 Open Access Article
Open Access ArticleSelective and ATP-driven transport of ions across supported membranes into nanoporous carriers using gramicidin A and ATP synthase†
Vitaliy
Oliynyk‡
a,
Christian
Mille‡
ab,
Jovice B. S.
Ng
a,
Christoph
von Ballmoos
c,
Robert W.
Corkery
bd and
Lennart
Bergström
*a
aDepartment of Materials and Environmental Chemistry, Stockholm University, 10691 Stockholm, Sweden. E-mail: lennart.bergstrom@mmk.su.se
bYKI, Institute for Surface Chemistry, 11486, Stockholm, Sweden
cDepartment of Biochemistry and Biophysics, Stockholm University, 10691 Stockholm, Sweden
dDepartment of Chemistry, Division of Physical Chemistry, KTH Royal Institute of Technology, Stockholm, Sweden
First published on 4th January 2013
Abstract
We report a robust and versatile membrane protein based system for selective uptake and release of ions from nanoporous particles sealed with ion-tight lipid bilayers of various compositions that is driven by the addition of ATP or a chemical potential gradient. We have successfully incorporated both a passive ion channel-type peptide (gramicidin A) and a more complex primary sodium ion transporter (ATP synthase) into the supported lipid bilayers on solid nanoporous silica particles. Protein-mediated controlled release/uptake of sodium ions across the ion-tight lipid bilayer seal from or into the nanoporous silica carrier was imaged in real time using a confocal laser scanning microscope and the intensity changes were quantified. ATP-driven transport of sodium ions across the supported lipid bilayer against a chemical gradient was demonstrated. The possibility of designing durable carriers with tight lipid membranes, containing membrane proteins for selective ion uptake and release, offers new possibilities for functional studies of single or cascading membrane protein systems and could also be used as biomimetic microreactors for controlled synthesis of inorganic multicomponent materials.
Introduction
Transport across biological membranes is mediated by membrane proteins and underpins basic functions of the living cells such as energy transduction, sensory perception, cell adhesion and movement, and cell recognition.1–3 The controlled uptake of specific ions is, together with proteins, an integral part of biomineralization processes in living organisms.4,5 The use of lipid vesicles to study various types of cell membrane events in vitro is wide-spread6–9 but limited by inherent instabilities.10,11 Supporting the lipid bilayers on planar12–14 or curved15,16 (particulate) solid substrates offers enhanced stability and has been the most widely used experimental cell surface model.Silicon dioxide has been shown to be a useful surface for fusion of liposomes and the subsequent formation of supported lipid bilayers.17 Between these substrates and the bilayer, several monolayers of water are located – making it a suitable environment for incorporated proteins that can freely diffuse in the membrane.18,19 Porous silica spheres are particularly attractive because of their endoskeleton-like structure, tunable internal pores and surfaces.20–24 Recent works have shown that nanoporous silica spheres can be coated with lipid bilayers25–28 and used as delivery vehicles into cells.29,30 A common approach to functionalize supported membranes with transmembrane proteins is to spread vesicles incorporating proteins (proteoliposomes) onto supports.
In a first step towards the construction of systems that can also control the uptake of specific ions, proton pumps have been incorporated into lipid bilayers supported onto porous silica spheres.23,30 Previous work showed that it was possible to form a supported lipid bilayer containing a transmembrane redox-driven proton pump, Cytochrome c Oxidase (CytcO), by fusion of small unilamellar vesicles with reconstituted CytcO onto the surface of colloidal nanoporous silica particles.30 It was demonstrated that the CytcO retains its functionality while the supported membrane system was shown to be proton tight, defining an interior particle compartment that is separated from the surrounding aqueous media. Further, studies have been performed where ATP synthase and bacteriorhodopsin were used in biomimetic scaffolds (sol–gel) and artificial organelles (polymersomes), functioning as support for proton transport coupled to the synthesis of ATP.29,30 Indeed, simple compartmentalized modules with partial cellular functions may also give us insight into early or minimal life forms (protocells).31
In this study, we go beyond proton transporters and demonstrate uptake and release of sodium ions across a supported membrane using gramicidin A and ATP synthase, a peptide and a multi-subunit transmembrane protein, respectively, which utilize different transport mechanisms. Gramicidin A is a dimer that assembles from two monomers of 15 amino acids each, and forms a cationic-selective channel of 4 Å that spans the lipid bilayer.32 Passive ion transport through the channel is driven by either an ion gradient or an electrical membrane potential. This peptide has been extensively used in biomembrane research due to its formation of stable channels in supported lipid bilayers that have different affinities for monovalent cations. Gramicidin A is used by soil bacteria as an antibiotic, killing gram-negative bacteria.33 Na+-dependent F1F0 ATP synthase, on the other hand, is a primary sodium ion transporter, that – when supplied with energy in the form of ATP – moves sodium ions across the membrane.34
Here, we report the successful incorporation of both gramicidin A and the more complex ATP synthase into lipid bilayers, deposited on synthetic, nanoporous silica spheres. For gramicidin A, the functional incorporation was confirmed by studying the passive transport of monovalent cations from the solution into the particle. In the case of ATP synthase, the addition of ATP drives the transport of ions across the membrane. The activity of both the passive and the active transport was followed in real time using dye loaded particles imaged using confocal laser scanning microscopy.
