 Open Access Article
Open Access ArticlePolyion complex libraries possessing naturally occurring differentiation for pattern-based protein discrimination†
Shunsuke
Tomita
* and
Keitaro
Yoshimoto
*
Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, 3-8-1 Komaba, Meguro, Tokyo 153-8902, Japan. E-mail: s_tomita@bio.c.u-tokyo.ac.jp; ckeitaro@mail.ecc.u-tokyo.ac.jp; Tel: +81-3-5454-6580
First published on 13th September 2013
Abstract
Polyion complexes with naturally occurring differentiation of enzymes serve to create receptor libraries with high differentiability and lower synthetic demands for pattern-based protein discrimination.
Current protein sensing techniques are largely based on a “lock-and-key” approach using antibodies.1 Application of this technique is however restricted because of the need for laborious development to obtain specific antibody–antigen pairs. In recent years, an alternative approach has been proposed, in which proteins are identified by “cross-reactive” sensor arrays consisting of differential receptors.2 In this approach, differential receptors in a sensor array differentially interact with an analyte protein, generating a response pattern. Statistical analysis of the patterns enables discrimination of individual analytes and possibly the bulk properties of complex biochemical mixtures. The individual differential receptors do not need to be specific to the analyte. Two features are necessary for sensor design: (i) structural diversity of receptors that interact with an analyte to respond differently to a variety of analytes; and (ii) the ability to transduce the binding event into a readable signal.
Over the past several years, this approach has been successfully applied for protein identification using various artificial receptors.3 Rotello and co-workers constructed receptor libraries of synthetic gold nanoparticles, in which differentiation of the receptors was generated by changes in the terminal motif of charged alkane thiol coating the gold nanoparticle surface, and binding events were transduced into fluorescent signals by Förster resonance energy transfer (FRET)4 or enzyme activity.5 Use of common low molecular weight chromophores and fluorophores has suggested one possibility to reduce synthetic efforts for generating receptor differentiation.6 While excellent receptor libraries with organic chemistry-based differentiation were reported, the construction of receptor libraries with high levels of structural diversity with lower synthetic demands still remains a challenge.
Here, we propose a novel strategy to develop biomacromolecule-based receptor libraries possessing high levels of naturally occurring differentiation with lower synthetic efforts. In this strategy, enzyme–polyelectrolyte complexes (EPCs), which are a type of polyion complex,7 were used as receptors, the differentiation of which was generated by unique heterogeneous surface properties of enzymes derived from distinct electrostatic, hydrogen bonding, hydrophobic, and morphological characteristics. Recently, we showed that EPC formation enables reversible inhibition of enzyme activity.8 Briefly, enzymes are inactivated by oppositely charged polyelectrolytes, followed by reactivation of enzyme activity by the addition of polyelectrolytes with the same charge as that of the enzyme through exchange reactions. It was expected that enzyme activity would be recovered not only by polyelectrolytes but also by proteins, and hence the changes in catalytic activity of enzymes in EPCs could be utilized to transduce receptor–analyte binding events, the signal intensity of which depends on naturally occurring differentiation of enzymes. The details of the pattern-based sensing system using EPCs are as follows. First, anionic enzyme and a cationic poly(ethylene glycol)-conjugated (PEGylated) polyelectrolyte are mixed for EPC formation predominantly by multiple electrostatic interactions, resulting in a decrease in enzyme activity (Fig. 1A). The PEG segment in the polymer plays a crucial role in preventing irreversible inactivation of enzymes responsible for aggregate formation.8d The addition of analyte proteins to EPCs causes competitive interactions between enzymes, PEGylated polyelectrolytes, and analyte proteins. These interactions may recover a portion of the enzyme activity, generating unique patterns of changes in enzyme activity as “fingerprints” for individual proteins (Fig. 1B).
