 Open Access Article
Open Access ArticleA ferrofluid-based homogeneous assay for highly sensitive and selective detection of single-nucleotide polymorphisms†
Wei
Shen
,
Cai Le
Lim
and
Zhiqiang
Gao
*
Department of Chemistry, National University of Singapore, Singapore 117543. E-mail: chmgaoz@nus.edu.sg; Fax: +65 6779-1691; Tel: +65 6516-3887
First published on 6th August 2013
Abstract
A simple and low-cost colorimetric assay utilizing ferrofluidic nanoparticulate probes (FNPs) and a ligase for single-nucleotide polymorphism genotyping is described. Excellent sensitivity and selectivity were accomplished through the engagement of the FNPs and a ligase chain reaction.
Nucleic acids provide the genomic templates that encode for proteins essential for all cellular functions.1 Ample evidence has revealed that diseases like cancer are closely associated with mutations in gene sequences and single-nucleotide polymorphisms (SNPs) in particular.2 Therefore, SNPs have become an important class of biomarkers in diagnosis after the completion of the Human Genome Project. Conventional techniques for SNP detection rely mostly on polymerase chain reaction (PCR) and fluorescence detection. Unfortunately, these techniques are highly time-consuming and cost intensive as the fluorescence readout requires expensive instrumentation and sophisticated algorithms.3 Owing to the desire for SNP detection devices with high specificity, high sensitivity and low-cost operation, enormous efforts have been invested in research to improve analytical performance of SNP assays.
With the advancement in nanotechnology over the past decade, materials with dimensions in the scale of 1 to 100 nm are playing a significant role in the development of SNP assays.4 With unique size-dependent physical and chemical properties, e.g. high surface-to-volume, surface plasmon resonance and quantum effect, nanoparticles allow stable immobilization of bioreceptors and coupling to many new signal transduction and amplification schemes.5,6 To date, many heterogeneous SNP screening assays using nanoparticles in conjunction with colorimetry, fluorometry or electrochemical methods have been proposed.7 Heterogeneous detection, however, has low hybridization efficiency due to steric hindrance, and a limited number of probes coated on the solid support further restrict the performance of the assays.7 To realize SNP assays that offer the simplicity of the heterogeneous approach along with high hybridization efficiency and reproducibility of homogeneous assays, magnetic nanoparticles (MNs) were used to facilitate homogeneous hybridization and provide simple and quick separation of the hybridized products.8 The ease of magnetic separation is frequently used to overcome the shortcoming of homogeneous ligation by separating reacted probes from unreacted ones which would otherwise pose a threat to the detection.8 In view of the urgent need for simple and inexpensive SNP assays, this report presented a novel SNP assay that for the first time exploited the intrinsic properties of ferrofluids (colloidal MNs). Efficient homogeneous hybridization was carried out by oligonucleotide-coated ferrofluidic nanoparticulate probes (FNPs). The amplification power of the assay was demonstrated by the incorporation of a ligase chain reaction (LCR) in which a minute amount of target crosslinks millions of FNPs together forming an aggregate of the FNPs. Separation of the reacted (aggregated) from the unreacted (free) FNPs was achieved simply based on their drastic difference in magnetic field strength under an external magnetic field. Finally, SNP is conveniently quantified by colorimetry.
As illustrated in Fig. 1, the SNP assay comprises of three steps: (i) capturing of target SNP strands via hybridization and LCR with the FNPs to form a FNP aggregate, (ii) magnetic separation of the FNP aggregate from unreacted FNPs and (iii) amplified colorimetric detection of the unreacted FNPs through the catalytic oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) by the FNPs.9 Highly water-soluble FNPs were prepared and discretely functionalized with the two amine-terminated oligonucleotides (FNP-1 and FNP-2) via amide coupling reaction with the carboxylic groups of polyacrylic acid (PAA) that encapsulated the FNPs. The FNP-1 will align with FNP-2 when both are annealed onto a target SNP template i.e. KRAS, a mutation (MT-C), as depicted in Fig. 1. Amplification is attained by the LCR in which multiple hybridizations occur among the FNPs with a minute amount of the target to form the FNP aggregate. Under an external magnetic field, each FNP acts as a single magnetic domain and when numerous FNPs aggregate during hybridization, the large FNP aggregate behaves much more strongly towards the magnetic field. Separation of the FNP aggregate from the unreacted FNPs is achieved by using a permanent magnet to pull and pack the FNP aggregate to the bottom of the reaction vessel.
