. Biomol. Chem., 2005, 3, 1832–1834 CAS.
- S. J. Coles, J. G. Frey, M. B. Hursthouse, M. E. Light, A. J. Milsted, L. A. Carr, D. DeRoure, C. J. Gutteridge, H. R. Mills, K. E. Meacham, M. Surridge, E. Lyon, R. Heery, M. Duke and M. Day, J. Chem. Inf. Model., 2006, 46, 1006–1016 CrossRef CAS.
- P. Murray-Rust and H. S. Rzepa, J. Cheminf., 2011, 3, 44 Search PubMed.
- R. Todeschini and V. Consonni, Molecular Descriptors for Chemoinformatics, 2nd Revised and Enlarged Edition, Series ed. R. Maimund, H. Kubinyi and G. Folkers, Wiley, 2009.
- A. R. Katritzky, V. S. Lobanov and M. Karelson, Chem. Soc. Rev., 1995, 24, 279 RSC.
- L. L. Chepelev and M. Dumontier, J. Cheminf., 2011, 3, 20 CAS.
- J. Downing, M. J. Harvey, P. B. Morgan, P. Murray-Rust, H. S. Rzepa, D. C. Stewart, A. P. Tonge and J. a. Townsend, J. Chem. Inf. Model., 2010, 50, 251–261 CrossRef CAS.
- B. B. Masek, L. Shen, K. M. Smith and R. S. Pearlman, J. Chem. Inf. Model., 2008, 48, 256–261 CrossRef CAS.
- D. Filimonov and V. Poroikov, J. Comput.-Aided Mol. Des., 2005, 19, 705–713 CrossRef CAS.
- L. L. Haak, D. Baker, D. K. Ginther, G. J. Gordon, M. A. Probus, N. Kannankutty and B. A. Weinberg, Science, 2012, 338, 196–197 CrossRef CAS.
- D. J. Wild, Expert Opin. Drug Discovery, 2009, 4, 995–1004 CrossRef CAS.
- M. Hohman, K. Gregory, K. Chibale, P. J. Smith, S. Ekins and B. Bunin, Drug Discovery Today, 2009, 14, 261–270 CrossRef CAS.
- C. R. Groom and F. H. Allen, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1, 368–376 CrossRef CAS.
- R. T. Kouzes, J. D. Myers and W. A. Wulf, IEEE Comput., 1996, 29, 40–46 CrossRef.
- C. Pancerella, J. Hewson, W. Koegler, D. Leahy, M. Lee, L. Rahn, C. Yang, J. D. Myers, B. Didier and R. McCoy, and others, in Proceedings of the 2003 international conference on Dublin Core and metadata applications: supporting communities of discourse and practice—metadata research & applications, Dublin Core Metadata Initiative, 2003, p. 13 Search PubMed.
- J. D. Myers, Cluster Comput., 2005, 5, 253–287 Search PubMed.
- National Research Council, Transforming Combustion Research through Cyberinfrastructure, Washington, DC, The National Academies Press, 2011.
- K. Schuchardt, C. Pancerella, L. A. A. Rahn, B. Didier, D. Kodeboyina, D. Leahy, J. D. D. Myers, O. O. O. Oluwole, W. Pitz and B. Ruscic, and Others, Concurrency and Computation: Practice and Experience, 2007, vol. 19, pp. 1703–1716 Search PubMed.
- E. T. Yu, A. Hawkins, I. D. Kuntz, L. a. Rahn, A. Rothfuss, K. Sale, M. M. Young, C. L. Yang, C. M. Pancerella and D. Fabris, J. Proteome Res., 2008, 7, 4848–4857 CrossRef CAS.
- T. Peachey, E. Mashkina, C.-Y. Lee, C. Enticott, D. Abramson, A. M. Bond, D. Elton, D. J. Gavaghan, G. P. Stevenson and G. F. Kennedy, Philos. Trans. R. Soc. London, Ser. A, 2011, 369, 3336–3352 CrossRef CAS.
- K. L. Schuchardt, B. T. Didier, T. Elsethagen, L. Sun, V. Gurumoorthi, J. Chase, J. Li and T. L. Windus, J. Chem. Inf. Model., 2007, 47, 1045–1052 CrossRef CAS.
- J. Hunter, M. Henderson and I. Khan, J. Chem. Inf. Model., 2007, 47, 2475–2484 CrossRef CAS.
