 Open Access Article
Open Access ArticleBranched peptideboronic acids (BPBAs): a novel mode of binding towards RNA†
Wenyu
Zhang‡
,
David I.
Bryson‡
,
Jason B.
Crumpton‡
,
Jessica
Wynn
and
Webster L.
Santos
*
Department of Chemistry and Virginia Tech Center for Drug Discovery, Virginia Tech, Blacksburg, Virginia 24061, USA. E-mail: santosw@vt.edu; Fax: +1 540 231 3255; Tel: +1 540 231 5041
First published on 7th February 2013
Abstract
We report branched peptideboronic acids (BPBAs) that bind to RRE IIB from an on-bead high-throughput screening of a 3.3.4-library (46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656 compounds). We demonstrate that boronic acids are tunable moieties that afford a novel binding mode towards RNA.
656 compounds). We demonstrate that boronic acids are tunable moieties that afford a novel binding mode towards RNA.
The human immunodeficiency virus type 1 (HIV-1) is regarded as the prototype member of the lentivirus subfamily of retroviruses, and it is the causative agent of the worldwide health problem, Acquired Immune Deficiency Syndrome (AIDS). In the past few decades, more than 20 drugs have become available for treatment of HIV-1 infection;1 at present, favored treatment focuses on combination therapy using HIV-1 reverse transcriptase (RT), protease, and gp41 inhibitors. Despite the fact that highly active antiretroviral therapy (HAART) has obtained notable successes in reducing plasma viral loads to undetectable levels, HAART fails to completely eliminate the virus from the body due to the remaining chronically HIV-infected CD4+ T cells, which contain the integrated but transcriptionally dormant HIV provirus.2 In addition, the emergence of drug-resistant viruses has been reported in patients receiving HAART.1 In order to keep pace with the rapidly evolving HIV-1, there is a great need for development of drugs that target novel viral mechanisms that are genetically well-conserved and less prone to mutation under selective pressure.
The highly structured HIV-1 Rev response element (RRE), which is a span of ∼240-nucleotides located in the env gene of all singly spliced and unspliced HIV-1 transcripts, is an example of an extremely well-conserved sequence of RNA across different HIV-1 isolates and plays an essential role in RNA replication by interaction with the Rev protein.3 It has been demonstrated that proviral colonies without the rev gene have no replicative abilities, and in the absence of rev protein, the stability of unspliced mRNA is decreased.4 While the details of the RRE–Rev export pathway is being investigated, some steps have been identified. It is known that of the singly spliced transcripts and multiply spliced transcripts, only multiply spliced transcripts can be exported to the cytoplasm and translated to their corresponding proteins including Rev.5 Once Rev is expressed, it is imported into the nucleus where it binds cooperatively to RRE.3 In particular, the stem-loop IIB of RRE (RRE IIB) has been recognized as the high affinity site where Rev initially binds.6 The resulting Rev–RRE ribonucleoprotein complex binds the host Crm1 and is then shuttled out of the nucleus through the nuclear pore after the larger complex binds to Ran-GTP.7 Since this cooperative binding allows for the export of full-length and singly spliced transcripts, the Rev–RRE export pathway has become a high profile drug target for its critical role in proliferation of HIV-1.8
In continuation of our effort toward developing molecules that target the tertiary structure of RNA, we focused our attention on RRE and envisioned utilizing RNA–ligand interactions that are outside the typical canonical mode of binding. We previously demonstrated that branching in peptideligands provides strong multivalent interactions with another HIV-1 related RNA, the transactivation response element (TAR).9 These branched peptides (BPs) displayed no cytotoxicity, provided excellent cell permeability, and bound to TAR in the submicromolar regime. Herein, we report the discovery and biophysical characterization of branched peptideboronic acids (BPBAs) as medium-sized ligands that bind to the tertiary structure of HIV-1 RRE IIB. Our investigations suggest that the boronic acid moiety plays a pivotal role in increasing binding affinity.