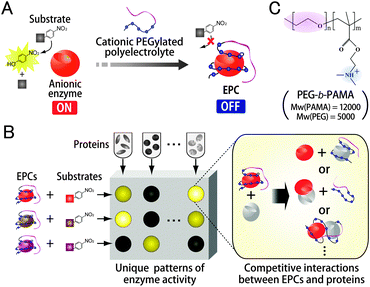 | ||
| Fig. 1 Components and possible mechanism of the EPC sensor array. (A) Decrease in activity of anionic enzyme by EPC formation with a cationic PEGylated polyelectrolyte. (B) Generation of enzyme activity patterns through differential competitive interactions between EPCs and proteins. (C) Molecular structure of PEG-b-PAMA. | ||
Following preliminary experiments regarding the inhibition of various anionic enzymes, EPCs of three anionic enzymes [β-galactosidases from Aspergillus oryzae (GALAO) and Escherichia coli (GALEC), and α-amylase from Aspergillus oryzae (AMYAO)] (Fig. S1, ESI†) with poly(ethylene glycol)-block-poly(N,N-dimethylaminoethyl methacrylate) (PEG-b-PAMA, Fig. 1C) were selected. These enzymes are commonly used in biological experiments because of their stability and commercial availability. The activities of these enzymes can be quantified colorimetrically from the time course of the increase in absorbance at 400 nm with a microplate reader using substrates that are enzymatically hydrolyzed to o- or p-nitrophenol. The optimum ratios between enzymes and PEG-b-PAMA were determined from titration experiments for enzymes with PEG-b-PAMA in 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) at pH 7.0. The activities of the selected enzymes against the corresponding substrates decreased significantly with increasing PEG-b-PAMA concentration (Fig. S2, ESI†).
To evaluate whether the competitive interactions between the prepared EPCs and proteins generate diverse responses, five human plasma proteins—albumin (ALB), immunoglobulin G (IMM), α1-antitrypsin (ANT), fibrinogen (FIB), and transferrin (TRA)—were chosen (Fig. S1, ESI†). Despite slight differences in pI between enzymes and proteins, different changes in enzyme activity were successfully obtained in the titration experiments with proteins (Fig. 2). These results suggested that activity changes of enzymes in EPCs were induced not only by exchange reactions in which enzymes were displaced by proteins through electrostatic interactions but also by various interactions between enzymes, PEGylated polyelectrolytes, and proteins, as shown in Fig. 1B. As enzymes and proteins had a variety of naturally induced hydrophobicity and tertiary structure (Fig. S1, ESI†), these interactions likely included hydrophobic interactions, steric interference, and hydrogen bonds in addition to electrostatic interactions.
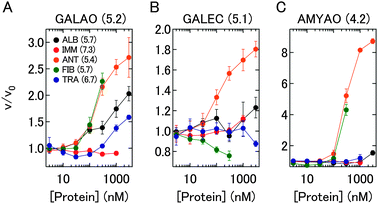 | ||
| Fig. 2 Changes in the activities of three EPCs. The y-axis indicates that the ratio of velocities of absorbance increases before (v0) and after (v) the addition of proteins. pI of enzymes and proteins are shown in parentheses. | ||
The performance of our sensor array consisting of the optimized concentrations of EPCs (see Methods in ESI†) was then investigated using seven analytes composed of five proteins. In addition to five individual proteins at 100 nM (ALB, IMM, ANT, FIB, and TRA), two mixtures of four proteins at 25 nM (A1 and A2) were prepared (Table S1, ESI†).
Individual analytes were tested using a sensor array including three EPCs for six replicates, providing a data set matrix (3 EPCs × 7 analytes × 6 replicates). As shown in Fig. 3A, the analytes generated distinct and reproducible enzyme activity patterns on the EPC sensor array (raw data are provided in Table S2, ESI†). The data were then analyzed by linear discriminant analysis using SYSTAT 13 software to identify analytes. This statistical analysis created discriminant functions that were linear combinations of the original data (enzyme activities in this case) weighted by coefficients producing the greatest analyte discrimination.9 Discriminant functions were calculated with the objective of maximizing the distance between analyte classes relative to the variation within analyte classes in the data set. After the analysis, three discriminant functions were obtained. Significantly, only the first discriminant function that provided the best discrimination among the classes accounted for 96.1% of the total variance. Discriminant scores calculated from the first two discriminant functions were visualized in a two-dimensional plot as shown in Fig. 3B, where each dot represents an enzyme activity pattern of individual analytes. Relative distances between the scores were correlated with similarities in the patterns. The scores were clustered to seven distinct groups according to the analytes, with very slight overlap between the 95% confidence ellipses of IMM and TRA, suggesting a significant difference between the patterns. Interestingly, the first discriminant scores were roughly correlated with pI of five proteins. It should be noted, however, that there is a large distance between ALB and ANT despite their similar pI and Mw, which is likely responsible for differences in the surface hydrophobicity (Φsurface) (ALB: 0.275 and ANT: 0.263) and tertiary structure of these proteins (Fig. S1, ESI†). Taken together, the EPC sensor array dominantly distinguished the electrostatic nature of proteins, but it is clear that other properties of proteins also affected the response patterns.
