 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A rapid screen for molecules that form duplex to duplex crosslinks in DNA†
María J. Marín‡a,
Benjamin D. Rackham‡b,
Andrew N. Roundb,
Lesley A. Howellb,
David A. Russell*a and
Mark Searcey*b
aSchool of Chemistry, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK. E-mail: d.russell@uea.ac.uk
bSchool of Pharmacy, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK. E-mail: m.searcey@uea.ac.uk; Fax: +44 (0)1603 592009; Tel: +44 (0)1603 592026
First published on 22nd August 2013
Abstract
We describe a gold nanoparticle based assay that can rapidly determine the crosslinking of DNA duplexes by ligands. Such compounds have potential in targeting highly compacted DNA such as that found in the nucleosome.
Studies of ligand–DNA interactions tend to focus on naked structures either through gel techniques1 or, more recently, through single molecule studies.2 In cells, DNA is tightly compacted into the nucleosome, where it is closely associated with the histone proteins and where it is tightly wound so that one duplex structure lies close to another.3 There are few studies of ligands binding to nucleosomal DNA, in part due to the complexity in forming the core particle and also due to the requirement for techniques such as footprinting4 and crystallography5 that allow the assessment of the results.
Surrogate techniques, that allow the study of histone–DNA and DNA–DNA interactions and the effects of these on molecule binding have not been disclosed and yet have the potential to change the way ligand–DNA interactions are viewed and new therapeutics are designed. One simple example is how small molecules with multiple functions are affected by the presence of both protein and other DNA strands when binding to a duplex sequence. There have been several recent descriptions of multinuclear platinum complexes, one of which has progressed into clinical trial6 and which could potentially have crosslinked protein to DNA or DNA duplex to DNA duplex.7 Pyrrolobenzodiazepine dimers such as the clinically tested SJG-1368 could potentially form crosslinks within the nucleosome that run along a minor groove from one duplex strand to another. These studies suggest that a simple test for this novel higher order interaction is required. Gold nanoparticles (ca. 16 nm in diameter) exhibit an intense red colour in aqueous solution that changes upon aggregation of the particles.9 The colour change is due to the coupling interactions between the surface plasmon fields of the particles. Various studies have focussed on the combination of single stranded DNA with gold nanoparticles to investigate sequence recognition properties10 and even the ability of small molecules to stabilise the duplex.11 However, to our knowledge, there are no studies of double stranded DNA (dsDNA) on nanoparticles to investigate duplex to duplex interactions that may be crucial in nucleosome targeting. In this paper, we describe a rapid colorimetric technique based on the aggregation of dsDNA functionalised gold nanoparticles that allows the identification of ligands that can crosslink two duplex strands – a DNA to DNA crosslink (Fig. 1a). We have applied this technique to the identification of a classical bisintercalator that has previously only been shown to bind in a two base pair sandwich fashion (bisacridine 1, Fig. 1b)12 and to a second molecule designed to form DNA to DNA crosslinks (bisacridine 2, Fig. 1b).13 The monointercalator 9-aminoacridine (9-AA, Fig. 1b) was used as a control since it does not have the ability to crosslink the duplex and therefore, it should not produce aggregation of the dsDNA functionalised gold nanoparticles (Fig. 1a).
 | ||
| Fig. 1 (a) Schematic representation of: (i) the aggregation of the dsDNA functionalised gold nanoparticles in the presence of a bisintercalator and (ii) non-aggregation of the dsDNA functionalised gold nanoparticles in the presence of the monointercalator; (b) chemical structure of compound 1, compound 2 and 9-aminoacridine. | ||
Water soluble gold nanoparticles were prepared via the citrate reduction method14 and were functionalised with thiolated DNA using a pH-assisted method.15 The thiolated DNA was added as a pre-formed duplex. The dsDNA functionalised nanoparticles formed a red suspension in phosphate buffer (10 mM, pH 7.4) (Fig. 2) and were stable in the presence of NaCl as compared with non-functionalised nanoparticles (Fig. S1 in ESI†).
