 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein β-interfaces as a generic source of native peptide tectons†
Céline
Valéry
*a,
Rishi
Pandey
a and
Juliet A.
Gerrard
*abcd
aBiomolecular Interaction Centre, University of Canterbury, Private Bag 4800, Christchurch 8140, New Zealand. E-mail: celine.valery@canterbury.ac.nz; juliet.gerrard@canterbury.ac.nz; Tel: +64 33642987
bRiddet Institute, Massey University, Palmerston North, New Zealand
cMacDiarmid Institute, University of Canterbury, New Zealand
dCallaghan Innovation Research Limited, P.O. Box 31310, Lower Hutt 5040, New Zealand
First published on 27th February 2013
Abstract
Motifs of 7–8 amino acids were designed from the β-continuous interfaces of non-related homo-oligomeric proteins. These peptides intrinsically self-assembled into nanoarchitectures in water, while retaining some properties of their parent interfaces, especially reversibility of assembly. These results reveal a novel source of native peptide tectons.
Proteins commonly function as homo-oligomers, with many of these complexes able to dissociate and associate readily in response to changing biological conditions.1 Such subtle equilibria rely on finely tuned protein–protein interfaces, which provide the necessary structural versatility through sets of complementary non-covalent interactions.2 These protein subunits appear to be optimised by evolution to drive protein self-assembly in a reversible and controlled manner.1,3 We hypothesised that the corresponding isolated interface sequences could serve as a source of native self-assembling motifs, or tectons, while retaining some of the parent interface properties as they self-assemble.
Peptide tectons are defined as simple or minimal sequences able to spontaneously self-associate into well-defined nanoscale assemblies.4 For the past two decades, peptide chemistry has focused on exploring design principles and sources of such motifs, towards downstream applications in bionanotechnology.5 In the specific case of β-sheet-based motifs, tectons still remain difficult to design outside a few discrete families of bioinspired sequences, such as the cyclic D,L-peptides designed from the pore-forming gramicidin,6 the diphenylalanine motif inspired from the high aromatic content in amyloid sequences,7 the fragments 16–22 from the Aβ amyloid peptide,8,9 the lanreotide peptide family designed as analogues of the self-assembling peptide hormone somatostatin-14,10,11 or the surfactant peptides inspired from lipid structures.12 Novel generic sources of tectons will therefore widen the current set of available sequences with which to explore specific applications. To design the peptides, we focused on one of the simplest interface motifs, the β-continuous interface, previously estimated to represent about 15% of the reported protein oligomeric interfaces.1 β-Continuous interfaces consist of antiparallel non-covalent close contacts of two identical β-strands from the two interacting protein units (Fig. 1).
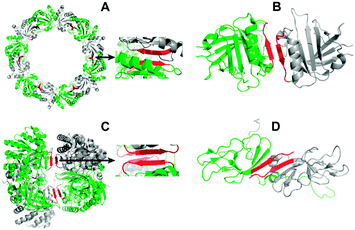 | ||
| Fig. 1 Protein homo-oligomers (pdb crystal structures for A–C, pdb solution structure for D, red: interface contacts according to the software PISA). (A) Peroxiredoxin III homo-dodecamer (1ZYE) (with zoom into the homodimer interface); (B) β-lactoglobulin homodimer (2Q39); (C) diaminopimelate decarboxylase homo-tetramer (2O0T) (with zoom into the tetramer interface); (D) umud' homodimer (1I4V). | ||
From a survey of the entire protein data bank for the key word ‘homodimer’ that resulted in >800 entries, we downsized the set of proteins to about 20 structures using the following unbiased criteria: (i) selecting native homo-oligomers with β-continuous interfaces, while excluding structures with ligands, to limit the complexity of the sets of interactions involved; (ii) excluding fragmented and complex interfaces, the latter being defined as involving multiple different secondary structure motifs; (iii) selecting only one representative interface from families of proteins with high sequence homology, to ensure sequence variety and a valid test of the hypothesis among different protein families. A statistical analysis performed on the interface sequences given by the PISA software for the final set of structures revealed that 90% contained 11 or fewer residues, 80% included at least one aromatic residue, all of the sequences contained at least 30% of hydrophobic residues, and 15% of charged residues (ESI†). The finally selected capped sequences (both N-acetylated and C-amidated to conserve the net charge of the corresponding interfaces) were chosen as representative of the entire set of homodimers examined.