 | ||
| Fig. 3 Pattern-based sensing of protein analytes using three EPCs (GALAO, GALEC, and AMYAO). (A) Enzyme activity patterns for seven analytes. Each value represents the average of six parallel measurements with the standard deviation. (B) Discriminant score plot of the first two discriminant functions of enzyme activity patterns analyzed by linear discriminant analysis. The 95% confidence ellipses for the individual protein analytes are shown. pI of proteins are shown in parentheses. | ||
The classification accuracy of the EPC sensor array was quantitatively evaluated by linear discriminant analysis with the jackknifed classification procedure.10 Here, linear discriminant analysis was based on the shortest Mahalanobis distance to the centroids of seven groups in a three-dimensional space. In the jackknifed classification, one replicate in an analyte was omitted from linear discriminant analysis and treated as an unknown. This unknown case was then classified by the linear discriminant analysis model generated from the remaining cases in the data set. This leave-one-out procedure was repeated for each case, giving 98% classification accuracy (Table 1). Analyzing the same data set using only one EPC showed classification accuracies of 57%, 19%, and 67% for GALAO, GALEC, and AMYAO, respectively (Table 1). The high accuracies of GALAO and AMYAO were partly due to the high efficiency of PEG-b-PAMA upon decreasing the activity levels of these enzymes (Fig. S2A and C, ESI†). As the discriminative ability between GALAO and AMYAO was highly complementary (Table 1), the combination of only GALAO and AMYAO generated 93% accuracy (Fig. S3, ESI†), indicating that naturally occurring differentiation, i.e., the structural diversity and complexity of enzymes, was effective for pattern-based protein sensing. Furthermore, with the addition of the EPC between amyloglucosidase from Aspergillus niger (AMYAN) (Fig. S1, ESI†) and PEG-b-PAMA to the three EPCs, classification accuracy for discrimination of analytes increased up to 100%, with no overlap between the 95% confidence ellipses (Fig. S4, ESI†).
| Analytes | % Correct | |||
|---|---|---|---|---|
| Three EPCs | GALAO | GALEC | AMYAO | |
| ALB | 100 | 100 | 33 | 67 |
| ANT | 100 | 67 | 0 | 100 |
| FIB | 100 | 17 | 83 | 100 |
| IMM | 83 | 100 | 17 | 0 |
| TRA | 100 | 83 | 0 | 50 |
| A1 | 100 | 0 | 0 | 83 |
| A2 | 100 | 33 | 0 | 67 |
| Total | 98 | 57 | 19 | 67 |
The robustness of the EPC sensor array system was finally tested using 21 unknown analytes in a blind experiment. The enzyme activity patterns of unknown cases obtained by three EPCs (GALAO, GALEC, and AMYAO) were classified into analyte classes according to their Mahalanobis distances. Twenty analytes were correctly classified, affording a discrimination accuracy of 95% (raw data are provided in Table S3, ESI†). Therefore, use of only three EPCs of commercially available enzymes and PEG-b-PAMA was sufficiently sensitive to discriminate between seven analytes, and was more sensitive than previously proposed cross-reactive protein sensors that used 36 commercially available organic compounds as receptor libraries.6b
In summary, we have shown that receptor libraries possessing high levels of structural diversity for pattern-based protein discrimination are successfully constructed with lower synthetic efforts using polyion complexes with naturally occurring differentiation of enzymes. In addition to the high differentiability, use of the catalytic activities of enzymes against substrates with the same chromophore for transducing binding events allowed simultaneous measurement and provided good sensitivity despite colorimetric assay. While EPCs have recently been applied in various fields, such as electrochemical sensors11 and enzyme delivery into cells or animals,12 to our knowledge, this is the first study to develop a cross-reactive sensor array using EPCs. Various anionic enzymes are commercially available, allowing the expansion of EPC libraries to select appropriate enzymes for sensing targets with lower synthetic demands. Although we have developed a new platform that dominantly discriminates electrostatic properties of proteins, the use of cationic PEGylated polyelectrolytes with different physicochemical characteristics would also demonstrate further diverse response patterns for proteins.
This work was supported by a Grant-in-Aid for JSPS Fellows from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Foundation for Interaction in Science & Technology.
Notes and references
- (a) I. E. Tothill, Semin. Cell Dev. Biol., 2009, 20, 55 CrossRef CAS PubMed; (b) M. I. Mohammed and M. P. Desmulliez, Lab Chip, 2011, 11, 569 RSC.
- D. Margulies and A. D. Hamilton, Curr. Opin. Chem. Biol., 2010, 14, 705 CrossRef CAS PubMed.
