 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Kinetic trapping of the host–guest association intermediate and its transformation into a thermodynamic inclusion complex†
Oksana
Danylyuk
*a,
Vladimir P.
Fedin
b and
Volodymyr
Sashuk
a
aInstitute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warszawa, Poland. E-mail: odanylyuk@ichf.edu.pl
bNikolaev Institute of Inorganic Chemistry, Siberian Branch of the Russian Academy of Sciences, 3 Acad. Lavrentiev. Ave., 600090 Novosibirsk, Russian Federation
First published on 22nd January 2013
Abstract
The molecular recognition and self-assembly between host cucurbit[6]uril and guest adrenaline led to kinetic trapping and crystallization of the intermediate exclusion complex, which was characterized by X-ray diffraction. The crystalline kinetic complex undergoes slow spontaneous dissolution and subsequently recrystallizes as a thermodynamic inclusion complex.
In supramolecular host–guest systems it is generally assumed that the self-assembly process proceeds under thermodynamic control, quickly leading from the kinetic intermediates to the single thermodynamic product.1 The kinetic control in self-assembly of host–guest systems is quite a rarity,2 although it is omnipresent in the biological world.3 The slow kinetics of host–guest complexation may enable detection and studies of individual supramolecular transformation steps and provide valuable insight into the mechanism of molecular recognition and self-assembly phenomena. The kinetic control and trapping of intermediate products have also been observed in the self-assembly of some metallo-supramolecular,4 hydrogen-bonded5 and helicate systems.6 The isolation and identification of different meta-stable organic species within inclusion compounds have been extensively studied by Toda et al.7 Herein, we report unexpected crystallization of the kinetically controlled association complex between macrocyclic host cucurbit[6]uril (CB6) and guest adrenaline (Fig. 1). This kinetically controlled association complex undergoes slow spontaneous dissolution and subsequently recrystallizes as a thermodynamic inclusion complex. While most assembling of host–guest complexes occur too fast to be monitored without stopped flow kinetics, here the really slow complexation on the human time scale allows revelation of the structural nuances of the host–guest intermediate that preludes the inclusion. The present work provides unique crystallographic insight into host–guest systems obtained from the same building blocks (CB6 and adrenaline) on different time scales or different conditions of an assembly process.
![Chemical structure of cucurbit[6]uril (CB6) and adrenaline.](/image/article/2013/CC/c3cc37868c/c3cc37868c-f1.gif) | ||
| Fig. 1 Chemical structure of cucurbit[6]uril (CB6) and adrenaline. | ||
In the course of our investigation of the potential of cucurbiturils to act as co-crystal formers towards active pharmaceutical ingredients, we discovered that it is possible to ‘catch’ the host–guest association intermediate between CB6 and protonated adrenaline in the crystalline state. The slow kinetics of the inclusion of the adrenaline methyl group into the host hydrophobic cavity and crystallization of the kinetic intermediate allow direct insight into the complexation mechanism. Surprisingly, the crystallization of the intermediate does not act as a ‘dead-end’ in the host–guest assembly process. The needle-like crystals of the supramolecular intermediate left in the mother solution slowly dissolve and recrystallize to form prismatic crystals of the thermodynamically stable product (Fig. 2). The low solubility of both intermediate associate 1 and final inclusion complex 2 enables their rapid isolation from solution (crystallization). The different crystal morphologies of kinetic and thermodynamic products allow direct observation of the system evolution in time – the visual observation of the rapid crystallization of the kinetic intermediate, then appearance of the second crystalline phase, and finally full conversion into a thermodynamic product.