We embarked on a strategy to improve the selectivity and binding affinity to the RNA target through the incorporation of unnatural amino acid side chains featuring the boronic acid functional group. Boronic acids have been used in various applications including biomolecules. For example, boronic acids anchored to a cellulose polymer support was first used to separate and purify RNA.10Peptides displaying boronic acid moieties have been demonstrated to form reversible covalent bonds with alizarin and glucose,11 in addition to being utilized as potent protease inhibitors.12 Furthermore, boron-containing compounds are well tolerated in vivo as is evident from the FDA approval of the first boron-containing drug, Bortezomib (Velcade, Fig. 1A).13 Another boron-containing small molecule, Tavaborole (AN2690), is currently in phase III clinical trials for treatment of onychomycosis, and its mode of action involves trapping the 2′- and 3′-oxygen atoms of the terminal adenosine in leucyl-tRNA synthetase as a boronate adduct (Fig. 1A).14 Since peptidyl boronic acids have not been investigated to target RNA, we hypothesized that we can capitalize on the empty p-orbital of boron by forming a reversible covalent bond to the Lewis bases in RNA. In particular, the 2′-hydroxyl group in RNA is well-suited as an electron donor towards boron.14 As a result, the strategy could promote selectivity for RNA over DNA and also boost affinity to the RNA target due to the formation of a reversible covalent bond (Fig. 1B). The boronic acid moiety as an unnatural side chain in peptides increases the complexity and diversity of peptide libraries. Further, the boronic acid functional group is an excellent candidate for probing RNA–ligand binding interactions that are atypical of canonical mode of binding.
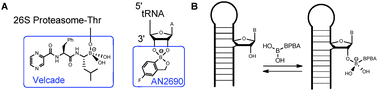 | ||
| Fig. 1 (A) Therapeutically relevant boronic acid derivatives that bind to protein or RNA targets. (B) Possible reversible covalent bond formation between Lewis bases in RNA and boronic acid moiety of branched peptides. | ||
A BPBA peptide library was synthesized on Tentagel beads by split and pool synthesis. The library was prepared9b such that there were three variable amino acid positions at both the N- and C-termini (A1–A3 and A4–A6, respectively), and each variable position was composed of six possible side chains (Fig. 2A). Each of the six possible side chains was chosen for its potential to interact with the RRE IIB target RNA. The specific amino acids for each type of binding interaction were chosen by random assignment in order to preclude bias in the library design. In position A1, for example, we selected amino acids with functional groups that can interact with the RNA through hydrophobic interaction (Val), electrostatic attraction (Lys), hydrogen bonding (Ser), pi-stacking (Trp), and reversible covalent bonding between boron (KBBA/FBPA) and a Lewis base presented by the RNA target (Fig. 2B). We chose to incorporate two boron-containing side chains at each variable position in the library to examine whether RRE IIB had a preference in the side chain length or boron Lewis acidity. Hence, the 3.3.4 branched peptideboronic acid library was composed of 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656 possible amino acid sequences linked to the bead by a photocleavable linker (3-amino-3-(2-nitrophenyl) propionic acid, ANP). Tyr was included at position A7 as a convenient spectroscopic handle in quantifying peptide concentrations.
656 possible amino acid sequences linked to the bead by a photocleavable linker (3-amino-3-(2-nitrophenyl) propionic acid, ANP). Tyr was included at position A7 as a convenient spectroscopic handle in quantifying peptide concentrations.
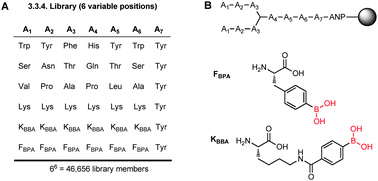 | ||
| Fig. 2 (A) 3.3.4-Branched peptideboronic acid library, (B) structure of BPBA library and unnatural amino acids bearing boronic acid groups. | ||
The BPBA library was subjected to on-bead high throughput screening9a against DY547 labeled HIV-1 RRE IIB RNA. During the screening process, the beads were first pretreated with an excess of both bovine serum albumin and competitor tRNA to minimize non-specific binding. Specific binding of the target RNA to peptide resulted in increased fluorescence of the bead, which was monitored by fluorescence microscopy. Eleven beads were selected as possible hits. These peptides were photocleaved via UV irradiation and then sequenced by MALDIMS-MS analysis.15 Hit compounds were resynthesized for further biophysical characterizations.