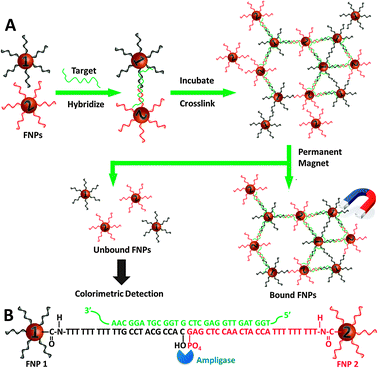 | ||
| Fig. 1 Schematic illustration of the working principle of the SNP assay. | ||
To ensure the success of the assay, it is crucial that there is a substantial difference between individual FNPs and the FNP aggregate so that the unreacted FNPs stay in the reaction mixture when a magnetic field is applied through a permanent magnet. In other words, the FNP aggregate should set out easily in a magnetic field whereas the free FNPs remain in solution. It is well known that when MNs are sufficiently small (≤10 nm) and each MN is thoroughly covered with an anti-clumping coating, the magnetic attraction of MNs is weak enough that the van der Waals force of the anti-clumping coating is sufficient to prevent magnetic clumping or agglomeration – forming a ferrofluid.10 It was reported that PAA coated MNs (6–8 nm) show ferrofluidic behavior when the colloidal solution of the PAA coated MNs is subjected to an external magnetic field, due to their small size and superparamagnetic properties.11 Therefore, the PAA coated MNs were employed in this work. Two types of FNPs were prepared by functionalizing the PAA coated MNs with two types of oligonucleotide probes, respectively (see ESI† for experimental details). Moreover, functionalization of the MNs with anionic oligonucleotides further stabilized the MNs, forming a very stable ferrofluid (Fig. 2, inset). Apart from the formation of ferrofluid, another critical factor is the functionality of the FNPs. The FNPs must be freely accessible to both the SNP target and the ligase, and particularly to the ligase as it is much bulkier than the SNP target. The two amine-terminated capture probes (CPs) were therefore designed with a (T)9 spacer to eliminate any possible steric effect. Furthermore, surface densities of the probes were kept low by controlling the concentrations of the oligonucleotide probes. It was estimated that on average 20 oligonucleotide probes were attached onto each FNP.
 | ||
| Fig. 2 UV-vis spectra of TMB solution treated with 10 nM MNPs after hybridizing with (1) control, (2) 2.0 and (3) 6.0 nM SNP target. Inset: images of the FNP solution without/with a magnet. | ||
To demonstrate the accessibility of the immobilized probes on the FNPs, a complementary KRAS MT-C target and a control were introduced into the FNPs, respectively. As shown in Fig. 2, the control showed practically identical UV-vis responses before and after hybridization and magnetic separation, thus suggesting that there is no FNP aggregate formation and the free FNPs remain in solution even when subjected to the permanent magnet. This is evidently due to their ultra-small size and high dispensability after functionalization.11 On the other hand, after hybridization with the KRAS MT-C target, a significant change in absorbance was observed after magnetic separation, indicating that a large amount of FNPs was removed using the magnet. The results suggest that the functionalized FNPs are capable of capturing the SNP target and hybridizing into a FNP aggregate that can be separated out using the permanent magnet.
To achieve the highest possible analytical signal, there should be efficient hybridization of FNPs with the SNP target to form the FNP aggregate that will be first separated out by the permanent magnet and leave the unreacted FNPs in the solution. One key factor that greatly influences the hybridization efficiency is the presence of cations. In general, cations are added into the hybridization buffer to compensate for the negatively charged phosphate groups on the oligonucleotide backbone and stabilize the hybridized duplexes.12 However, an increase in ionic strength of the buffer often leads to colloid coagulation (self-aggregation) as the double layer thickness decreases. Self-aggregation of the FNPs would pose a threat in distinguishing the unreacted (free) and hybridized (aggregated) FNPs. Furthermore, pH-induced conformational changes in the oligonucleotide probes may hinder the accessibility and binding affinity of the target to the immobilized probes.13 Optimization of the hybridization buffer is, therefore, a critical step to establish good stability of the FNPs and at the same time promote hybridization with the SNP target.