- B. K. Alsberg and A. Clare, J. Chemom., 2010, 24, 408–417 CrossRef CAS.
- A. Llinàs, R. C. Glen and J. M. Goodman, J. Chem. Inf. Model., 2008, 48, 1289–1303 CrossRef.
- A. J. Hopfinger, E. X. Esposito, A. Llinàs, R. C. Glen and J. M. Goodman, J. Chem. Inf. Model., 2009, 49, 1–5 CrossRef CAS.
- R. Guha, M. T. Howard, G. R. Hutchison, P. Murray-rust, H. Rzepa, C. Steinbeck, J. Wegner and E. L. Willighagen, J. Chem. Inf. Model., 2006, 46, 991–998 CrossRef CAS.
- D. P. Martinsen, Science, 2007, 2, 2–5 Search PubMed.
- S. M. Bachrach, J. Cheminf., 2009, 1, 2 Search PubMed.
- N. M. O'Boyle, R. Guha, E. L. Willighagen, S. E. Adams, J. Alvarsson, J.-C. Bradley, I. V. Filippov, R. M. Hanson, M. D. Hanwell, G. R. Hutchison, C. A. James, N. Jeliazkova, A. S. Lang, K. M. Langner, D. C. Lonie, D. M. Lowe, J. Pansanel, D. Pavlov, O. Spjuth, C. Steinbeck, A. L. Tenderholt, K. J. Theisen and P. Murray-Rust, J. Cheminf., 2011, 3, 37 Search PubMed.
- P. Murray-Rust, J. Cheminf., 2011, 3, 48 Search PubMed.
- P. Murray-Rust, Ser. Rev., 2008, 34, 52–64 CrossRef.
- C. L. Palmer, N. M. Weber and M. H. Cragin, Proceedings of the American Society for Information Science and Technology, 2011, 48, 1–10 CrossRef.
- B. Losoff, Issues in Science and Technology Librarianship, 2009 Search PubMed.
- J. A. Evans and J. Reimer, Science, 2009, 323, 1025 CrossRef CAS.
- C. Borgman, Learned Publishing, 2008, 21, 29–38 CrossRef.
- M. Samwald, A. Jentzsch, C. Bouton, C. S. Kallesøe, E. Willighagen, J. Hajagos, M. S. Marshall, E. Prud'hommeaux, O. Hassenzadeh, E. Pichler and S. Stephens, J. Cheminf., 2011, 3, 19 Search PubMed.
- A. J. Williams, L. Harland, P. Groth, S. Pettifer, C. Chichester, E. L. Willighagen, C. T. Evelo, N. Blomberg, G. Ecker, C. Goble and B. Mons, Drug Discovery Today, 2012, 17, 1188–1198 CrossRef.
- B. Chen, X. Dong, D. Jiao, H. Wang, Q. Zhu, Y. Ding and D. J. Wild, BMC Bioinf., 2010, 11, 255 CrossRef.
- C. Steinbeck, Y. Han, S. Kuhn, O. Horlacher, E. Luttmann and E. Willighagen, J. Chem. Inf. Comput. Sci., 2003, 43, 493–500 CrossRef CAS.
- T. Kuhn, E. L. Willighagen, A. Zielesny and C. Steinbeck, BMC Bioinf., 2010, 11, 159 CrossRef.
- S. M. Russell, G. Boulton, P. Clarke, D. Eyton and J. Norton, The Independent Climate Change E-mails Review, 2010.
- J. G. Frey, International Journal of Digital Curation, 2008, 3, 44–62 CrossRef.
- K. W. Boyack, Scientometrics, 2009, 57, 7–60 Search PubMed.
- J. G. Frey, D. De Roure and L. Carr, in Euroweb 2002 the Web and the GRID: from e-Science to e-Business, 2002, pp. 15–17 Search PubMed.
- J. G. Frey, Drug Discovery Today, 2009, 14, 552–561 CrossRef.
- A. Reese, Significance, 2007, 4, 184–186 CrossRef.
- S. Kuhn, T. Helmus, R. J. Lancashire, P. Murray-Rust, H. S. Rzepa, C. Steinbeck and E. L. Willighagen, J. Chem. Inf. Model., 2007, 47, 2015–2034 CrossRef CAS.
- N. Beagrie, J. Chruszcz and B. Lavoie, Keeping Research Data Safe A Cost Model and Guidance for UK Universities, 2008.