Experimental section
Materials
Positively charged DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) or neutral (zwitterionic) DLPC lipids (1,2-dilauroyl-sn-glycero-3-phosphatidylcholine) and soy PC (L-α-phosphatidylcholine) were purchased from Avanti Polar Lipids (Alabaster, AL). Texas Red® 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine and triethylammonium salt (Texas Red® DHPE) lipid were purchased from Molecular Probes®. Gramicidin from Bacillus aneurenolyticus as a powder, adenosine 5′-triphosphate disodium salt hydrate (Na2-ATP), Rhodamine 6G and fluorescein were purchased from Sigma-Aldrich. The dye Oregon Green 488 and the Sodium Green™ (SG) tetra(tetramethylammonium) salt were purchased from Molecular Probes Europe BV. The spherical, nanoporous silica (SNS) particles were produced by an aerosol-assisted technique using a vibrating-orifice aerosol generator following previously established procedures.35,36 The templating amphiphilic polymer, Brij 56, was removed by calcination, which yields monodisperse, hydrophilic, nanoporous silica spheres with a particle diameter of 6 μm, characterized by a interconnected network of pores with a diameter of 2.7 nm and a specific surface area of 778 m2 g−1. Details of synthesis and characterization are available in Fig. S1 (ESI†).Dye loading of nanoporous particles
The SNS particles (10–15 mg) were loaded with dyes, Rhodamine 6G, Oregon Green 488, and fluorescein, through incubation (at least 8 h) in 300 μl of a PBS buffer (pH 7.4) or an aqueous solution of the selected dye. The final dye concentrations in the particles are 0.01–0.05 mM. Sodium Green™ (SG) was used to monitor Na+-uptake across the lipid membrane. It was loaded into SNS particles at a concentration of 10 μM. Dye loading and lipid sealing were performed in 100 mM KCl (aq.). Unincorporated dye was removed through repeated centrifugation of the particles in a PBS buffer or 100 mM KCl at least 3 times.Preparation of DOTAP and soy PC vesicles
Dispersion of multilamellar vesicles (MLV) was prepared by dissolving the lipids in Dulbecco's phosphate buffered saline solutions (PBS, 135 mM, pH 7.2, Sigma) to a final concentration of 2.5 mg ml−1.Preparation of DLPC-gramicidin A vesicles
Gramicidin A, DLPC and in some experiments DHPE-Texas Red were first separately dissolved in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of chloroform and methanol, mixed together, dried under a weak flow of nitrogen gas, and placed under vacuum overnight. The dried peptide–lipid mixtures (containing 1 mol% gramicidin A and 0–3% Texas red tagged lipids) were dispersed in Milli-Q water, in Dulbecco's phosphate buffered saline solutions (PBS, 135 mM, pH 7.2, Sigma), or in 100 mM KCl, to a final concentration of 2.5 mg ml−1, and homogenized by vortexing. Small unilamellar vesicles (SUVs) were prepared from the respective MLV dispersions by sonication in an ultrasound bath until the solution became transparent.
1 mixture of chloroform and methanol, mixed together, dried under a weak flow of nitrogen gas, and placed under vacuum overnight. The dried peptide–lipid mixtures (containing 1 mol% gramicidin A and 0–3% Texas red tagged lipids) were dispersed in Milli-Q water, in Dulbecco's phosphate buffered saline solutions (PBS, 135 mM, pH 7.2, Sigma), or in 100 mM KCl, to a final concentration of 2.5 mg ml−1, and homogenized by vortexing. Small unilamellar vesicles (SUVs) were prepared from the respective MLV dispersions by sonication in an ultrasound bath until the solution became transparent.
ATP synthase purification and reconstitution
The protein purification and membrane reconstitution were done as described previously.37 In short, 5 g of E. coli DK8 cells38 containing the heterogeneously expressed I. tartaricus ATP synthase was resuspended in 25 ml French press buffer II (200 mM Tris–HCl pH 7.8, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2.5% glycerol, 0.1 mM diisopropylfluorophosphate, 50 μg DNase I restriction enzyme) and disrupted in a French pressure cell followed by centrifugation for 10 min at 8000 g. The membranes were isolated by centrifugation at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 45 min and the pellets were solubilized in 20 mL extraction buffer (50 mM Tris–HCl pH 7.5, 100 mM KCl, 250 mM sucrose, 40 mM aminocaproic acid, 15 mM p-aminobenzamidine, 5 mM MgCl2, 0.1 mM EDTA, 0.2 mM dithiothreitol, 0.8% soy PC 1.5% octyl glucoside, 0.5% sodium cholate, 0.5% sodium deoxycholate, 0.3% soy PC, 2.5% glycerol and 30 mM imidazole). The mixture was stirred for 90 minutes at 4 °C and centrifuged at 200 000 g for 45 min. The supernatant containing the solubilized fraction was sterile filtered and loaded on a HisTrap HP 1 ml column (GE Healthcare). The fractions containing active ATP synthase were pooled and stored in liquid nitrogen.
000 g for 45 min and the pellets were solubilized in 20 mL extraction buffer (50 mM Tris–HCl pH 7.5, 100 mM KCl, 250 mM sucrose, 40 mM aminocaproic acid, 15 mM p-aminobenzamidine, 5 mM MgCl2, 0.1 mM EDTA, 0.2 mM dithiothreitol, 0.8% soy PC 1.5% octyl glucoside, 0.5% sodium cholate, 0.5% sodium deoxycholate, 0.3% soy PC, 2.5% glycerol and 30 mM imidazole). The mixture was stirred for 90 minutes at 4 °C and centrifuged at 200 000 g for 45 min. The supernatant containing the solubilized fraction was sterile filtered and loaded on a HisTrap HP 1 ml column (GE Healthcare). The fractions containing active ATP synthase were pooled and stored in liquid nitrogen.
