 Open Access Article
Open Access ArticleA new platform for a convenient genotyping system†
Keum-Soo
Song
a,
Satish Balasaheb
Nimse
b,
Junghoon
Kim
a,
Danishmalik Rafiq
Sayyed
b and
Taisun
Kim
*b
aBiometrix Technology, Inc., 202 BioVenture Plaza, Chuncheon, 200-161, Korea
bInstitute for Applied Chemistry and Department of Chemistry, Hallym University, Chuncheon, 200-702, Korea. E-mail: tskim@hallym.ac.kr; Fax: +82-33-256-3421
First published on 1st February 2013
Abstract
The high SNP discrimination ratio of 360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100% target-specific hybridization at 25 °C, detection limit of 101 copies, and differentiation of 101 to 107 copies of the PCR product of high-risk HPV genotypes in clinical samples ensure the application of the 9G membrane in a convenient platform for DNA genotyping.
1, 100% target-specific hybridization at 25 °C, detection limit of 101 copies, and differentiation of 101 to 107 copies of the PCR product of high-risk HPV genotypes in clinical samples ensure the application of the 9G membrane in a convenient platform for DNA genotyping.
The challenges presented by global health-care issues and emerging diseases of natural origin lend urgency to the development of rapid, field-deployable pathogen detection and diagnostic tools.1 Ideally, to be of general field utility, a diagnostic tool must be capable of sensitive and specific pathogen detection while retaining simplicity of use and independence from complex laboratory instrumentation.2
The DNA microarray technology has revolutionized the field of medical diagnostics in terms of identification and discrimination of pathogens in infectious diseases.3 However, tedious experimental conditions such as the high temperature hybridizations, 40–60 °C, long hybridization and incubation times required for microarray assays increase sample-to-answer times.4 Moreover, transfer of the DNA chips from solution to solution during the several washing steps, the unusual drying in the centrifuge, costly instrumentation, and the need for highly trained professionals severely limit the utility of microarrays in field applications where a laboratory infrastructure is limited or unavailable.
In a step toward addressing these potential limitations, the use of a lateral flow strip membrane (LFSM) is an economical and simple alternative approach that has recently attracted considerable attention.5 The LFSM-based methods possess several benefits, including a user-friendly format, long-term stability, short assay time, and low-cost analysis.6,7 Moreover, these strip membranes eliminate complex analysis procedures that involve expensive instrumentation and minimize the requirement for highly trained professionals.
The disadvantages of the reported LFSM-based methods are a pre-hybridization reaction time of 10 to 30 min at desired temperatures, hybridization with the target sequence at 55 °C,8 detection of one analyte at a time.9 Moreover, some of the methods require special and expensive hardware for quantitative detection.10,11 The clinical applications of LFSM based methods are still questionable due to their low clinical sensitivity and specificity in the detection of pathogens in clinical samples.12,13 The reasons behind this include low signal intensity, signal to background ratio (SBR) and low SNP discrimination ratio, which can compromise detection and discrimination of pathogens.14 Current methods based on the LFSM are limited in their infield and DNA genotyping applications.
Recently we have reported the 9G DNAChip technology15 for the genotyping of highly pathogenic viruses such as human papillomavirus (HPV)16 and human influenza virus (H1N1).17 Though this technology is highly reproducible and allows the fabrication of DNA chips for 100% clinically sensitive and specific genotyping of pathogens,18 the use of costly instrumentation limits its use in the laboratory settings.
Therefore, to address these problems a 9G membrane technology was developed for genotyping the pathogens.
The 9G membrane (Scheme 1) was developed by using the Cy5 labeled synthetic single stranded target DNA probes complementary to the immobilized probes. The optimized 9G membrane was tested for its performance by using the Cy5 labeled PCR products of the HPV genotypes. The feasibility of application of the 9G membrane in a rapid genotyping was tested by the detection and discrimination of the HPV genotypes in standard clinical samples. The 9G membrane has the following features: high sensitivity and 100% target-specific hybridization at 25 °C, and no complex and expensive instrumentation is needed. The 9G membrane can discriminate the SNP among the genotypes with a ratio of 360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which is the highest ever reported. Moreover, it is for the first time that the genotyping results are obtained by the direct hybridization of the PCR products to the immobilized probes.