- G. L. Holliday, P. Murray-Rust and H. S. Rzepa, J. Chem. Inf. Model., 2006, 46, 145–157 CrossRef CAS.
- L. Moreau, J. Freire, J. Futrelle, R. E. McGrath, J. Myers and P. Paulson, in IPAW 2008, LNCS 5272, 2008, pp. 323–326 Search PubMed.
- M. Borkum, C. Lagoze, J. Frey and S. Coles, in 2010 IEEE Sixth International Conference on e-Science, IEEE, 2010, pp. 316–323.
- T. Margaritopoulos, M. Margaritopoulos, I. Mavridis and A. Manitsaris, in Proc. Int'l Conf. on Dublin Core and Metadata Applications, 2008, pp. 104–113.
- J. Hunter and K. Cheung, Int. J. Digit. Libr., 2007, 7, 99–107 CrossRef.
- P. Murray-Rust and H. S. Rzepa, J. Digit. Inf., 2004, 5 Search PubMed.
- A.-L. Barabási, Linked: The New Science of Networks, Perseus Books Group, Cambridge, MA, 2002 Search PubMed.
- P. M. Davis, J. Am. Soc. Inf. Sci. Technol., 2004, 43, 332–357 Search PubMed.
- C. Brown, J. Am. Soc. Inf. Sci. Technol., 2007, 58, 2055–2065 CrossRef CAS.
- L. Bonetta, Cell, 2009, 139, 452–453 CrossRef CAS.
- Research Information Network, Patterns of Information Use and Exchange: Case Studies of Researchers in the Life Sciences, 2009.
- JISC and British Library, Researchers of Tomorrow: the research behaviour of Generation Y doctoral students Acknowledgements, 2012.
- J. Hur and D. J. Wild, Chem. Cent. J., 2008, 2, 11 CrossRef.
- Y. Zhou, B. Zhou, S. Jiang and F. J. King, J. Chem. Inf. Model., 2010, 50, 47–54 CrossRef CAS.
- P. Murray-Rust and H. S. Rzepa, Internet J. Chem., 2003, 6 CAS.
- D. Banfi and L. Patiny, CHIMIA Int. J. Chem., 2008, 62, 2 CrossRef.
- D. Jiao and D. J. Wild, J. Chem. Inf. Model., 2009, 49, 263–269 CrossRef CAS.
- Z. B. Miled, Y. Liu, D. Powers, O. Bukhres, M. Bem, R. Jones and R. J. Oppelt, J. Chem. Inf. Comput. Sci., 2003, 43, 25–35 CrossRef CAS.
- S. F. Yan, F. J. King, Y. He, J. S. Caldwell and Y. Zhou, J. Chem. Inf. Model., 2006, 46, 2381–2395 CrossRef CAS.
- S. B. Singh, R. D. Hull and E. M. Fluder, J. Chem. Inf. Comput. Sci., 2003, 43, 743–752 CrossRef CAS.
- S. Ranu and A. K. Singh, J. Chem. Inf. Model., 2009, 49, 2537–2550 CrossRef CAS.
- J. Kazius, S. Nijssen, J. Kok, T. Bäck and A. P. Ijzerman, J. Chem. Inf. Model., 2006, 46, 597–605 CrossRef CAS.
- C. L. Bruce, J. L. Melville, S. D. Pickett and J. D. Hirst, J. Chem. Inf. Model., 2007, 47, 219–227 CrossRef CAS.
- P. Gedeck, B. Rohde and C. Bartels, J. Chem. Inf. Model., 2006, 46, 1924–1936 CrossRef CAS.
- A. M. Richard, Chem. Res. Toxicol., 2006, 19, 1257–1262 CrossRef CAS.
- D. Botstein, Mol. Biol. Cell, 2010, 21, 4–6 CrossRef CAS.
- O. Casher and H. S. Rzepa, J. Chem. Inf. Model., 2006, 46, 2396–2411 CrossRef CAS.
- G. V Gkoutos, P. Murray-Rust, H. S. Rzepa and M. Wright, J. Chem. Inf. Comput. Sci., 2001, 41, 1124–1130 CrossRef.
- P. Sefton, J. Downing and N. Day, J. Digit. Inf., 2010, 11, 1–19 Search PubMed.
- P. Murray-Rust and H. S. Rzepa, J. Chem. Inf. Comput. Sci., 2003, 43, 757–772 CrossRef CAS.