 | ||
| Fig. 2 Photographs of the crystals of CB6 complexes with adrenaline showing the system evolution in time from starting the crystallization experiment: (a) 1 hour – needles of complex 1; (b) 2 days – both needles of 1 and prisms of 2; (c) 1 week – only prisms of 2. | ||
The self-assembly of CB6 with adrenaline in aqueous solution leads to the isolation and characterization of several distinct products with different stoichiometries, depending on the time scale and crystallization conditions. The needle-like crystals immediately start growing at the liquid–liquid interface upon careful layering of aqueous hydrochloric solution of adrenaline on the aqueous solution of CB6. The magnesium chloride salt was used to improve CB6 solubility in water, as the presence of metal cations or protons is required to enhance CB6 aqueous solubility. Single-crystal X-ray diffraction revealed these first formed crystals to be the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 association complex [(C36H36N24O12)·(H2O)3·(C9H14NO3)]·Cl·11H2O (1) between the host and the cationic guest. The crystal structure of 1 shows that protonated adrenaline interacts with CB6 through one portal only without entry of its methyl group into the host cavity which is occupied by three water molecules (Fig. 3).‡ The protonated amino group of adrenaline is in close proximity to two carbonyl oxygen atoms of the host portal (N–H⋯O, 2.687 and 3.068 Å) suitable for ion–dipole and hydrogen bonding interactions. Additionally, the hydrogen bond is donated to one of the water molecules inside the cavity (N–H⋯O, 2.910 Å). The methyl group of the guest perches above the portal and is unfavorably exposed to portal carbonyl oxygen atoms and water molecules in the crystal lattice. Evidently, the ion–dipole and hydrogen bonding anchoring of the protonated amino group provides enough stabilization to overcome the penalty of one methyl group exposure to the aqueous phase. This stabilization is sufficient to enable the crystallization of the intermediate. Surprisingly, in the solid state another portal of CB6 interacts neither with protonated adrenaline nor magnesium ions present in the mother solution, but is closed with the adjacent CB6 by means of weak CH⋯O interactions between methine and methylene groups of one CB6 skeleton and carbonyl oxygen atoms from the free portal of its neighbor (Fig. S1, ESI†).
1 association complex [(C36H36N24O12)·(H2O)3·(C9H14NO3)]·Cl·11H2O (1) between the host and the cationic guest. The crystal structure of 1 shows that protonated adrenaline interacts with CB6 through one portal only without entry of its methyl group into the host cavity which is occupied by three water molecules (Fig. 3).‡ The protonated amino group of adrenaline is in close proximity to two carbonyl oxygen atoms of the host portal (N–H⋯O, 2.687 and 3.068 Å) suitable for ion–dipole and hydrogen bonding interactions. Additionally, the hydrogen bond is donated to one of the water molecules inside the cavity (N–H⋯O, 2.910 Å). The methyl group of the guest perches above the portal and is unfavorably exposed to portal carbonyl oxygen atoms and water molecules in the crystal lattice. Evidently, the ion–dipole and hydrogen bonding anchoring of the protonated amino group provides enough stabilization to overcome the penalty of one methyl group exposure to the aqueous phase. This stabilization is sufficient to enable the crystallization of the intermediate. Surprisingly, in the solid state another portal of CB6 interacts neither with protonated adrenaline nor magnesium ions present in the mother solution, but is closed with the adjacent CB6 by means of weak CH⋯O interactions between methine and methylene groups of one CB6 skeleton and carbonyl oxygen atoms from the free portal of its neighbor (Fig. S1, ESI†).