The sequence and dissociation constant of the hit BPBAs are shown in Table 1. Using standard dot blot assay techniques, 32P-labeled RRE IIB was titrated with increasing concentration of BPBAs (see ESI† for details).9b Three peptides that showed no binding were identified as false positives. The hits contained varying numbers of boronic acid residues ranging from 0 to 5. In particular, three peptides (BPBA1, BPBA2 and BPBA3) had low micromolar binding affinities (1.4, 3.3 and 8.7 μM, respectively) and were investigated further. To further confirm that the dissociation constants determined from the dot blot assay were reliable, electrophoretic mobility shift assay (EMSA) was performed with BPBA1, BPBA2 and BPBA3. The dissociation constants determined through EMSA (0.3 ± 0.1, 0.6 ± 0.2 and 4.6 ± 2.4 μM, respectively) were comparable to the low micromolar results obtained via dot blot assay and were consistent in rank order of affinity (ESI†). The slight discrepancy with the Kd values may be a result of nonspecific interactions between the peptide and nitrocellulose. Nevertheless, these sequences contained boronic acid residues with boron containing amino acids present in positions A1, A3 and A4. Interestingly, no boron containing amino acids were found at positions A5 or A6 in any hit sequence nor was there any correlation between the number of boronic acid moieties and the resulting binding affinities. Further, it was observed that the longer, more Lewis acidic residue of KBBA was the more preferred boron-containing amino acid over the less electron demanding FBPA residue. This result suggests that boron containing side chains can provide non-canonical, alternative, and favorable binding interaction with the RNA target (vide infra).
| Peptide | Sequencea | K d (μM) | MW (g mol−1) |
|---|---|---|---|
| a * = lysine branching unit. Each value is an average of at least three experiments. | |||
| BPBA1 | (WKK)2 * KBBAYWY | 1.4 ± 0.4 | 1817.99 |
| BPBA2 | (KBBAKFBPA)2 * KBBAKKY | 3.3 ± 1.2 | 2031.10 |
| BPBA3 | (FBPAYFBPA)2 * NKSY | 8.7 ± 2.3 | 1727.78 |
| BPBA4 | (KKKBBA)2 * FBPATSY | 26.8 ± 4.4 | 1751.98 |
| BPBA5 | (KKBBAF)2 * KKWY | 27.2 ± 6.9 | 1853.04 |
| BPBA6 | (WYK)2 * PTWY | 28.5 ± 4.4 | 1646.34 |
| BPBA7 | (KKBBAK)2 * KLKY | 58.4 ± 4 | 1742.09 |
| BPBA8 | (KBBAYK)2 * HKKY | 86.5 ± 10 | 1836.04 |
Further sequence analysis indicated that a Lys residue was most preferred in a majority of the positions A1–A6. A preference for positively charged side chains, however, is not unexpected because the Rev protein binds RRE IIB through the polyarginine nuclear localization signal (NLS) located in its N-terminal region.16 The positively charged Lys side chains in our hits may function similarly to the Arg residues of the Rev NLS and likely provide the necessary electrostatic attraction to the negatively charged phosphate backbone of RRE IIB. However, it is noteworthy that the number of Lys residues in the hit sequences did not have a significant influence in the binding affinity. For example, an increase in the net positive charge did not result in increased binding affinity (compare BPBA1 to BPBA7 and BPBA8). Taken together, the data suggests that the dissociation constants of the hit peptides are not entirely due to electrostatic attraction. Further analysis shows that aliphatic residues were the least preferred side chains in our pool of hit compounds. This suggests that modes of binding other than hydrophobic interactions are generally more beneficial for strong binding between these BPBAs and RRE IIB.