1, which is the highest ever reported. Moreover, it is for the first time that the genotyping results are obtained by the direct hybridization of the PCR products to the immobilized probes.
 | ||
| Scheme 1 A 9G DNA membrane for a convenient genotyping system. | ||
The 9G membranes were obtained by slight modification of the 9G DNAChip technology used for the fabrication of the 9G DNAChips. Unlike the nitrocellulose membranes19 in other methods, a glass membrane was used to produce the 9G membrane because the glass membranes are similar to glass slides in physicochemical properties. Therefore the immobilization mechanism of oligonucleotide probes on the DNA chips can be reproduced on the glass membranes. By lining an 18 pmol μL−1 solution of the oligonucleotide probes (Probe1–Probe8) appended with 9 consecutive guanines (see the ESI† Table S1) the oligonucleotides can be immobilized on the AMCA (aminocalix[4]arene)20 membrane in 4 h. After immobilization, the membranes were soaked in the blocking solution and then dried to generate the 9G membrane. These 9G membranes were assembled into strips to obtain the final 9G membrane. The optimal conditions for the effective detection of the Cy5-labeled complementary target oligonucleotide probes were determined by time dependent hybridization and time dependent washing after hybridization, all at 25 °C. The HC (Probe6), PC (Probe7), and PCR (Probe8) probes were used as the internal standards, for hybridization control, positive control, and PCR control, respectively. Hence, after hybridization of the immobilized probes with the Cy5 labeled PCR product of the HPV16, the peaks corresponding to the HC, PC, and PCR appeared along with the peak corresponding to the HPV16 (Scheme 1, also see the ESI† Scheme S1).
The optimum washing time was determined and found to be 20 min (see the ESI† Fig. S2). To determine the optimum hybridization time, the immobilized probes on the 9G membrane were allowed to hybridize at different time intervals (5, 10, 15, and 20 min) by loading the solution containing 5, 10, 20, and 40 fmol μL−1 of the single stranded target probe10 complementary to Probe1 (HPV16) in the sample port. After hybridization for 5, 10, 15, and 20 min, the 9G membranes were washed by loading the washing solution in the washing port. After washing, the 9G membranes were scanned by a BMT membrane reader to obtain the results (Fig. 1a, also see the ESI† Fig. S3). Hybridization with 40 fmol μL−1 of Probe10 shows the highest fluorescence intensity. Fig. 1a clearly depicts that the fluorescence intensity increases with an increase in time with respect to the increase in concentrations. The hybridization signal increases with the increase in hybridization time and reaches a steady state in 10 min and remains the same for the 20 min hybridization. After hybridization of Probe1 (HPV16) with 40 fmol μL−1 of Probe10 for 5 and 10 min, the fluorescent intensities were found to be 2615 and 3552, respectively. However, after 15 min hybridization the fluorescent intensity reaches 3696 and does not show a significant increase after the 20 min hybridization. It should be noted that the hybridization efficiency reaches 72% and 98% after hybridization for 5 min and 10 min, respectively. After 20 min hybridization the hybridization efficiency was found to be 100%. Similar results were also found upon hybridization of the target Probe11 (HPV18) with the probes immobilized on the 9G membrane (see the ESI† Fig. S4 and S5). Moreover, a similar pattern of hybridization was found for the hybridization of the probes on the 9G membrane with the Cy5 labeled PCR product of HPV16 (see the ESI† Fig. S4–S7). Therefore 20 min was considered as the optimum hybridization time and was used in further experiments.
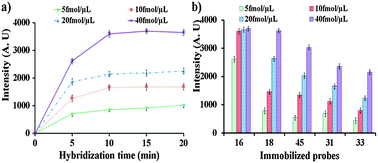 | ||
| Fig. 1 (a) Determination of optimum hybridization time, hybridization with target Probe10 complementary to HPV16 (Probe1) with the concentration of 5, 10, 20, and 40 fmol μL−1 for 5, 10, 15, and 20 min, and (b) optimum concentration, hybridization with single stranded target Probe10–14 complementary to HPV16, 18, 45, 31, and 33, respectively, with the concentrations of 5, 10, 20, and 40 fmol μL−1 for 20 min. | ||
To get insight into the performance of the 9G membrane, the immobilized probes were allowed to hybridize with the Cy5 labeled single stranded target probes Probe10–14 complementary to HPV16, HPV18, HPV45, HPV31, and HPV33, respectively, at various concentrations. The hybridization for 20 min was performed by loading the target Probe10–14 with concentrations ranging from 5 to 40 fmol μL−1 into the sample loading port on the 9G membranes. After hybridization, 9G membranes were washed and then scanned using the BMT membrane reader to obtain the results (Fig. 1b, also see the ESI† Fig. S8). For the hybridization of the target Probe10 (HPV16), the hybridization signals for the 5, 10, 20, and 40 fmol μL−1 were found to be 2612, 3596, 3652, and 3696, respectively. The hybridization signal increases with the increase in the concentration and reaches a steady state at 10 fmol μL−1 and remains the same for 40 fmol μL−1. Similar results were observed upon hybridization of the target probes Probe11, Probe12, Probe13, and Probe14 with the immobilized probes on the 9G membrane. It is interesting to note that the target probes Probe10–14 were specifically bound to their complementary probes on the 9G membrane without any non-specific hybridization and at low concentration such as 5 fmol μL−1 to high concentration of 40 fmol μL−1. These results indicate that the 9G membrane has high selectivity in terms of 100% target hybridization and high sensitivity as the target probes can be detected at a low concentration of 5 fmol μL−1. Therefore, the 9G membrane was evaluated for its performance in the detection and discrimination of the HR-HPV genotypes.