In this work, tris-based buffers were used to ensure that the hybridization process is solely influenced by the free cations of the added salts. Monovalent salts were found to be much more efficient in duplex stabilization in solution than divalent salts.12 Thus, the cationic shielding effect influencing the stability and hybridization efficiency of the FNPs was studied with concentrations of the monovalent sodium in the range of 50 mM to 2 M (Fig. 3a). The negligible change in absorbance for FNPs in the absence of the SNP target (trace 1) showed relatively good stability of the FNPs in the buffer. A slight increase in ΔAbs observed when sodium concentration increased above 1 M suggests that coagulation of the FNPs occurred due to the decrease in electrostatic repulsion between FNPs as ionic strength of buffer was further increased. Fortunately, this effect did not obstruct the hybridization process. The change in absorbance for the FNPs after reacting with the SNP target (trace 2) was significantly larger than that for the control (trace 1). This indicates that the FNPs have hybridized with the SNP target to form the FNP aggregate which is then separated out quickly from the solution using the magnet. Thus, the amount of free FNPs in solution decreases substantially and leads to the significant change in absorbance. It was also observed that the hybridization efficiency increases with increasing concentration of sodium, a trend found in several other DNA sensing systems.14,15 On the other hand, a reverse effect started to show when the salt concentration was increased beyond 1 M. Again, this could be due to the decrease in electrostatic repulsion, causing the FNPs to aggregate and hinder the hybridization process. Nonetheless, the hybridization was found to be most efficient at 1 M of sodium concentration.
 | ||
| Fig. 3 The effect of (a) monovalent cations (Na+) and (b) pH on the stability and hybridization efficiency of the FNPs (direct UV-vis tests of the FNPs without TMB amplification). | ||
The accessibility and binding affinity of the target to the FNPs were studied in 50 mM tris buffers containing 1.0 M NaCl in the pH range from 5.5 to 9.5 (Fig. 3b). The FNPs remained considerably stable in the pH range studied (trace 1) while the hybridization process was significantly affected by the acidity of the hybridization buffer (trace 2). The largest response was observed at pH 7.5, thus implying that there is minimal steric hindrance between neighboring FNPs and the hybridization process was most efficient. At lower pH, protonation of the PAA coating decreased the electrostatic repulsion between the FNPs and hindered the hybridization processes. Similarly, low hybridization efficiency observed at higher pH could be due to extensive deprotonation of the CPs such that the probes bend away from each other. Therefore, the optimal conditions that allow efficient hybridization and differentiation of the hybridized FNP aggregate from the unreacted FNPs were 1 M of sodium at pH 7.5.
With this distinct change associated with the SNP target and the catalytic properties of the FNPs toward the oxidation of TMB,9 additional experiments were conducted to establish the relationship between the colorimetric signal and the target concentration. It was found that the FNPs can be directly utilized for the detection of SNPs in the range of 1.0 to 90 nM. However, this sensitivity is far below the requirement for SNP detection and an in situ amplification mechanism is therefore inevitable to make this assay more appealing.
To improve the assay sensitivity, the minute amount of target captured by the FNPs has to be amplified and this could be fulfilled by LCR. This allele-specific enzymatic ligation provides similar amplification power to that of PCR and is able to amplify the target and ligate the MNP probes simultaneously under specific thermal cycling conditions.8,16 More importantly, through judicious design of the two probes with SNPs located at the ligation sites, LCR is much more specific than PCR for SNP genotyping.17 To significantly improve the sensitivity of the assay, LCR was utilized to amplify minute amounts of the target and ligate the FNPs to form the FNP aggregate. As depicted in Fig. 4, the assay was able to distinguish the SNP target at a concentration as low as 3 pM after only 15 thermal cycles. This sensitivity was three orders of magnitude higher than that without target amplification. About 40% of the FNPs were cross-linked at the high concentration end of the calibration curve. Femtomolar concentrations of SNP targets were detected when extensive thermal cycles (≥20 cycles) were performed (Fig. S4, ESI†).