To determine the role of the boronic acid functional group in binding to RRE IIB RNA, we designed control peptide variants of BPBA1 and BPBA3 wherein the boronic acid moiety was removed. As much as possible, modifications were performed with minimal structural perturbation. First, the boronic acid moiety in the para position of BPBA1 was replaced with a hydrogen atom to afford BPBA1.1. The change was accompanied by an approximate 6-fold increase in the observed Kd value (1.4 vs. 8.2 μM, Fig. 3). Although the precise role of the boronic acid moiety in binding is currently unknown, the decrease in affinity suggests that the boronic acid functional group interacts with RRE. The single boronic acid moiety in BPBA1 contributes a modest fraction of binding affinity with RRE IIB. In this case, electrostatic attraction accounts for the majority of interaction with the RNA provided by the high density of Lys residues. In order to investigate the hypothesis that the Lewis acidity of boron can increase binding affinity, we synthesized a fluorinated analog, BPBA1.2, wherein fluorine was placed ortho to the boronic acid at position A4. The electron withdrawing ability of fluorine results in increased Lewis acidity and is expected to facilitate complexation. To our delight, when the Kd was determined by dot blot assay, the binding affinity improved to 0.8 ± 0.1 μM, suggesting an induction of stronger interaction with RRE through the boronic acid moiety. This result supports the notion that the binding affinity of BPBAs can be tuned by manipulating the Lewis acidity of the boronic acid and is consistent with the observation that more electrophilic boronic acids are more acidic as they can form stable Lewis acid–base complexes.17
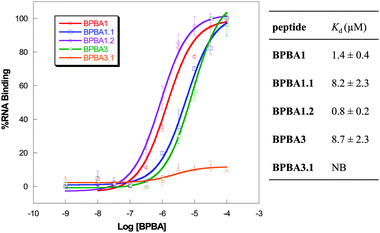 | ||
| Fig. 3 Binding curves and dissociation constants of boron and non-boron containing branched peptides using dot blot assay with RRE IIB RNA. NB = no binding. | ||
We also investigated the effect of the number of boronic acid moieties in hit peptideBPBA3, which contains four boron atoms. When a boronic acid-free analog of BPBA3 was synthesized (BPBA3.1), nearly all binding was abolished (Fig. 3). The drastic loss of binding with BPBA3.1 is likely due to the high density of boron containing side chains because the major source of binding interaction, presumably Lewis acid–base complexation, is lost. These results indicate that boronic acid side chains can be utilized in peptides to boost binding affinity with a highly structured RNA target. Further, such interaction represents an additional and unique mode of binding that increases the repertoire of RNA binding motifs.
In conclusion, we generated a BPBA library that was designed to interrogate the effect of boronic acids when screened against the tertiary structure of an RNA target: RRE IIB. High throughput screening and biophysical characterization of hit compounds resulted in peptides with binding affinities in the low micromolar range, wherein the presence of boronic acid groups introduced a novel, alternative mode of interaction. Structure–activity relationship studies demonstrated that the binding affinity of BPBAs can be tuned by changing the electronic property of the boron center. This result highlights that boronic acid moieties can impact binding towards RNA, and this strategy is applicable to other systems since boronic acids can be readily engineered into peptides.
We thank the Virginia Tech mass spectrometry incubator for the use of MALDI-TOF instrumentation and Dr Keith Ray for his invaluable help with the acquisition of mass spectra. This work was supported by NIH (RO1 GM093834).
Notes and references
- M. Baba, Antiviral Res., 2006, 71, 301–306 CrossRef CAS.
- M. Stevens, E. D. Clercq and J. Balzarini, Med. Res. Rev., 2006, 26, 595–625 CrossRef CAS.