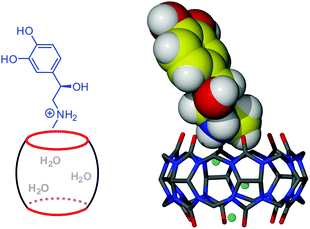 | ||
| Fig. 3 Schematic view and crystal structure of the kinetically controlled association intermediate complex 1 between CB6 and protonated adrenaline (three water molecules inside the cavity are shown as blue balls). | ||
While ion–dipole anchoring of the guest ammonium group at the CB6 portal is sufficient to drive the rapid formation and trapping of the intermediate exclusion complex, the hydrophobic effect slowly pulls the intermediate state into a stable inclusion complex. The crystals of the kinetically trapped 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 exclusion complex did not persist in solution and were slowly transformed upon dissolving and again crystallization into stable inclusion complex [(C36H36N24O12)·(C9H14NO3)2]·2Cl·15H2O (2) of 1
1 exclusion complex did not persist in solution and were slowly transformed upon dissolving and again crystallization into stable inclusion complex [(C36H36N24O12)·(C9H14NO3)2]·2Cl·15H2O (2) of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 host–guest stoichiometry.§ The main driving force for the inclusion of the organic residue is the hydrophobic effect due to removal of three high-energy water molecules from the CB6 cavity.8 In the thermodynamically stable inclusion complex 2 both CB6 portals complex adrenaline in a similar manner, namely protonated amino groups interact with CB6 carbonyl oxygen atoms (N–H⋯O, 2.867 and 2.869 Å for one adrenaline molecule, 2.836 and 2.695 Å for another) and two methyl groups are included into the host cavity instead of three water molecules (Fig. 4). The appearance of the prismatic crystals of complex 2 among needles of 1 can be noticed from several hours to several days after starting the crystallization experiment, depending on the crystallization conditions such as the rate of mixing the solutions and the type of crystallization vial or mechanical stress (shaking or stirring accelerates transformation). The full transformation of crystalline complex 1 into crystalline complex 2 takes from several days to several weeks, also depending on the crystallization conditions. It should be remembered that crystallization itself is a highly specific and complex event during which kinetics competes with thermodynamics.9 So, the time needed for transformation from kinetic complex 1 into thermodynamic complex 2 includes not only time for ingression of the organic residue into the host cavity but also dissolution and recrystallization steps that can be time-consuming. We tried to monitor the composition of the mother solution during the transformation by 1H NMR spectroscopy. The main signals observed in the spectra, taken at different times, come from uncomplexed adrenaline and CB6 (ESI†). However, some additional signals of very low intensity appear upfield that may indicate the formation of inclusion complexes of different stoichiometries. Obviously, the transformation 1–2 occurs at very low concentrations almost non-detectable at the NMR scale by partial slow dissolving of the form 1 followed by recrystallization of form 2. Presumably, between dissolution of the exclusion complex and crystallization of the inclusion complex the elusive ‘flip-flop’ entry of methyl groups into the macrocyclic cavity takes place.
2 host–guest stoichiometry.§ The main driving force for the inclusion of the organic residue is the hydrophobic effect due to removal of three high-energy water molecules from the CB6 cavity.8 In the thermodynamically stable inclusion complex 2 both CB6 portals complex adrenaline in a similar manner, namely protonated amino groups interact with CB6 carbonyl oxygen atoms (N–H⋯O, 2.867 and 2.869 Å for one adrenaline molecule, 2.836 and 2.695 Å for another) and two methyl groups are included into the host cavity instead of three water molecules (Fig. 4). The appearance of the prismatic crystals of complex 2 among needles of 1 can be noticed from several hours to several days after starting the crystallization experiment, depending on the crystallization conditions such as the rate of mixing the solutions and the type of crystallization vial or mechanical stress (shaking or stirring accelerates transformation). The full transformation of crystalline complex 1 into crystalline complex 2 takes from several days to several weeks, also depending on the crystallization conditions. It should be remembered that crystallization itself is a highly specific and complex event during which kinetics competes with thermodynamics.9 So, the time needed for transformation from kinetic complex 1 into thermodynamic complex 2 includes not only time for ingression of the organic residue into the host cavity but also dissolution and recrystallization steps that can be time-consuming. We tried to monitor the composition of the mother solution during the transformation by 1H NMR spectroscopy. The main signals observed in the spectra, taken at different times, come from uncomplexed adrenaline and CB6 (ESI†). However, some additional signals of very low intensity appear upfield that may indicate the formation of inclusion complexes of different stoichiometries. Obviously, the transformation 1–2 occurs at very low concentrations almost non-detectable at the NMR scale by partial slow dissolving of the form 1 followed by recrystallization of form 2. Presumably, between dissolution of the exclusion complex and crystallization of the inclusion complex the elusive ‘flip-flop’ entry of methyl groups into the macrocyclic cavity takes place.