The ATP synthase was then reconstituted into SUVs. The SUVs were prepared by dissolving 20 mg soy PC in 2 ml liposome buffer (20 mM Tris–phosphate, pH 7, 5 mM MgCl2) followed by sonication for 3 × 30 s on ice using a tip-sonicator (MSE Soniprep 150, Labtec AG, Wohlen, Switzerland). Sodium cholate (1% final concentration) and purified ATP synthase in a lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein ratio 200
protein ratio 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) were added to the suspension of SUVs. The protein/lipid solution was kept at 4 °C for 30 min followed by loading on a PD-10 gel filtration column (GE Healthcare), pre-equilibrated with liposome buffer. The final concentration of ATP synthase was approximately 0.3 ATP synthase/vesicle (i.e. two out of three liposomes are empty, not containing any enzyme).
1 (w/w) were added to the suspension of SUVs. The protein/lipid solution was kept at 4 °C for 30 min followed by loading on a PD-10 gel filtration column (GE Healthcare), pre-equilibrated with liposome buffer. The final concentration of ATP synthase was approximately 0.3 ATP synthase/vesicle (i.e. two out of three liposomes are empty, not containing any enzyme).
ATP synthesis measurement
The enzymatic activity of the reconstituted ATP synthase was verified as described previously.37 In short, the proteoliposomes were loaded with NaCl (50 mM) and KCl (0.5 mM) during the reconstitution process. The proteoliposomes were then dispersed into a buffer containing 1 mM NaCl and 100 mM KCl (together with the chemicals for luminescence detection of ATP). The addition of valinomycin causes K+ to flow into the proteoliposomes according to its concentration gradient, creating an electrical Nernst potential across the membrane (where inside is positive).Preparing supported bilayers onto spherical nanoporous silica particles
Coating the SNS particles with lipid bilayers with or without membrane proteins was done through direct fusion of small unilamellar lipid vesicles (SUVs). Freshly prepared lipid SUVs were incubated with the SNS particle suspension for ∼1 hour and mixed during incubation by in–out pipetting followed by washing 3 times. Excess SUVs were removed by 3 times centrifugation with buffer exchange. The pellet collected after removing the supernatant was vortexed in 100 mM KCl, or in Millipore-grade water to resuspend the coated particles.Confocal laser scanning microscope (CLSM) imaging
The CLSM experiments for the lipid bilayer coated SNS with fluorescent dyes encapsulated in the pores of the silica particles were performed in either Dulbecco's phosphate buffered saline solutions (Sigma-Aldrich, PBS, 135 mM, pH 7.2), 100 mM KCl, or in Millipore-grade water. An inverted Zeiss Axiovert Observer Z1 microscope equipped with an LSM 5 Exciter scanner was used for CLSM imaging. The Texas Red-tagged lipid and Rhodamine 6G-containing SNS particles were imaged using a 543 nm HeNe laser together with a long-pass 560 nm filter. The Oregon Green 488, fluorescein or Sodium Green™-containing SNS particles were imaged using a 488 nm argon laser together with a long-pass 505 nm filter. A 63 × 1.4 NA oil-immersion objective lens was used for all imaging. The image profiles were stored as eight- or twelve-bit line scans with a resolution of 512 pixels × 512 pixels representing an area of 146.2 μm × 146.2 μm. Control experiments were conducted to evaluate the photobleaching of the dyes and the light attenuation during the experiment. (See ESI† for details). Unless otherwise stated, specimens were prepared for imaging by placing a drop (50–200 μl) of a dispersion containing SNS particles onto a glass-bottom microwell dish (35 mm Petri dish, 14 mm microwell, Mat-Tek Corporation, USA). The microwell dish was then covered with a cap to avoid liquid evaporation. The dye-containing mesoporous spheres were allowed to equilibrate for 5–15 min prior to CLSM imaging. The dye-containing or fluorescently labelled lipid coated spheres were optically sectioned into 10–20 slices of equal thinness separated by approximately 1 μm, using z-axis scans. The slice with the largest diameter (nearly identical to the diameter of the sphere) is defined as the equatorial slice. In experiments where the intensity of the fluorescence was monitored over extended time (over 2 hours), the pinhole was fully opened to minimize photobleaching.Na+ transport across gramicidin containing lipid bilayers
The SNS particles were loaded with SG and sealed with gramicidin A-containing DLPC lipid bilayers. Transport of Na+ was triggered by increasing the bulk Na+ concentration through media exchange with 100 mM NaCl as described above.Na+ transport across ATP synthase containing supported lipid bilayers