For the detection and discrimination of the HR-HPV genotypes, hybridization solutions containing 101 to 107 copies of Cy5 labeled DNA were prepared from the PCR product of the respective HR-HPV genotypes. The number of copies of DNA in the hybridization solutions was cross checked using gel electrophoresis which confirms that the respective solutions contain 101 to 107 copies (see the ESI† Fig. S10–S14).
Hybridization solutions containing 101 to 107 copies of Cy5 labeled DNA were loaded into the sample port and allowed to hybridize. After 20 min hybridization, the 9G membranes were washed and scanned to obtain the final results (Fig. 2, also see the ESI† Fig. S9). The results show that there is a linear increase in the fluorescence intensity upon increase in the number of copies of the Cy5 labeled PCR product of HPV16 (Fig. 2). Similar results were also observed for the other genotypes (see the ESI† Fig. S10–S14). The tentative idea of the number of copies of the HPV genotypes in the unknown samples can be obtained by interpolating the fluorescence intensities in the standard curve (Fig. 2h).
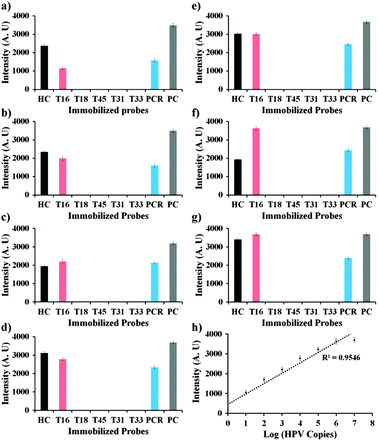 | ||
| Fig. 2 (a)–(g) Graphs representing the respective fluorescence intensities after the hybridization of immobilized probes with 101–107 copies of the Cy5 labeled PCR product of HPV16, respectively; (h) standard curve of fluorescence intensity corresponding to the number of copies of HPV DNA. | ||
After hybridization of the probes on the 9G membrane with 106 copies of Cy5 labeled PCR products of the respective HR-HPV genotypes, the respective genotype can be efficiently detected (see the ESI† Fig. S15). The sequences of the probes corresponding to the 5 HR-HPV genotypes are almost similar to each other with 2 to 3 mismatches. It is important to note that the HR-HPV genotypes were efficiently detected without any cross-hybridization or non-specific binding indicated by the sharp peaks and a baseline with a very low signal to noise ratio.
After hybridization of the 106 copies of the Cy5 labeled PCR product of HPV16 with the immobilized probes the peak intensity for HPV16 was 3596. However, the probes corresponding to HPV18, HPV45, HPV31, and HPV33 showed peak intensities in the range of 8 to 10. Similar results were also observed for the other genotypes. The SNP discrimination ratio of HPV16, HPV18, HPV45, HPV31, and HPV33 is approximately 360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The newly developed 9G membrane has been successfully used as a diagnostic tool, since it discriminates HR-HPV genotypes in clinical samples (see the ESI† Tables S2–S11) in 40 min (see the ESI† Fig. S16).
1. The newly developed 9G membrane has been successfully used as a diagnostic tool, since it discriminates HR-HPV genotypes in clinical samples (see the ESI† Tables S2–S11) in 40 min (see the ESI† Fig. S16).