- S. Stephens, D. Lavigna, M. Dilascio and J. Luciano, Web Semant.: Sci., Serv. Agents World Wide Web, 2006, 4, 216–221 CrossRef.
- N. Adams, J. Winter, P. Murray-Rust and H. S. Rzepa, J. Chem. Inf. Model., 2008, 48, 2118–2128 CrossRef CAS.
- P. Murray-Rust and H. S. Rzepa, J. Cheminf., 2011, 3, 44 Search PubMed.
- P. Murray-Rust, J. A. Townsend, S. E. Adams, W. Phadungsukanan and J. Thomas, J. Cheminf., 2011, 3, 43 CAS.
- H. Chen, J. Ma, Y. Wang and Z. Wu, Comput. Informat., 2008, 27, 5–20 Search PubMed.
- N. Adams, E. O. Cannon and P. Murray-Rust, in International Conference on Biomedical Ontology, Nature Publishing Group, 2009, vol. 1, p. 2.
- P. Sankar, K. Alain and G. Aghila, J. Chem. Inf. Model., 2010, 50, 755–770 CrossRef CAS.
- E. Muñoz, G. M. Kopanos, A. Espuña and L. Puigjaner, 19th European Symposium on Computer Aided Process Engineering, Elsevier, 2009, vol. 26.
- J. Park, G. R. Rosania and K. Saitou, J. Chem. Inf. Model., 2009, 49, 1993–2001 CrossRef CAS.
- G. V Gkoutos, H. Rzepa, R. M. Clark, O. Adjei and H. Johal, J. Chem. Inf. Comput. Sci., 2003, 43, 1342–1355 CrossRef.
- G. J. Postma, J. Chem. Inf. Model., 1996, 36, 770–785 CrossRef CAS.
- J. Frey, G. Hughes, H. Mills, M. C. Schraefel, G. Smith and D. De Roure, The UK e-Science All Hands Meeting, EPSRC, Nottingham, UK, 2004.
- R. Bramley, K. Chiu, T. Devadithya, N. Gupta, C. Hart, J. C. Huffman, K. Huffman, Y. Ma and D. F. McMullen, J. Chem. Inf. Model., 2006, 46, 1017–1025 CrossRef CAS.
- D. F. McMullen and K. Huffman, Concurrency Comput.: Pract. Exp., 2007, 19, 1621–1631 CrossRef.
Footnotes |
| † Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions, Available from: http://ec.europa.eu/research/horizon2020/index_en.cfm?pg=home&video=none |
| ‡ Isaac Newton, 1676, in a letter to Robert Hooke, although Newton did not originate the notion. |
| § Application Programming Interface. http://en.wikipedia.org/wiki/Application_programming_interface |
| ¶ The Master Chemical Mechanism. http://mcm.leeds.ac.uk/MCM/home.htt |
| || Introduction to the eMinerals project. http://www.eminerals.org/ |
| ** N. Adams, Semantic Chemistry, 2009. http://www.semanticuniverse.com/articles-semantic-chemistry.html |
| †† http://www.ccdc.cam.ac.uk/Solutions/CSDSystem/Pages/CSD.aspx |
| ‡‡ http://www.rcsb.org/pdb/home/home.do |
| §§ B. Cate, Access to Scientific Data: Some Legal Considerations, 2007. http://www.crystalgrid.org/cate.pdf |
| ¶¶ N. Chakravarti, Content management vs. knowledge management, 2008. http://www.knowledgeboard.com/item/2932/23/5/3 |
| |||| The IUPAC International Chemical Identifier (InChI). http://www.iupac.org/inchi/ |
| *** http://www.jcheminf.com/series/InChI |
| ††† Standard InChI-based Representation of Chemical Reactions. http://iupac.org/nc/home/projects/project-db/project-details.html?tx_wfqbe_pi1[project_nr]=2009-043-2-800 |
| ‡‡‡ Uniform Resource Identifier (URI). The W3C provides a range of materials relating to naming and addressing, for which a starting point is at http://www.w3.org/Addressing/ |
| §§§ ChemSpider. http://www.chemspider.com/ |
| ¶¶¶ eCrystals. http://ecrystals.chem.soton.ac.uk/ |
| |||||| E. Willighagen, InChI's in CML, 2006. http://cmlexplained.blogspot.co.uk/2006/09/inchis-in-cml.html |
| **** M. Dehmer, K. Varmuza, D. Bonchev. Statistical Modelling of Molecular Descriptors in QSAR/QSPR, volume 2. DOI: 10.1002/9783527645121 |