The selected peptides of 7–8 residues, respectively, correspond to the β-continuous interfaces of (i) the bovine peroxiredoxin III homodimer within the dodecameric ring,13 (ii) the bovine β-lactoglobulin homodimer,14,15 (iii) the Mycobacterium tuberculosis diaminopimelate decarboxylase homotetramer,16 and (iv) the E. coli umud' protein homodimer17 (Fig. 1 and 2, Table 1).
 | ||
| Fig. 2 Left panel: capped peptides designed from the β-interfaces of bovine peroxiredoxin III (peptide 1), β-lactoglobulin (peptide 2), TB diaminopimelate decarboxylase (peptide 3), and umud' protein (peptide 4). Right panel: liquid crystalline optical textures for (A) peptide 1 at 20 mg mL−1 in water, (B) peptide 2 at 40 mg mL−1, and (C) peptide 4 at 150 mg mL−1. Magnification ×100. | ||
All the corresponding oligomers exist in solution.16,18–20 To our knowledge, none of these peptides or similar sequences have been previously probed for assembly under any condition, except for a β-lactoglobulin motif similar to peptide 2, reported to assemble under denaturing conditions involving urea.21
All four peptides formed birefringent hydrogels within the concentration range of 20–200 mg mL−1 in pure water, with liquid crystalline properties observed for at least three of them (Fig. 2A–C, ESI† for peptide 3). Electron microscopy revealed that all four peptides spontaneously self-assemble in pure water into nanostructures with elongated morphologies (Fig. 3): nanoribbons for peptide 1, bundles of nanofibrils for peptide 2, and single nanofibrils for peptides 3 and 4. Similar morphologies were obtained in high pH buffer (ESI†). These elongated nanostructures fall within the spectrum of morphologies previously reported for β-sheet self-assembling peptides.22
 | ||
| Fig. 3 Transmission electron micrographs of negatively stained (A) peptide 1 at 40 mg mL−1 in water, (B) peptide 2 at 30 mg mL−1 in water, (C) peptide 3 at 200 mg mL−1 in water, (D) peptide 4 at 100 mg mL−1 in water. Scale bars correspond to 200 nm. | ||
The four peptides exhibited an expected propensity for β-sheet secondary structures at low concentrations in pure water (0.008 mg mL−1), as shown by the far-UV circular dichroism spectra with negative minima around 215–220 nm (Fig. 4A and B). For the peptide hydrogels, FT-Raman spectra showed sharp amide I vibrations centered at 1660–1670 cm−1, which correspond to extended β-sheet networks23 (Fig. 4C).
 | ||
| Fig. 4 Peptide conformation. CD spectra at 0.008 mg mL−1 in water of peptides 1 and 2 (A), and peptides 3 and 4 (B). Panel C: FT-Raman amide I vibrations of peptides 1 and 2 at 100 mg mL−1 in water, peptides 3 and 4 at 200 mg mL−1. | ||
In all four cases, the β-sheet networks were found to dissociate upon dilution, as shown by the ThT binding assays performed by dilution from an initial concentrated sample (1 day old) (Fig. 5). TEM analysis confirmed nanostructure reversibility (ESI†). Reversibility has been previously reported for a few peptide nanostructures.5,11,24 The ThT curves further suggest a higher critical concentration of assembly for peptide 3 than for the other peptides. Interestingly, this result correlates with the type of interface from which the peptides were designed. Peptides 1, 2 and 4 correspond to homo-dimeric interfaces, with Kd in the μM range for both the peroxiredoxin18 and β-lactoglobulin homodimers,19 but in the pm range for the umud' homodimer.20 However, peptide 3 was designed from a tetrameric interface, with a Kd dimer–tetramer still under debate, although suggested in the μM range.16
 | ||
| Fig. 5 ThT binding assays upon dilution, for peptides 1 and 2 (A), and peptides 3 and 4 (B). Fluorescence intensity values just after dilution (white scatters), and 24 h after dilution (black scatters) of an initial sample equilibrated for 24 h (highest concentration). | ||
The correlation between the peptide relative behaviour in solution and the type of oligomeric interface supports that these sequences are tectons with native intrinsic assembly properties derived from the parent interfaces. The results presented herein augur well for other peptide sequences derived from protein interfaces providing a rich source of useful self-assembling tectons.