- M. H. Malim, J. Hauber, S. Y. Le, J. V. Maizel and B. R. Cullen, Nature, 1989, 338, 254–257 CrossRef CAS.
- B. K. Felber, M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland and G. N. Pavlakis, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 1495–1499 CrossRef CAS.
- V. W. Pollard and M. H. Malim, Annu. Rev. Microbiol., 1998, 52, 491–532 CrossRef CAS.
- (a) M. H. Malim, L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber and B. R. Cullen, Cell, 1990, 60, 675–683 CrossRef CAS; (b) X. J. Huang, T. J. Hope, B. L. Bond, D. McDonald, K. Grahl and T. G. Parslow, J. Virol., 1991, 65, 2131–2134 CAS.
- (a) M. Fornerod, M. Ohno, M. Yoshida and I. W. Mattaj, Cell, 1997, 90, 1051–1060 CrossRef CAS; (b) P. Askjaer, T. H. Jensen, J. Nilsson, L. Englmeier and J. Kjems, J. Biol. Chem., 1998, 273, 33414–33422 CrossRef CAS.
- A. I. Dayton, J. G. Sodroski, C. A. Rosen, W. C. Goh and W. A. Haseltine, Cell, 1986, 44, 941–947 CrossRef CAS.
- (a) D. I. Bryson, W. Zhang, W. K. Ray and W. L. Santos, Mol. BioSyst., 2009, 5, 1070–1073 RSC; (b) D. I. Bryson, W. Zhang, P. M. McLendon, T. M. Reineke and W. L. Santos, ACS Chem. Biol., 2012, 7, 210–217 CrossRef CAS.
- E. C. Moore, D. Peterson, L. Y. Yang, C. Y. Yeung and N. F. Neff, Biochemistry, 1974, 13, 2904–2907 CrossRef CAS.
- G. Springsteen and B. Wang, Chem. Commun., 2001, 1608–1609 RSC.
- (a) D. H. Kinder and J. A. Katzenellenbogen, J. Med. Chem., 1985, 28, 1917–1925 CrossRef CAS; (b) E. S. Priestley, I. De Lucca, B. Ghavimi, S. Erickson-Viitanen and C. P. Decicco, Bioorg. Med. Chem. Lett., 2002, 12, 3199–3202 CrossRef CAS.
- (a) R. C. Kane, P. F. Bross, A. T. Farrell and R. Pazdur, Oncologist, 2003, 8, 508–513 CrossRef; (b) S. J. Baker, C. Z. Ding, T. Akama, Y. K. Zhang, V. Hernandez and Y. Xia, Future Med. Chem., 2009, 1, 1275–1288 CrossRef CAS.
- F. L. Rock, W. Mao, A. Yaremchuk, M. Tukalo, T. Crepin, H. Zhou, Y. K. Zhang, V. Hernandez, T. Akama, S. J. Baker, J. J. Plattner, L. Shapiro, S. A. Martinis, S. J. Benkovic, S. Cusack and M. R. Alley, Science, 2007, 316, 1759–1761 CrossRef CAS.
- J. B. Crumpton, W. Zhang and W. L. Santos, Anal. Chem., 2011, 83, 3548–3554 CrossRef CAS.
- (a) J. L. Battiste, H. Mao, N. S. Rao, R. Tan, D. R. Muhandiram, L. E. Kay, A. D. Frankel and J. R. Williamson, Science, 1996, 273, 1547–1551 CAS; (b) M. D. Daugherty, B. Liu and A. D. Frankel, Nat. Struct. Mol. Biol., 2010, 17, 1337–1342 CrossRef CAS.
- D. Hall, Boronic Acids: Preparation, Applications in Organic Synthesis, Medicine and Materials, Wiley-VCH GmbH & Co., Weinheim, 2nd edn, 2011 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures, dot blot assays, EMSA, HPLC purification, characterization and MALDI/MS of BPBAs. See DOI: 10.1039/c3cc00243h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2013 |