SG-loaded SNS particles were sealed with ATP synthase proteoliposomes. ATP synthase-mediated transport of Na+ was triggered by adding Na2ATP of known concentration (final concentration of 0.4 mM). A control study was performed by buffer exchange with 4.0 mM NaCl to check for Na+ permeability through the ATP synthase-containing soy PC lipid bilayers. Additional control experiments were conducted using SG-loaded SNS particles sealed by soy PC-only bilayers and adding 8 mM NaCl solution. Changes in the fluorescence intensity as a result of interactions of the transported Na+ ions with the SG dye molecules loaded in the SNS particles were followed over time (40 s, 1 s interval).Ionic quenching of fluorescein by iodide
Mesoporous silica particles were incubated overnight in a 1 mM fluorescein solution in 100 mM KCl. Excess dye was removed by repeated centrifugation and supernatant exchange. The pellet collected after removing the supernatant was divided into two fractions. One fraction of the fluorescein-loaded particles was kept as it was, while the second fraction was coated using gramicidin A-containing DLPC proteoliposomes, followed by centrifugation to remove excess SUVs. The resulting pellets obtained from both fractions were separately resuspended in 50 μl of 100 mM KCl. 20 μL of the resuspended dye-containing particles (with and without gramicidin A-containing DLPC bilayer coatings) were deposited on a glass-bottom Petri dish and 1.7 ml Millipore water was added to each. The particles were allowed to equilibrate for 10–15 minutes prior to imaging. Images of the equatorial slice of the sphere were recorded before and after addition of 0.3 ml of freshly prepared 2 M KI. Time series images were recorded at either 5 or 10 minute intervals for up to 60 minutes. All images were recorded using the same experimental conditions and microscope settings.Results and discussion
Creating ion-tight membranes by supported lipid bilayers onto silica nanoporous spheres
The silica nanoporous spheres (SNS) were coated with bilayers of different types of lipids following a well-established vesicles fusion technique.39,40 By adding 1–3% of Texas Red-tagged lipids into the lipid bilayer it was possible to directly image the lipid-coated spheres by confocal laser scanning microscopy (CLSM). Fig. 1 shows that the liposome fusion technique produces continuous bilayers on the outer surfaces of the SNS particles for both the positively charged DOTAP lipids and for the zwitterionic DLPC lipids.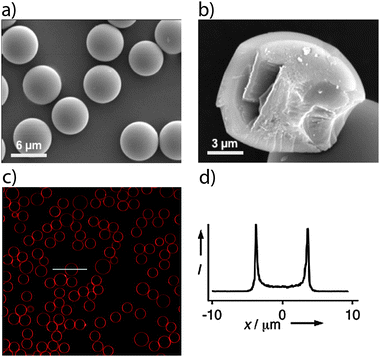 | ||
| Fig. 1 Characterization of lipid bilayer coated nanoporous silica spheres. SEM images of the (a) monodisperse nanoporous silica (SNS) spheres and (b) their internal fractured surface that shows that the SNS is solid without a hollow core. (c) CLSM image of the SNS spheres sealed with DOTAP lipid bilayers containing 1–3% Texas Red® DHPE. (d) Intensity graph obtained along the white horizontal line representing the size of the particle. The CLSM images were obtained 7 days after sealing and subsequent storage at 5 °C. | ||
About 1–3% of Texas Red® DHPE lipid were mixed with the lipids to facilitate fluorescence imaging of the lipid coating. CLSM images obtained immediately after coating (not shown) and after 7 days of storage at +5 °C (Fig. 1c and d) show that the supported lipid bilayers (red rings) exhibit long-term stability. The relatively small pore size of the SNS used in this study is well below the critical pore size of 10 nm that was shown by Davis et al.23 to be the upper limit for the formation of stable lipid bilayers onto the outer surface of mesoporous silica particles. The durability and permeability of the supported-DOTAP or -DLPC lipid bilayers were evaluated by following the diffusion of fluorescent dyes that were preloaded into the SNS particles prior to lipid bilayer sealing.
Fig. 2 shows that the intensity of fluorescein inside the lipid bilayer-sealed particles remains virtually unchanged while the intensity of the uncoated SNS drops dramatically over time. This observed drop in fluorescence intensity is consistent with the results obtained in a previous study where small negatively charged dye molecules were released from uncoated mesoporous spheres within 5–7 hours.36 Similar experiments on negatively charged Oregon Green and positively charged Rhodamine 6G show that DOTAP and DLPC supported lipid bilayers are able to effectively seal the molecules inside the porous carriers.