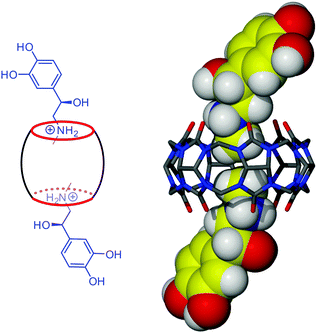 | ||
| Fig. 4 Schematic view and crystal structure of the thermodynamic inclusion complex 2 between CB6 and protonated adrenaline. | ||
When adrenaline is added to magnesium chloride CB6 solution in non-protonated form the direct crystallization of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 inclusion complex is observed; however, the resulting complex [(C36H36N24O12)·(C9H13NO3)2]·18H2O (3)¶ of CB6 with neutral adrenaline crystallizes in a different space group and is characterized by a different supramolecular framework than complex 2 (ESI†). In order to explore the influence of the supramolecular reaction medium on the host–guest assembly process, an attempt to get the CB6–adrenaline complex in the absence of metal ions has been made. When using 5 M hydrochloric acid instead of magnesium chloride solution for dissolving CB6 and adrenaline, the inclusion complex [(C36H36N24O12)·(C18H26N2O5)2]·6Cl·2H3O·20H2O (4)|| of CB6 with ‘dimerized’ adrenaline crystallized after standing for several days at room temperature. Obviously, in strongly acidic conditions, the condensation reaction via an electrophilic substitution mechanism between two adrenaline molecules takes place10 and the product of this reaction, ‘dimerized’ adrenaline, is complexed by CB6 present in the solution (Fig. 5).
2 inclusion complex is observed; however, the resulting complex [(C36H36N24O12)·(C9H13NO3)2]·18H2O (3)¶ of CB6 with neutral adrenaline crystallizes in a different space group and is characterized by a different supramolecular framework than complex 2 (ESI†). In order to explore the influence of the supramolecular reaction medium on the host–guest assembly process, an attempt to get the CB6–adrenaline complex in the absence of metal ions has been made. When using 5 M hydrochloric acid instead of magnesium chloride solution for dissolving CB6 and adrenaline, the inclusion complex [(C36H36N24O12)·(C18H26N2O5)2]·6Cl·2H3O·20H2O (4)|| of CB6 with ‘dimerized’ adrenaline crystallized after standing for several days at room temperature. Obviously, in strongly acidic conditions, the condensation reaction via an electrophilic substitution mechanism between two adrenaline molecules takes place10 and the product of this reaction, ‘dimerized’ adrenaline, is complexed by CB6 present in the solution (Fig. 5).
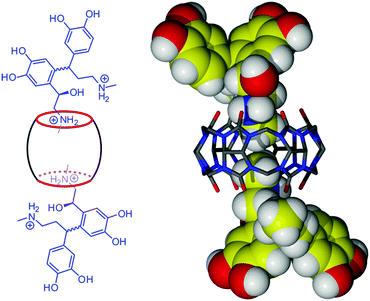 | ||
| Fig. 5 Schematic view and crystal structure of the inclusion complex 4 between CB6 and dimerized adrenaline. | ||
In summary, the supramolecular reaction between host cucurbit[6]uril and guest adrenaline in aqueous medium proceeds through different pathways and leads to diverse assembled solid-state structures, depending on the conditions of the self-assembly process. The kinetic control was observed in the case of CB6 assembly with protonated adrenaline in magnesium chloride aqueous solution due to strong cation–dipole interaction between the host and the cationic guest. This kinetically regulated pathway involves the sequential crystallization of the trapped intermediate exclusion complex and the final thermodynamic inclusion complex. The isolation and structural characterization of both complexes enabled us to reproduce each step of the inclusion process, thus, to ultimately confirm the predicted two-step complexation model for cucurbit[6]uril host–quest systems with organic cations.11 In the absence of strong non-covalent interactions between the host and the guest, the self-assembly of CB6 with neutral adrenaline proceeds under thermodynamic control, leading directly to inclusion complex formation. In strongly acidic conditions necessary for CB6 solubilization in the absence of metal ions, the obtained crystalline compound was found to be the inclusion complex of CB6 with dimerized adrenaline – the condensation product of two adrenaline molecules. We expect that the high degree of host–guest structural diversity achieved through simple manipulation of building blocks and/or self-assembly pathways will inspire new applications of supramolecular chemistry in non-equilibrium systems,12 resembling those of the biological world.
The project was funded by the National Science Centre (SONATA grant DEC-2011/03/D/ST5/05486).