Notes and references
- I. M. A. Nooren and J. M. Thornton, J. Mol. Biol., 2003, 325, 991–1018 CrossRef CAS.
- C. Chothia and J. Janin, Nature, 1975, 256, 705–708 Search PubMed.
- S. Dey, A. Pal, P. Chakrabarti and J. Janin, J. Mol. Biol., 2010, 398, 146–160 Search PubMed.
- E. H. C. Bromley, K. Channon, E. Moutevelis and D. N. Woolfson, ACS Chem. Biol., 2008, 3, 38–50 CrossRef CAS.
- C. Valery, F. Artzner and M. Paternostre, Soft Matter, 2011, 7, 9583–9594 RSC.
- M. R. Ghadiri, J. R. Granja, R. A. Milligan, D. E. McRee and N. Khazanovich, Nature, 1993, 366, 324–327 CrossRef CAS.
- M. Reches and E. Gazit, Science, 2003, 300, 625–627 CrossRef CAS.
- K. Lu, J. Jacob, P. Thiyagarajan, V. P. Conticello and D. G. Lynn, J. Am. Chem. Soc., 2003, 125, 6391–6393 CrossRef CAS.
- I. W. Hamley, G. Cheng, V. Castelletto, S. Handschin and R. Mezzenga, Chem. Commun., 2012, 48, 3757–3759 RSC.
- C. Valery, M. Paternostre, B. Robert, T. Gulik-Krzywicki, T. Narayanan, J. C. Dedieu, G. Keller, M. L. Torres, R. Cherif-Cheikh, P. Calvo and F. Artzner, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10258–10262 CrossRef CAS.
- W. Van Grondelle, C. L. Iglesias, E. Coll, F. Artzner, M. Paternostre, F. Lacombe, M. Cardus, G. Martinez, M. Montes, R. Cherif-Cheikh and C. Valery, J. Struct. Biol., 2007, 160, 211–223 CrossRef CAS.
- S. Vauthey, S. Santoso, H. Y. Gong, N. Watson and S. G. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5355–5360 CrossRef CAS.
- Z. B. Cao, A. W. Roszak, L. J. Gourlay, J. G. Lindsay and N. W. Isaacs, Structure, 2005, 13, 1661–1664 Search PubMed.
- J. J. Adams, B. F. Anderson, G. E. Norris, L. K. Creamer and G. B. Jameson, J. Struct. Biol., 2006, 154, 246–254 Search PubMed.
- L. Vijayalakshmi, R. Krishna, R. Sankaranarayanan and M. Vijayan, Proteins: Struct., Funct., Bioinf., 2008, 71, 241–249 Search PubMed.
- S. Weyand, G. Kefala, D. Svergun and M. Weiss, J. Struct. Funct. Genomics, 2009, 10, 209–217 Search PubMed.
- A. E. Ferentz, G. C. Walker and G. Wagner, EMBO J., 2001, 20, 4287–4298 CrossRef CAS.
- S. Barranco-Medina, J. J. Lazaro and K. J. Dietz, FEBS Lett., 2009, 583, 1809–1816 Search PubMed.
- R. K. O. Apenten and D. Galani, Thermochim. Acta, 2000, 359, 181–188 Search PubMed.
- S. M. Simon, F. J. R. Sousa, R. Mohana-Borges and G. C. Walkers, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 1152–1157 CrossRef CAS.
- D. Hamada, T. Tanaka, G. G. Tartaglia, A. Pawar, M. Vendruscolo, M. Kawamura, A. Tamura, N. Tanaka and C. M. Dobson, J. Mol. Biol., 2009, 386, 878–890 CrossRef CAS.
- A. Mitraki and E. Kasotakis, in Self-Assembled Peptide Nanostructures, ed. J. Castillo, L. Sasso and W. E. Svendsen, Pan Stanford Publishing, 2013 Search PubMed.
- S. Krimm and J. Bandekar, Adv. Protein. Chem., 1986, 38, 181–364 CAS.
- K. B. Andersen, J. Castillo-Leon, M. Hedstrom and W. E. Svendsen, Nanoscale, 2011, 3, 994–998 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Acknowledgements, full experimental details, PDB survey details, birefringence of peptide 3 samples, pH influence on peptide assemblies, TEM analysis of nanostructure reversibility. See DOI: 10.1039/c3cc39052g |
| This journal is © The Royal Society of Chemistry 2013 |