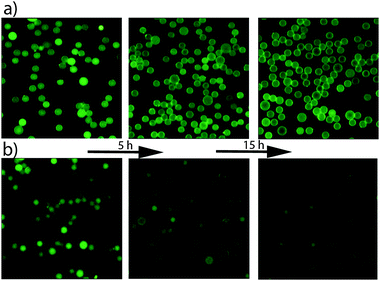 | ||
| Fig. 2 Time evolution of the release of fluorescein from; (a) SNS particles sealed with DLPC lipid bilayers; (b) unsealed SNS. | ||
Na+ uptake and release through gramicidin A channels driven by concentration gradients
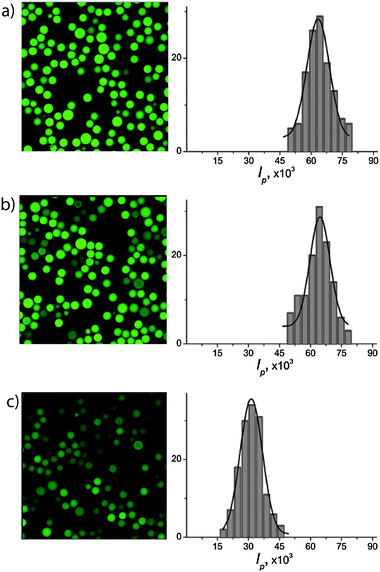
We incorporated 1 mol% of gramicidin A into DLPC liposomes (tagged with 1–3% Texas Red-labeled lipid) and subsequently deposited these liposomes as lipid bilayer coatings on the SNS particles (a) via a liposome fusion procedure.30 In order to directly observe membrane protein-mediated transport of Na+ using CLSM, we had loaded the SNS particles (prior to lipid coating) with Sodium Green™ (SG), a Na+-sensitive dye that increases in fluorescence upon interaction with Na+. The SG provides spatial and temporal resolution of the Na+ concentrations with sufficient selectivity in the presence of other monovalent cations exhibiting e.g. a 41-fold higher affinity for binding Na+versus K+ and thus displays much higher quantum yield in Na+-containing solutions.39Fig. 3b shows a CLSM image of the membrane protein-containing lipid bilayer coated porous particles where the SG-loaded interior sphere (green) and the Texas Red-labeled lipid bilayers coating (red ring) are clearly distinguished. The CLSM images in c show that SNS particles sealed with DLPC lipid bilayers, containing 1 mol% of gramicidin A, are only weakly fluorescent (low quantum yield). Exchange of the external media from 100 mM KCl to 100 mM NaCl results in a significant increase in fluorescence intensity of the particles (c, compare with Fig. 3e) suggesting that Na+ is transported into the porous spheres. Integration of the fluorescence intensities inside the particles before (f) and after (d) the increase of the bulk Na+ concentration shows that the average intensity increases by a factor of two. To show that gramicidin A-containing DLPC-bilayers are defect free and do not allow unrestricted passage of ions, an ionic quenching test of fluorescein by iodide was performed. Iodide is known to be an efficient quencher of fluorescence for fluorescein while it does not penetrate lipid bilayers in the fluid phase.41
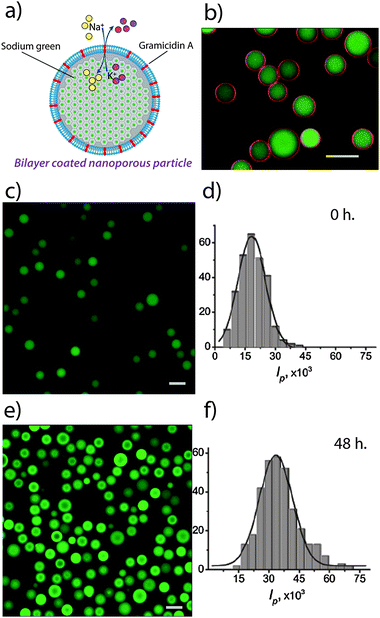 | ||
| Fig. 3 Gramicidin-mediated uptake of Na+ into lipid bilayer-coated particles. (a) Schematic illustration. (b) A two-channel CLSM image of fluorescein-loaded particles sealed with a fluorescently labeled lipid bilayers containing 1 mol% of gramicidin A, (c) and (e) are the CLSM images of SG-loaded SNS particles (sealed with DLPC lipid bilayers containing 1 mol% of gramicidin A) with and without the addition of Na+ respectively, (d) and (f) are the respective histograms of the integral intensities per particle measured from the CLSM images. The data in (e) and (f) were obtained 48 hours after addition of Na+. Scale bars are 10 μm. | ||
We find that the fluorescence of the uncoated fluorescein-loaded SNS particles (Fig. 4a) is effectively quenched after addition of I− (b). The particles sealed with DLPC-gramicidin A bilayers (c) on the other hand did not show observable fluorescence quenching one hour after I− addition (d) indicating an ion-tight membrane and a Na+ selectivity for gramicidin A. This shows that the transport of Na+ into the DLPC-coated SNS illustrated in Fig. 3c is dominated by transport through the gramicidin A channels and that the transport through possible defects in the bilayer is insignificant. As a reference for unrestricted uptake, we find that adding 10 mM NaCl to unsealed SNS loaded with 10 μM of SG results in a 3-fold increase in the intensity (see Fig. S2, ESI†). In contrast, the fluorescence remained unchanged when a solution with the same concentration of Na+ was added to SG-loaded SNS coated with a pure DLPC lipid bilayer (no gramicidin) (see Fig. S3, ESI†). This process can also be reversed, i.e. DLPC-coated SNS preloaded with SG in Na+ buffer can release the Na+ ions through gramicidin A channels into a K+-containing (but Na+ free) buffer (Fig. 5).