Notes and references
- J. M. Lehn, Supramolecular Chemistry: Concepts and Perspectives, VCH, Weinheim, 1995 Search PubMed.
- J. D. Badjic, S. J. Cantrill and J. F. Stoddart, J. Am. Chem. Soc., 2004, 126, 2288 CrossRef CAS; A. S. M. Dyck, U. Kisiel and C. Bohne, J. Phys. Chem. B, 2003, 107, 11652 CrossRef; P. Mukhopadhyay, P. Y. Zavalij and L. Isaacs, J. Am. Chem. Soc., 2006, 128, 14093 CrossRef; T. Oshikiri, Y. Takashima, H. Yamaguchi and A. Harada, J. Am. Chem. Soc., 2005, 127, 12186 CrossRef; J. C. Chapin, M. Kvasnica and B. W. Purse, J. Am. Chem. Soc., 2012, 134, 15000 CrossRef.
- W.-J. Chung, J.-W. Oh, K. Kwak, B. Y. Lee, J. Meyer, E. Wang, A. Hexemer and S.-W. Lee, Nature, 2011, 478, 364 CrossRef CAS; C. M. Dobson, Nature, 2003, 426, 884 CrossRef; P. A. Jennings and P. E. Wright, Science, 1993, 262, 892 Search PubMed.
- S. Tashira, M. Tominaga, T. Kusukawa, M. Kawano, S. Sakamoto, K. Yamaguchi and M. Fujita, Angew. Chem., Int. Ed., 2003, 42, 3267 CrossRef CAS; V. M. Cangelosi, T. G. Carter, L. N. Zakharov and D. W. Johnson, Chem. Commun., 2009, 5606 RSC.
- T. F. A. de Greef, G. B. W. L. Ligthart, M. Lutz, A. L. Spec, E. W. Meijer and R. P. Sijbesma, J. Am. Chem. Soc., 2008, 130, 5479 CrossRef CAS.
- A. Lohr, M. Lysetska and F. Wurthner, Angew. Chem., Int. Ed., 2005, 44, 5071 CrossRef CAS; Q. Gan, Y. Ferrand, C. Bao, B. Kauffmann, A. Greland, H. Jiang and I. Huc, Science, 2011, 331, 1172 CrossRef; Q. Gan, Y. Ferrand, N. Chandramouli, B. Kauffmann, C. Aube, D. Dubreuil and I. Huc, J. Am. Chem. Soc., 2012, 134, 15656 CrossRef.
- Z. Urbanczyk-Lipkowska, K. Yoshizawa, S. Toyota and F. Toda, CrystEngComm, 2003, 5, 114 RSC; H. Hosomi, S. Ohba, K. Tanaka and F. Toda, J. Am. Chem. Soc., 2000, 122, 1818 CrossRef CAS; R. Boese, J. Benet-Buchholz, A. Stanger, K. Tanaka and F. Toda, Chem. Commun., 1999, 319 RSC.
- W. M. Nau, M. Florea and K. I. Assaf, Isr. J. Chem., 2011, 51, 559 CrossRef CAS; F. Biedermann, V. D. Uzunova, O. A. Scherman, W. M. Nau and A. D. Simone, J. Am. Chem. Soc., 2012, 134, 15318 CrossRef.
- G. R. Desiraju, Nat. Mater., 2002, 1, 77 CrossRef CAS.
- J. E. Forrest, S. Kasparek, R. A. Heacock and T. P. Forrest, Can. J. Chem., 1969, 47, 2118 CrossRef CAS.
- R. Hoffmann, W. Knoche, C. Fenn and H. J. Buschmann, J. Chem. Soc., Faraday Trans., 1994, 90, 1507 RSC; C. Marquez and W. M. Nau, Angew. Chem., Int. Ed., 2001, 40, 3155 CrossRef CAS; C. Marquez, R. R. Hudgins and W. M. Nau, J. Am. Chem. Soc., 2004, 126, 5806 CrossRef.