To determine the ability of selectively detecting the SNP target in a sample matrix containing a large excess of mismatch sequences, SNP detection was carried out with KRAS mutation-to-wild type target (MT-to-WT) ratios ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 1
0 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. Compared to the control set (MT-to-WT = 1
1000. Compared to the control set (MT-to-WT = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), no significant change in the signal was observed even when the WT concentration was 1000 times higher (Fig. S5, ESI†). This implies that the nonspecific binding of the mismatch sequence to the FNPs is negligible. The excellent analytical performance of this assay strongly supports the fact that the specific-designed FNPs are highly sensitive and selective in the screening for the SNP target even in complex matrices. Ongoing efforts are devoted to applying this assay to screen for SNPs in real samples and expanding its capability to detect methylated DNAs and microRNAs.
0), no significant change in the signal was observed even when the WT concentration was 1000 times higher (Fig. S5, ESI†). This implies that the nonspecific binding of the mismatch sequence to the FNPs is negligible. The excellent analytical performance of this assay strongly supports the fact that the specific-designed FNPs are highly sensitive and selective in the screening for the SNP target even in complex matrices. Ongoing efforts are devoted to applying this assay to screen for SNPs in real samples and expanding its capability to detect methylated DNAs and microRNAs.
In summary, the proposed assay revealed the versatility and power of FNPs as the target capturer and the signal generator for SNP genotyping. The assay was able to discriminate one copy of the SNP target in 1000 copies of the WT gene. The demonstrated attractive characteristics of simple, rapid and low cost colorimetric SNP genotyping could therefore open new prospects in the development of SNP genotyping tools for uses at point-of-care.
This work was supported by the A*STAR-ANR program.
Notes and references
- L. Stryer, Biochemistry, Freeman, San Francisco, CA, 4th edn, 1995 Search PubMed.
- L. B. Jorde, Genome Res., 2000, 10, 1435 CrossRef CAS.
- N. C. Tansil and Z. Q. Gao, Nano Today, 2006, 1, 28 CrossRef.
- K. K. Jain, Clin. Chem., 2007, 53, 2002 CAS.
- C.-Y. Zhang, H.-C. Yeh, M. T. Kkuroki and T.-H. Wang, Nat. Mater., 2005, 4, 826 CrossRef CAS.
- Y. Zhang, Y. Guo, P. Quirkeb and D. J. Zhou, Nanoscale, 2013, 5, 5027 RSC.
- J. K. K. Ng and W. T. Liu, Anal. Bioanal. Chem., 2006, 386, 427 CrossRef CAS.
- X. Chen, A. Ying and Z. Q. Gao, Biosens. Bioelectron., 2012, 36, 89 CrossRef CAS.
- L. Gao, et al. , Nat. Nanotechnol., 2007, 2, 577 CrossRef CAS.
- C. Holm and J. J. Weis, Curr. Opin. Colloid Interface Sci., 2005, 10, 133 CrossRef CAS.
- J. Ge, Y. Hu, M. Biasini, C. Dong, J. Guo, W. P. Beyermann and Y. Yin, Chem.–Eur. J., 2007, 13, 7153 CrossRef CAS.
- T. Špringer, H. Šípová, H. Vaisocherová, J. Štépánek and J. Homola, Nucleic Acids Res., 2010, 38, 7343 CrossRef.
- J. Zhang, H. P. Lang and C. Gerber, Langmuir, 2012, 28, 6494 CrossRef CAS.
- A. W. Peterson, R. J. Heaton and R. M. Georgiadis, Nucleic Acids Res., 2001, 29, 5163 CrossRef CAS.
- Y. Okahata, M. Kawase, K. Niikura, F. Ohtake, H. Furusawa and Y. Ebara, Anal. Chem., 1998, 70, 1288 CrossRef CAS.
- A. J. Lowe, Y. S. Huh, A. D. Strickland and C. A. Batt, Anal. Chem., 2010, 82, 5810 CrossRef CAS.
- W. Shen, H. Deng, A. Teo and Z. Gao, Chem. Commun., 2012, 48, 10225 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and characterization of the FNPs, the SNP assay procedure and comparison of the proposed assay with other SNP assays. See DOI: 10.1039/c3cc43281e |
| This journal is © The Royal Society of Chemistry 2013 |