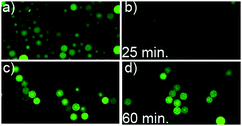 | ||
| Fig. 4 Iodide quenching of fluorescein-loaded silica nanoporous spheres. Representative CLSM images of the fluorescein-loaded mesoporous silica particles; with coatings of gramicidin A-containing (1 mol%) DLPC lipid bilayers (c and d), and without any coating (a and b). Images recorded before (a and c) and after (b and d) addition of I− to the bulk media (final I− concentration 300 mM). The time indicated is the time after addition of I. | ||
 | ||
| Fig. 5 Gramicidin-mediated release of Na+ from lipid bilayer-coated particles. CLSM images of monodisperse nanoporous silica spheres loaded with SG and 100 mM of Na+, and sealed with DLPC lipid bilayers containing 1 mol% of gramicidin A. The left panel shows a representative CLSM image and the histograms to the right show the integral intensities per particle (a) before, (b) after exchange of the bulk solution with Na+-containing fresh buffer (control sample) and (c) after exchange of the Na+-based with a K+-based bulk solution. Images in panels (b) and (c) were recorded 20 h after buffer exchange. The significant decrease in the fluorescence intensity after the addition of the K+-containing bulk solution in (c) shows that there is a significant uptake of potassium and release of sodium ions into/from the gramicidin containing lipid bilayer coated SNS. At the same time, unchanged fluorescence of the control sample (b) after 20 h shows that there are no major leaks. | ||
The fluorescence intensity after sodium uptake varies between individual particles. This is not surprising considering a spatial variation in the concentration and/or activity of the bilayer-bound gramicidin, variations in the pore volume and pore structure, both within and between particles, which is a known characteristic of porous spheres synthesized using the aerosol-assisted process,35 or by the different concentrations of the correctly oriented protein molecules in the lipid membrane of different particles.26
Na+ transport through ATP synthase channels driven by ATP
ATP synthase (Fig. 6) is an enzyme from the marine bacterium Ilyobacter tartaricus that converts the electrochemical energy of the sodium gradient across the membrane to catalyze the reaction of ADP to ATP.42 The ATP synthase can be viewed as a miniature engine composed of two opposing rotary motors, F1 and F0, which are connected in series. The F1 represents the water soluble head of the complex that utilizes the torque generated by the F0 motor to synthesize ATP from ADP by sequential conformational changes in the nucleotide binding sites.34 When the concentration of the ATP is high compared to the Na+ gradient, ATP synthase runs in the reverse direction and hydrolyses ATP to ADP and pumps Na+ ions across the membrane, thus working as a primary sodium pump.43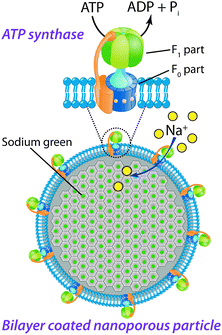 | ||
| Fig. 6 Schematic illustration of the ATP synthase incorporated into a lipid bilayer that covers a nanoporous particle loaded with SG. The ATP synthase comprises a membrane soluble F0 part that uses the Na+ gradient to rotate the F0 which subsequently generates a toque in the F1 part through a mechanical coupling between them. This conformational change results in a catalytic site for the reaction of ATP from ADP. In the reverse ATP hydrolysis mode, ATP is instead used up and ions are actively transported through the channel. | ||
Purified ATP synthase from I. tartaricus, heterogeneously expressed in E. coli was reconstituted in proteoliposomes. Functional reconstitution was verified by ATP synthesis measurements driven by a K+/valinomycin induced diffusion potential and a Na+-gradient (Fig. 7). The Na+ gradient is established by dispersing the proteoliposomes (loaded with 50 mM NaCl) into a solution containing 1 mM NaCl, while the electrical potential (inside positive) is created by valinomycin mediated K+ influx into the liposomes. Together, they build up a potential of approximately 230 mV, which is sufficient to initiate ATP synthesis.44 With time, the K+ concentration inside the liposomes rapidly increases and the electrical potential, and thus the ATP synthesis rate decreases.
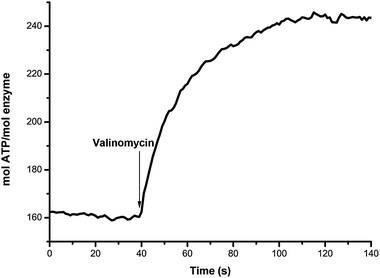 | ||
| Fig. 7 Time dependent activity of the ATP synthase reconstituted in proteoliposomes. The synthesis of ATP by transportation of Na+ ions is measured after applying a K+/valinomycin induced diffusion potential and a Na+-gradient. | ||
The SNS particles were preloaded with SG before sealing with soy PC bilayers containing ATP synthase through liposomal fusion of reconstituted proteoliposomes. We have evaluated transmembrane transport of Na+ into membrane-sealed SNS particles by subjecting the particles to sodium ion solutions, and compared the uptake in the absence and the presence of ATP. Fig. 8 shows the change in fluorescence intensity (normalized to the intensity before adding Na2ATP or NaCl) over time for a selected number of particles. The stable fluorescence intensity in the control experiment with soy PC bilayer coated particles (without ATP synthase, Fig. 8a) shows that the lipid bilayer coating itself is ion-tight. Fig. 8b shows that the SG-loaded SNS particles sealed with soy PC bilayers that contain ATP synthase display a rapid increase in the fluorescence intensity of up to 60% when 0.4 mM Na2ATP is added to the bulk solution. The result shows a spread in the maximum uptake that is likely to be related to varying number of ATP synthase molecules per particle. A much smaller (∼20% increase in fluorescence intensity) uptake is also observed when the system is supplied with 4 mM NaCl in the absence of ATP (Fig. 8c). Hence, while the transport of Na+ into the lipid bilayer-coated SNS is dominated by ATP-driven transport, there is also a smaller passive Na+-influx in the absence of ATP. The rapid saturation of the intensity increase suggests that the passive transport is occurring through defective ATP synthases that lack the F1 head group after reconstitution, i.e. functioning as a passive channel that will transport Na+ ions due to the chemical gradient (4 mM outside, <200 μM inside). Indeed, dissociation of F1 from F0 leaving behind the two parts as separate functional entities, during reconstitution, has been reported previously.44