- S. C. Warren, O. Guney-Altay and B. A. Grzybowski, J. Phys. Chem. Lett., 2012, 3, 2103 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, NMR spectra, additional structural plots and X-ray crystallographic files. CCDC 889594 (1), 889593 (2), 908048 (3) and 908049 (4). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c3cc37868c |
‡ Crystal data for 1: C45H78O29ClN25, Mr = 1468.8, colourless, 0.65 × 0.10 × 0.10, orthorhombic, space group P212121, a = 12.6226(2), b = 15.7655(3), c = 31.0112(6) Å, V = 6171.2 (2) Å3, Z = 4, ρcalc = 1.581 g cm−3, μ(MoKα) = 0.173 mm−1, θmax = 28.3°, 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 586 reflections measured, 14 586 reflections measured, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683 unique, 983 parameters, 22 restraints, R = 0.081, wR = 0.175 (R = 0.116, wR = 0.192 for all data), GooF = 1.03. Flack parameter = −0.05(14). CCDC 889594. 683 unique, 983 parameters, 22 restraints, R = 0.081, wR = 0.175 (R = 0.116, wR = 0.192 for all data), GooF = 1.03. Flack parameter = −0.05(14). CCDC 889594. |
§ Crystal data for 2: C54H94O33Cl2N26, Mr = 1706.3, colourless, 0.40 × 0.30 × 0.20, orthorhombic, space group P212121, a = 12.8019(2), b = 20.4511(4), c = 27.5592(5) Å, V = 7215.3(2) Å3, Z = 4, ρcalc = 1.556 g cm−3, μ(MoKα) = 0.200 mm−1, θmax = 27.9°, 81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 552 reflections measured, 17 552 reflections measured, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 147 unique, 1107 parameters, R = 0.078, wR = 0.193 (R = 0.084, wR = 0.198 for all data), GooF = 1.11. Flack parameter = 0.10(10). CCDC 889593. 147 unique, 1107 parameters, R = 0.078, wR = 0.193 (R = 0.084, wR = 0.198 for all data), GooF = 1.11. Flack parameter = 0.10(10). CCDC 889593. |
¶ Crystal data for 3: C54H98O36N26, Mr = 1687.6, yellow, 0.60 × 0.20 × 0.15, monoclinic, space group P21, a = 12.4610(2), b = 15.7685(2), c = 19.4788(3) Å, β = 102.946(2)°, V = 3730.1(1) Å3, Z = 2, ρcalc = 1.463 g cm−3, μ(CuKα) = 1.082 mm−1, θmax = 71.5°, 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685 reflections measured, 14 685 reflections measured, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 194 unique, 1396 parameters, R = 0.077, wR = 0.223 (R = 0.086, wR = 0.236 for all data), GooF = 1.05. Flack parameter = −0.1(2). CCDC 908048. 194 unique, 1396 parameters, R = 0.077, wR = 0.223 (R = 0.086, wR = 0.236 for all data), GooF = 1.05. Flack parameter = −0.1(2). CCDC 908048. |
|| Crystal data for 4: C72H134O44Cl6N28, Mr = 2308.9, colourless, 0.40 × 0.35 × 0.20, triclinic, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 12.7909(2), b = 14.3424(2), c = 15.1838(3) Å, α = 79.138(1)°, β = 72.676(1)°, γ = 73.236(1)°, V = 2530.04(1) Å3, Z = 1, ρcalc = 1.526 g cm−3, μ(MoKα) = 0.275 mm−1, θmax = 27.9°, 36 , a = 12.7909(2), b = 14.3424(2), c = 15.1838(3) Å, α = 79.138(1)°, β = 72.676(1)°, γ = 73.236(1)°, V = 2530.04(1) Å3, Z = 1, ρcalc = 1.526 g cm−3, μ(MoKα) = 0.275 mm−1, θmax = 27.9°, 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 319 reflections measured, 11 319 reflections measured, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 915 unique, 787 parameters, R = 0.071, wR = 0.184 (R = 0.085, wR = 0.195 for all data), GooF = 0.96. CCDC 908049. 915 unique, 787 parameters, R = 0.071, wR = 0.184 (R = 0.085, wR = 0.195 for all data), GooF = 0.96. CCDC 908049. |
| This journal is © The Royal Society of Chemistry 2013 |