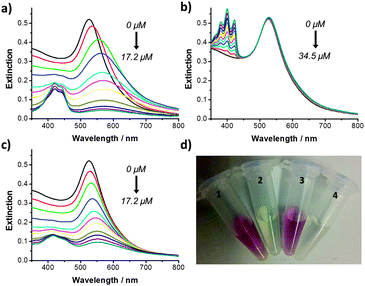 | ||
| Fig. 2 UV-Vis extinction spectrum of dsDNA functionalised gold nanoparticles (PB 10 mM, pH 7.4) after addition of increasing concentrations of: (a) 1; (b) 9-AA; and (c) 2 and (d) Eppendorf tubes containing dsDNA functionalised gold nanoparticles after addition of: (1) no compound; (2) 1 (17.2 μM); (3) 9-AA (34.5 μM); and (4) 2 (17.2 μM). All of the samples were shaken by hand for 1 min prior to measurement. | ||
9-Aminoacridine-4-carboxamides, such as compounds 1 and 2, are known DNA intercalating agents. The bisintercalator linked through an eight methylene chain, compound 1, has previously been shown to bind to and unwind supercoiled DNA and to have a potent antiproliferative effect on tumour cells in culture.12 We have previously shown that the compound with a six methylene linker is able to crosslink DNA duplexes,16 but the longer chain allows for bisintercalation as a “staple” like structure similar to natural products such as echinomycin.17 9-Aminoacridine-4-carboxamide exhibits some sequence selectivity, with a preference for GC rich sequences.18 The double stranded oligonucleotide used to functionalise the gold nanoparticles (5′-CTACGTGGACCTGGAGAGAGGAAGGAGACTGCCTG) contains multiple sites for intercalation with compounds 1 and 2. Increasing concentrations of compound 1 (from 0 to 17.2 μM) were added to a sample of the dsDNA functionalised gold nanoparticles. The samples were slowly shaken by hand for 1 min. The UV-Vis extinction spectrum of the functionalised gold nanoparticle solution was measured before and after addition of the corresponding compound 1 concentration (Fig. 2a). Upon addition of increasing concentrations of 1, the surface plasmon absorption band showed a significant red shift from 525 to 578 nm and a decrease in intensity. Incubation of the gold nanoparticles with 1 led to a rapid (<1 min) disappearance of the red colour (Fig. 2d). These results suggest that the bisintercalator 1 is causing aggregation of the dsDNA functionalised gold nanoparticles through duplex to duplex crosslinking. A similar titration was performed with phosphate buffer (PB) and no colour change was observed (Fig. S2a in ESI†). The increase in the extinction seen at 400–500 nm in Fig. 2a is due to the addition of increasing concentrations of 1 (Fig. S2b in ESI†).
The observed colour change could be a consequence of simple intercalation rather than crosslinking. In order to elucidate whether crosslinking between 1 and the DNA was taking place, the dsDNA functionalised gold nanoparticles were incubated with 9-AA. Titration with 9-AA from 0 to 34.5 μM (i.e. twice the final concentration of the bisintercalator) led to neither a change in the UV-Vis extinction spectrum of the nanoparticles (Fig. 2b) nor in the colour of the solution (Fig. 2d), suggesting that the aggregation of the nanoparticles in the presence of 1 is not simply a consequence of intercalation but rather a crosslinking. The increase in extinction seen at 350–450 nm is due to the addition of increasing concentrations of 9-AA (Fig. S2c in ESI†).