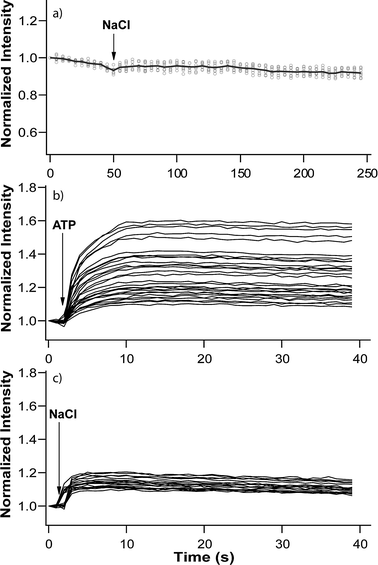 | ||
| Fig. 8 ATP synthase-mediated transport of Na+ into lipid bilayer-coated particles. Normalized intensity of fluorescence of SG-loaded porous silica spheres sealed by (a) only a soy PC bilayer after addition of 8 mM NaCl. Normalized intensity of fluorescence of SG-loaded porous silica spheres sealed by asoy PC bilayer containing 1 mol% ATP synthase after addition of (b) 0.4 mM Na2ATP, and (c) 4 mM NaCl. Specimens used in (b) and (c) were from the same batch and experimental conditions were kept the same. The first graph contains data for 5 spheres and the last 2 graphs contain data for 30 spheres. | ||
This versatile system allows for a solid and tunable platform for the utilization of large transmembrane proteins for both functional studies and biomimetic synthesis. It could for example be used with several different proteins, creating a chain reaction which further mimics the complex cascade reaction occurring in living cells. Also, it is envisaged that combinations of different ion transporters incorporated into supported lipid bilayers onto porous carriers could be used as biomimetic microreactors for controlled synthesis of inorganic multicomponent materials. A further uniqueness is that the membranes are supported by nanoporous particles, functioning as biomimetic proxies for cellular endoskeletons, which lends physical strength, dimensional stability and internal subdivision of space to these “protocells”.
Conclusion
In summary, we have shown that liposome fusion of cationic lipid DOTAP, zwitterionic phosphocholine lipid DLPC and soy PC lipid onto rigid, nanoporous silica spheres results in the formation of durable, ion-tight coatings that separate the water-filled interior of the porous endoskeleton from the bulk solution. We have incorporated two different types of biologically active molecules, the simple and well-studied peptide gramicidin A and the more complex primary sodium pump ATP synthase, into lipid bilayers through liposomal fusion of small unilamellar vesicles onto the nanoporous silica spheres. Using a confocal laser scanning microscope and a sodium sensitive dye, Sodium Green, we have successfully followed and imaged in real time chemical signal-triggered gramicidin A uptake and release and ATP synthase-mediated transmembrane transport of Na+ into the lipid bilayer coated porous carriers.The possibility of designing “protocells” with tunable porous endoskeletons offers a physically robust and biochemically versatile system, suitable for functional studies of complex membrane protein systems. Future prospects also include the exciting possibilities for biomimetic synthesis where membrane proteins control the transport of the ionic reactants into a nanoporous microreactor.
Acknowledgements
This work is supported by the Swedish Science Council (VR) and the Institute Excellence Centre CODIRECT, hosted by YKI and sponsored by Vinnova (the Swedish Government Agency for Innovation Systems), KK-stiftelsen (the Knowledge Foundation) and SSF (the Swedish Foundation for Strategic Research) and Industry. CvB is supported by a fellowship from the Swiss Science Foundation (SNF).Notes and references
- S. Hakomori, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 225–232 CrossRef CAS.
- R. J. Keenan, D. M. Freymann, R. M. Stroud and P. Walter, Annu. Rev. Biochem., 2001, 70, 755–775 CrossRef CAS.
- A. V. Koulov, T. N. Lambert, R. Shukla, M. Jain, J. M. Boon, B. D. Smith, H. Li, D. N. Sheppard, J.-B. Joos, J. P. Clare and A. P. Davis, Angew. Chem., Int. Ed., 2003, 42, 4931–4933 CrossRef CAS.
- L. Addadi and S. Weiner, Angew. Chem., Int. Ed.. Engl., 1992, 31, 153–169 CrossRef.
- Y. Cai and J. Yao, Nanoscale, 2010, 2, 1842–1848 RSC.
- C. G. Ferguson, R. D. James, C. S. Bigman, D. A. Shepard, Y. Abdiche, P. S. Katsamba, D. G. Myszka and G. D. Prestwich, Bioconjugate Chem., 2005, 16, 1475–1483 CrossRef CAS.
- W. H. Binder, V. Barragan and F. M. Menger, Angew. Chem., Int. Ed., 2003, 42, 5802–5827 CrossRef CAS.
- S. M. Butterfield, A. Hennig and S. Matile, Org. Biomol. Chem., 2009, 7, 1784–1792 CAS.
- A. Hennig, L. Fischer, G. Guichard and S. Matile, J. Am. Chem. Soc., 2009, 131, 16889–16895 CrossRef CAS.
- A. Jesorka and O. Orwar, Annu. Rev. Anal. Chem., 2008, 1, 801–832 CrossRef CAS.
- M. M. A. E. Claessens, B. F. Van Oort, F. A. M. Leermakers, F. A. Hoekstra and M. A. Cohen Stuart, Biophys. J., 2004, 87, 3882–3893 CrossRef CAS.
- E. Sackmann, Science, 1996, 271, 43–48 CAS.
- P. S. Cremer and S. G. Boxer, J. Phys. Chem. B, 1999, 103, 2554–2559 CrossRef CAS.
- M. L. Wagner and L. K. Tamm, Biophys. J., 2000, 79, 1400–1414 CrossRef CAS.