- (a) H. Zhou, L. Baldini, J. Hong, A. J. Wilson and A. D. Hamilton, J. Am. Chem. Soc., 2006, 128, 2421 CrossRef CAS PubMed; (b) H. Li and G. C. Bazan, Adv. Mater., 2009, 21, 964 CrossRef CAS; (c) X. Li, F. Wen, B. Creran, Y. Jeong, X. Zhang and V. M. Rotello, Small, 2012, 8, 3589 CrossRef CAS PubMed; (d) H. Pei, J. Li, M. Lv, J. Wang, J. Gao, J. Lu, Y. Li, Q. Huang, J. Hu and C. Fan, J. Am. Chem. Soc., 2012, 134, 13843 CrossRef CAS PubMed; (e) Y. Lu, H. Kong, F. Wen, S. Zhang and X. Zhang, Chem. Commun., 2013, 49, 81 RSC.
- (a) C. C. You, O. R. Miranda, B. Gider, P. S. Ghosh, I. B. Kim, B. Erdogan, S. A. Krovi, U. H. Bunz and V. M. Rotello, Nat. Nanotechnol., 2007, 2, 318 CrossRef CAS PubMed; (b) M. De, S. Rana, H. Akpinar, O. R. Miranda, R. R. Arvizo, U. H. F. Bunz and V. M. Rotello, Nat. Chem., 2009, 1, 461 CrossRef CAS PubMed.
- O. R. Miranda, H. T. Chen, C. C. You, D. E. Mortenson, X. C. Yang, U. H. Bunz and V. M. Rotello, J. Am. Chem. Soc., 2010, 132, 5285 CrossRef CAS PubMed.
- (a) D. C. Gonzalez, E. N. Savariar and S. Thayumanavan, J. Am. Chem. Soc., 2009, 131, 7708 CrossRef CAS PubMed; (b) C. Hou, J. Dong, G. Zhang, Y. Lei, M. Yang, Y. Zhang, Z. Liu, S. Zhang and D. Huo, Biosens. Bioelectron., 2011, 26, 3981 CrossRef CAS PubMed.
- (a) A. F. Thünemann, M. Müller, H. Dautzenberg, J. Joanny and H. A. Löwen, Adv. Polym. Sci., 2004, 116, 113 CrossRef; (b) B. Kayitmazer, D. Seeman, B. B. Minsky, P. L. Dubin and Y. Xu, Soft Matter, 2013, 9, 2553 RSC.
- (a) S. Ganguli, K. Yoshimoto, S. Tomita, H. Sakuma, T. Matsuoka, K. Shiraki and Y. Nagasaki, J. Am. Chem. Soc., 2009, 131, 6549 CrossRef CAS PubMed; (b) S. Tomita, L. Ito, H. Yamaguchi, G. Konishi, Y. Nagasaki and K. Shiraki, Soft Matter, 2010, 6, 5320 RSC; (c) S. Tomita and K. Shiraki, J. Polym. Sci., Part A, 2011, 49, 3835 CrossRef CAS; (d) T. Kurinomaru, S. Tomita, S. Kudo, S. Ganguli, Y. Nagasaki and K. Shiraki, Langmuir, 2012, 28, 4334 CrossRef CAS PubMed.
- (a) P. C. Jurs, G. A. Bakken and H. E. McClelland, Chem. Rev., 2000, 100, 2649 CrossRef CAS PubMed; (b) T. Zhang, N. Y. Edwards, M. Bonizzoni and E. V. Anslyn, J. Am. Chem. Soc., 2009, 131, 11976 CrossRef CAS PubMed.
- B. G. Tabachnick and L. S. Fidell, Using Multivariate Statistics, Allyn & Bacon, Needham Heights, MA, 4th edn, 2001 Search PubMed.
- (a) F. Lisdat, R. Dronov, H. Mohwald, F. W. Scheller and D. G. Kurth, Chem. Commun., 2009, 274 RSC; (b) K. Ariga, Q. Ji, T. Mori, M. Naito, Y. Yamauchi, H. Abe and J. P. Hill, Chem. Soc. Rev., 2013, 42, 6322 RSC.
- D. S. Manickam, A. M. Brynskikh, J. L. Kopanic, P. L. Sorgen, N. L. Klyachko, E. V. Batrakova, T. K. Bronich and A. V. Kabanov, J. Controlled Release, 2012, 162, 636 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3cc45640d |
| This journal is © The Royal Society of Chemistry 2013 |