In a recent study, we described the use of click chemistry to generate bisintercalators designed to bind to higher order DNA structures.13 Some of these compounds were designed to direct the second chromophore away from the duplex, one such molecule being the bisintercalator compound 2. When the dsDNA functionalised gold nanoparticles were incubated with increasing concentrations of 2 (0 to 17.2 μM) a red shift (from 525 to 554 nm) in the surface plasmon absorption band was observed accompanied by a decrease in the extinction intensity (Fig. 2c). The changes in the surface plasmon band led to the disappearance of the red colour (Fig. 2d). The increase in the extinction seen at 400–500 nm is due to the addition of increasing concentrations of 2 (Fig. S2d in ESI†). A summary of how the addition of compound 1, 2 and 9-AA affects the intensity of the extinction maxima can be seen in Fig. S3 in ESI.†
The surfactant sequestration technique has been utilised in kinetic studies of drug–DNA interactions in order to assess dissociation kinetics of ligands from the duplex.19 Surfactant sequestration was applied to demonstrate the reversibility of the ligand–DNA binding and, therefore, the reversibility of the aggregation of the dsDNA functionalised gold nanoparticles. In an initial step, dsDNA functionalised gold nanoparticles were treated with 1 (3.5 μM) to induce aggregation of the particles. A red shift in the surface plasmon absorption band of the nanoparticles from 525 to 560 nm (Fig. 3a) and a change in the colour of the solution (Fig. 3b) were observed upon addition of 1. Increasing concentrations of sodium dodecylsulfate (SDS) (from 34.8 to 442.9 μM) were added to the solution of gold nanoparticles containing 1. The samples were shaken by hand for 1 min and the UV-Vis extinction spectrum was recorded (Fig. 3a). After addition of SDS (442.9 μM), the extinction maximum shifted back to 525 nm (Fig. 3a) and the colour of the solution turned to the initial deep red (Fig. 3c). These results clearly demonstrate the reversibility of the DNA-compound 1 crosslinking reaction. Addition of increasing concentrations of SDS to a solution of DNA functionalised gold nanoparticles without compound 1 had no effect on the surface plasmon absorption band of the solution (Fig. S4 in ESI†). The reversibility process was also confirmed by dynamic light scattering that showed an increase in the hydrodynamic diameter of the dsDNA functionalised gold nanoparticles after addition of compound 1. The diameter of the aggregated particles decreased when SDS was added to the solution containing 1, i.e., when the aggregation was reversed (Table S1 in ESI†).
 | ||
| Fig. 3 (a) UV-Vis extinction spectrum of dsDNA functionalised gold nanoparticles (PB 10 mM, pH 7.4): (1) alone; (2) after addition of 1 (3.5 μM); and (3) after addition of 1 (3.5 μM) and SDS (442.9 μM). (b) and (c) Eppendorf tubes containing dsDNA functionalised gold nanoparticles (PB 10 mM, pH 7.4): (1) alone; (2) after addition of 1 (3.5 μM); and (3) after addition of 1 (3.5 μM) and SDS (442.9 μM). All of the samples were shaken by hand for 1 min prior to measurement. | ||
Further verification of the reversibility of the crosslinking between the DNA and compound 1 came from the titration of a solution of dsDNA functionalised gold nanoparticles containing 1 (3.5 μM, aggregated nanoparticles) with non-immobilised calf thymus DNA (CT-DNA). When the CT-DNA is in excess, it competes with the gold nanoparticles for ligand binding and should lead to disaggregation as 1 is sequestered onto the non-immobilised DNA. Upon addition of CT-DNA (97 μM base pairs), the surface plasmon absorption band shifted back to 525 nm and the extinction intensity increased (Fig. S5a in ESI†) confirming the reversibility of the ligand–DNA crosslinking. Addition of the same concentrations of CT-DNA to a solution of dsDNA functionalised nanoparticles without 1 produced no changes in the surface plasmon absorption band (Fig. S5b in ESI†).
Transmission electron microscopy (TEM) was used to further confirm the crosslinking between dsDNA functionalised gold nanoparticles and compound 1. TEM images of the dsDNA functionalised particles showed dispersed gold nanoparticles (Fig. 4a). In the presence of compound 1 the nanoparticles aggregated forming large clusters as observed in Fig. 4b.
 | ||
| Fig. 4 TEM images of dsDNA functionalised gold nanoparticles (a) before and (b) after addition of 1 (3.5 μM). Scale bars: (a) 20 nm and (b) 100 nm. | ||
The ability of small molecules to crosslink duplex DNA (from strand to strand) is well documented and studied – it forms the basis for the clinically used antitumour agents such as cyclophosphamide – whereas duplex to duplex crosslinking is largely unexplored and remains difficult to visualise by macroscopic means, such as gel electrophoresis. As the move towards personalised medicine and gene targeting continues, the ability to target DNA that is compacted into the nucleosome and to study its susceptibility to drugs and ligands is an imperative. In this paper, we have shown that a compound previously demonstrated to bind to one duplex structure is also able to form duplex to duplex crosslinks. We have also demonstrated that it is possible to design molecules with rigid linkers that can reach across from one DNA strand to another, a potential benefit in a compact structure in the cell. The use of gold nanoparticle conjugates to demonstrate duplex to duplex crosslinking is a rapid colorimetric approach to detection which could be extended to other molecules.