| †††† PubChem. http://pubchem.ncbi.nlm.nih.gov/ |
| ‡‡‡‡ Dial-a-Molecule Grand Challenge. http://www.dial-a-molecule.org/ |
| §§§§ ChemSpider. http://www.chemspider.com/ |
| ¶¶¶¶ J. C. Bradley et al. UsefulChem. http://usefulchem.wikispaces.com/ |
| |||||||| M. Nielsen, Opening Science, 2012. http://openingscience.org/post/17877811625 |
| ***** OpenTox. http://www.opentox.org |
| ††††† J. C. Bradley, Useful Chemistry Open Notebook Science. http://usefulchem.blogspot.com/ |
| ‡‡‡‡‡ C. Neyon, Cameron's LaBLog. http://biolab.isis.rl.ac.uk/camerons_labblog |
| §§§§§ M. Todd et al., Pictet–Spengler route to Praziquantel. http://www.ourexperiment.org/racemic_pzq |
| ¶¶¶¶¶ K. W. Boyack, Articles on Science Mapping, Accuracy, and Metrics. http://mapofscience.com/publication.html |
| |||||||||| JCAMP-DX. http://www.jcamp-dx.org/ |
| ****** Digital Curation Centre, Overview of funders' data policies. http://www.dcc.ac.uk/resources/policy-and-legal/overview-funders-data-policies |
| †††††† N. Beagrie et al., Keeping research data safe (Phase 2), 2010. http://www.jisc.ac.uk/publications/reports/2010/keepingresearchdatasafe2.aspx |
| ‡‡‡‡‡‡ Digital Curation Centre, What is digital curation? http://www.dcc.ac.uk/digital-curation/what-digital-curation |
| §§§§§§ Research Information Network, Patterns of information use and exchange: case studies of researchers in the life sciences. http://www.rin.ac.uk/our-work/data-management-and-curation/ |
| ¶¶¶¶¶¶ N. Beagrie et al., Keeping research data safe (Phase 2), 2010. http://www.jisc.ac.uk/publications/reports/2010/keepingresearchdatasafe2.aspx |
| |||||||||||| G. Siemens, Connectivism: A Learning Theory for the Digital Age, 2004. http://www.elearnspace.org/Articles/connectivism.htm |
| ******* JISC, Managing Research Data Programme (MRD) 2011–13. http://www.jisc.ac.uk/whatwedo/programmes/di_researchmanagement/managingresearchdata.aspx |
| ††††††† PubChem. http://pubchem.ncbi.nlm.nih.gov/ |
| ‡‡‡‡‡‡‡ Europa Press Release, Scientific data: open access to research results will boost Europe's innovation capacity, 2012. http://europa.eu/rapid/pressReleasesAction.do?reference=IP/12/790_en.htm |
| §§§§§§§ http://deposit.rcsb.org/depoinfo/PDB_deposition_release_policies.html |
| ¶¶¶¶¶¶¶ Pistoia Alliance. http://www.pistoiaalliance.org/ |
| |||||||||||||| Allotrope Foundation. http://www.allotrope.org |
| ******** N. Adams, Semantic Chemistry, 2009. http://www.semanticuniverse.com/articles-semantic-chemistry.html |
| †††††††† Chemical Entities of Biological Interest (ChEBI). http://www.ebi.ac.uk/chebi/ |
| ‡‡‡‡‡‡‡‡ T. Bhat, Chemical Taxonomies and Ontologies for Semantic Web, 2010. http://www.semanticuniverse.com/articles-chemical-taxonomies-and-ontologies-semantic-web.html |
| §§§§§§§§ L. Hawizy, The Semantification of Chemistry, 2010. http://www.semanticuniverse.com/articles-semantification-chemistry.html |
| ¶¶¶¶¶¶¶¶ S. Coles and J. Frey. The Relevance of Linking. Monograph. Available from: http://repository.jisc.ac.uk/419/ |
| |||||||||||||||| C. Samson. The burgeoning needs of biology. http://www.scientific-computing.com/features/feature.php?feature_id=147 |
| ********* Reproducibility Initiative. https://www.scienceexchange.com/reproducibility |
| ††††††††† Organic Syntheses. http://www.orgsyn.org/ |
| This journal is © The Royal Society of Chemistry 2013 |