The 9G membrane, a new platform for the convenient genotyping, was developed and elaborated. It is for the first time that the genotyping results are obtained by the direct hybridization of the PCR products with the immobilized probes. The 9G membranes give final results in 40 min (hybridization, washing, drying, and scanning) after the PCR. The SNP discrimination ratio of 360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 100% target-specific hybridization at 25 °C make the 9G membrane a promising diagnostic tool for accurate HR-HPV genotyping. The 9G membrane has been successfully used as a diagnostic tool, since it discriminates HR-HPV genotypes in clinical samples. However, to assess the clinical application of the 9G membrane, a clinical study to detect and discriminate the HR-HPV types in a large number of clinical samples is under investigation. We believe that these results, especially the hybridization and washing at 25 °C, can facilitate many practical applications of the 9G membrane. Further application of the 9G membranes in the biomarker detection kit is under investigation.
1 and 100% target-specific hybridization at 25 °C make the 9G membrane a promising diagnostic tool for accurate HR-HPV genotyping. The 9G membrane has been successfully used as a diagnostic tool, since it discriminates HR-HPV genotypes in clinical samples. However, to assess the clinical application of the 9G membrane, a clinical study to detect and discriminate the HR-HPV types in a large number of clinical samples is under investigation. We believe that these results, especially the hybridization and washing at 25 °C, can facilitate many practical applications of the 9G membrane. Further application of the 9G membranes in the biomarker detection kit is under investigation.
This work was supported by the Ministry of Knowledge and Economy.
Notes and references
- V. Gubala, L. F. Harris, A. J. Ricco, M. X. Tan and D. E. Williams, Anal. Chem., 2012, 84, 487–515 CrossRef CAS.
- S. Yang and R. E. Rothman, Lancet Infect. Dis., 2004, 4, 337–348 CrossRef CAS.
- E. Hodges, et al. , Nat. Genet., 2007, 39, 1522 CrossRef CAS.
- O. V. Matveeva, S. A. Shabalina, V. A. Nemtsov, A. D. Tsodikov, R. F. Gesteland and J. F. Atkins, Nucleic Acids Res., 2003, 31, 4211 CrossRef CAS.
- (a) Y. He, et al. , Biosens. Bioelectron., 2011, 26, 2018–2024 CrossRef CAS; (b) L. Wang, et al. , Chem. Commun., 2011, 47, 1574–1576 RSC.
- Y. Tsuda, et al. , Microbiol. Immunol., 2007, 51, 903–907 CAS.
- K. H. Chan, et al. , J. Clin. Virol., 2007, 38, 169–171 CrossRef CAS.
- T. Laurent, et al. , Diagn. Microbiol. Infect. Dis., 2009, 63, 173–181 CrossRef CAS.
- (a) X. Mao, et al. , Anal. Chem., 2009, 81, 1660–1668 CrossRef CAS; (b) J. Chen, Z. Fang, P. Lie and L. Zeng, Anal. Chem., 2012, 84, 6321–6325 CrossRef CAS.
- P. Corstjens, et al. , Clin. Chem., 2001, 47, 1885–1893 CAS.
- A. J. Baeumner, J. Pretz and S. Fang, Anal. Chem., 2004, 76, 888–894 CrossRef CAS.
- A. Pierik, et al. , J. Clin. Microbiol., 2011, 49, 1395–1402 CrossRef CAS.
- P. E. Gravitt, et al. , J. Clin. Microbiol., 2000, 38, 357–361 CAS.
- X. Xia, Y. Xu, X. Zhao and Q. Li, Clin. Chem., 2009, 55, 179–182 CAS.
- (a) K. Song, et al. , Chem. Commun., 2011, 47, 7104–7106 RSC; (b) S. B. Nimse, et al. , Chem. Commun., 2011, 47, 12444–12446 RSC.
- K. Song, et al. , J. Virol. Methods, 2012, 183, 132–138 CrossRef CAS.
- V. Nguyen, et al. , Chem. Commun., 2012, 48, 4582–4584 RSC.
- H. An and K. Song, et al. , J. Clin. Microbiol., 2012, 20, 562–568 CrossRef.
- B. Ngom, Y. Guo, X. Wang and D. Bi, Anal. Bioanal. Chem., 2010, 397, 1113–1135 CrossRef CAS.
- (a) S. B. Nimse, et al. , Tetrahedron Lett., 2010, 51, 6156–6160 CrossRef CAS; (b) S. B. Nimse, et al. , Tetrahedron Lett., 2010, 51, 2840–2845 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and method, Table S1, Fig. S1–S8. See DOI: 10.1039/c3cc39231g |
| This journal is © The Royal Society of Chemistry 2013 |