We thank the School of Chemistry (UEA) for the post-doctoral fellowship of M.J.M. and the School of Pharmacy (UEA) for a studentship for B.D.R. The authors are grateful to Dr Kim Find (John Innes Centre, Norwich) for assistance with the TEM.
Notes and references
- A. J. Hampshire, D. A. Rusling, V. J. Broughton-Head and K. R. Fox, Methods, 2007, 42, 128 CrossRef CAS PubMed.
- I. Mela, R. Kranaster, R. M. Henderson, S. Balasubramanian and J. M. Edwardson, Biochemistry, 2012, 51, 578 CrossRef CAS PubMed.
- (a) D. M. Lilley and J. F. Pardon, Annu. Rev. Genet., 1979, 13, 197 CrossRef CAS PubMed; (b) O. J. Rando and K. Ahmad, Curr. Opin. Cell Biol., 2007, 19, 250 CrossRef CAS PubMed.
- P. M. Brown and K. R. Fox, Methods Mol. Biol., 1997, 90, 81 CAS.
- R. K. Suto, R. S. Edayathumangalam, C. L. White, C. Melander, J. M. Gottesfeld, P. B. Dervan and K. Luger, J. Mol. Biol., 2003, 326, 371 CrossRef CAS.
- D. I. Jodrell, T. R. Evans, W. Steward, D. Cameron, J. Prendiville, C. Aschele, C. Noberasco, M. Lind, J. Carmichael, N. Dobbs, G. Camboni, B. Gatti and F. De Braud, Eur. J. Cancer, 2004, 40, 1872 CrossRef CAS PubMed.
- T. Muchova, S. M. Quintel, N. P. Farrell, V. Brabec and J. Kasparkova, JBIC, J. Biol. Inorg. Chem., 2012, 17, 239 CrossRef CAS PubMed.
- J. A. Hartley, Expert Opin. Invest. Drugs, 2011, 20, 733 CrossRef CAS PubMed.
- (a) C. L. Schofield, A. H. Haines, R. A. Field and D. A. Russell, Langmuir, 2006, 22, 6707 CrossRef CAS PubMed; (b) C. L. Schofield, R. A. Field and D. A. Russell, Anal. Chem., 2007, 79, 1356 CrossRef CAS PubMed.
- (a) C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607 CrossRef CAS PubMed; (b) D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, Angew. Chem., Int. Ed. Engl., 2010, 49, 3280 CrossRef CAS PubMed; (c) K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739 CrossRef CAS PubMed.
- S. J. Hurst, M. S. Han, A. K. Lytton-Jean and C. A. Mirkin, Anal. Chem., 2007, 79, 7201 CrossRef CAS PubMed.
- L. P. G. Wakelin, X. Bu, A. Eleftheriou, A. Parmar, C. Hayek and B. W. Stewart, J. Med. Chem., 2003, 46, 5790 CrossRef CAS PubMed.
- L. A. Howell, R. A. Bowater, M. A. O'Connell, A. P. Reszka, S. Neidle and M. Searcey, ChemMedChem, 2012, 7, 792 CrossRef CAS PubMed.
- B. V. Enüstün and J. Turkevich, J. Am. Chem. Soc., 1963, 85, 3317 CrossRef.
- X. Zhang, M. R. Servos and J. Liu, J. Am. Chem. Soc., 2012, 134, 7266 CrossRef CAS PubMed.
- N. H. Hopcroft, A. L. Brogden, M. Searcey and C. J. Cardin, Nucleic Acids Res., 2006, 34, 6663 CrossRef CAS PubMed.
- M. J. Waring and L. P. G. Wakelin, Nature, 1974, 252, 653 CrossRef CAS.
- M. Searcey, S. McClean, B. Madden, A. T. McGown and L. P. G. Wakelin, Anti-Cancer Drug Des., 1998, 13, 837 CAS.
- L. P. G. Wakelin, P. Chetcuti and W. A. Denny, J. Med. Chem., 1990, 33, 2039 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details for this work. See DOI: 10.1039/c3cc45600e |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2013 |