- S. Mornet, O. Lambert, E. Duguet and A. Brisson, Nano Lett., 2005, 5, 281–285 CrossRef CAS.
- A.-L. Troutier and C. Ladavière, Adv. Colloid Interface Sci., 2007, 133, 1–21 CrossRef CAS.
- J. T. Groves, N. Ulman and S. G. Boxer, Science, 1997, 275, 651 CrossRef CAS.
- S. J. Johnson, T. M. Bayerl, W. Weihan, H. Noack, J. Penfold, R. K. Thomas, D. Kanellas, A. R. Rennie and E. Sackmann, Biophys. J., 1991, 60, 1017–1025 CrossRef CAS.
- B. W. Koenig, S. Krueger, W. J. Orts, C. F. Majkrzak, N. F. Berk, J. V. Silverton and K. Gawrisch, Langmuir, 1996, 12, 1343–1350 CrossRef CAS.
- J. M. Rosenholm, C. Sahlgren and M. Linden, Nanoscale, 2010, 2, 1870–1883 RSC.
- R. Mortera, J. Vivero-Escoto, I. I. Slowing, E. Garrone, B. Onida and V. S.-Y. Lin, Chem. Commun., 2009, 3219–3221 RSC.
- Y. Sakamoto, T.-W. Kim, R. Ryoo and O. Terasaki, Angew. Chem., Int. Ed., 2004, 43, 5231–5234 CrossRef CAS.
- R. W. Davis, A. Flores, T. A. Barrick, J. M. Cox, S. M. Brozik, G. P. Lopez and J. A. Brozik, Langmuir, 2007, 23, 3864–3872 CrossRef CAS.
- J. M. Rosenholm, A. Penninkangas and M. Lindén, Chem. Commun., 2006, 3909–3911 RSC.
- K. Katagiri, M. Hashizume, J.-I. Kikuchi, Y. Taketani and M. Murakami, Colloids Surf., B, 2004, 38, 149–153 CrossRef CAS.
- M. M. Baksh, M. Jaros and J. T. Groves, Nature, 2004, 427, 139–141 CrossRef CAS.
- T. Buranda, J. Huang, G. V. Ramarao, L. K. Ista, R. S. Larson, T. L. Ward, L. A. Sklar and G. P. Lopez, Langmuir, 2003, 19, 1654–1663 CrossRef CAS.
- S. Trepout, S. S. Mornet, H. Benabdelhak, A. Ducruix, A. R. Brisson, O. Lambert and S. Trépout, Langmuir, 2007, 23, 2647–2654 CrossRef CAS.
- J. Liu, A. Stace-Naughton, X. Jiang and C. J. Brinker, J. Am. Chem. Soc., 2009, 131, 1354–1355 CrossRef CAS.
- G. Nordlund, J. B. S. Ng, L. Bergström and P. Brzezinski, ACS Nano, 2009, 3, 2639–2646 CrossRef CAS.
- S. Mann, Acc. Chem. Res., 2012, 45, 2131–2141 CrossRef CAS.
- A. Finkelstein and O. S. Andersen, J. Membr. Biol., 1981, 59, 155–171 CrossRef CAS.
- B. A. Wallace, J. Struct. Biol., 1998, 121, 123–141 CrossRef CAS.
- C. von Ballmoos, G. M. Cook and P. Dimroth, Annu. Rev. Biophys., 2008, 37, 43–64 CrossRef CAS.
- G. V. R. Rao, G. P. López, J. Bravo, H. Pham, A. K. Datye, H. F. Xu and T. L. Ward, Adv. Mater., 2002, 14, 1301–1304 CrossRef CAS.
- J. B. S. Ng, P. Kamali-Zare, H. Brismar and L. Bergstrom, Langmuir, 2008, 24, 11096–11102 CrossRef CAS.
- T. Vorburger, J. Z. Ebneter, A. Wiedenmann, D. Morger, G. Weber, K. Diederichs, P. Dimroth and C. von Ballmoos, FEBS J., 2008, 275, 2137–2150 CrossRef CAS.
- D. J. Klionsky, W. S. Brusilow and R. D. Simoni, J. Bacteriol., 1984, 160, 1055–1060 CAS.
- V. Oliynyk, U. Kaatze and T. Heimburg, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 236–245 CrossRef CAS.
- V. Oliynyk, M. Jäger, T. Heimburg, V. Buckin and U. Kaatze, Biophys. Chem., 2008, 134, 168–177 CrossRef CAS.
- M. Langner, Chem. Phys. Lipids, 1991, 60, 127–132 CrossRef CAS.
- W. Laubinger and P. Dimroth, Biochemistry, 1989, 28, 7194–7198 CrossRef CAS.
- S. Neumann, U. Matthey, G. Kaim and P. Dimroth, J. Bacteriol., 1998, 180, 3312 CAS.
- A. Wiedenmann, P. Dimroth and C. Von Ballmoos, Mol. Microbiol., 2009, 72, 479–490 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: 1.1 Synthesis of nanoporous silica particles, 1.2 characterization of the monodisperse mesoporous spheres, Fig. S1 characterization of mesoporous silica particles. Fig. S2 CLSM scans of non-sealed particles before and after Na+ uptake, Fig. S3 CLSM images of the particles sealed with DLPC lipid bilayers without gramicidin A. See DOI: 10.1039/c2cp43166a |
| ‡ These authors contributed equally to this work. |
| This journal is © the Owner Societies 2013 |
