 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecularly imprinted polymers for the extraction of imiquimod from biological samples using a template analogue strategy
Panagiotis
Manesiotis
*a,
Sakina
Kashani
b and
Peter
McLoughlin
b
aSchool of Chemistry and Chemical Engineering, Queen's University Belfast, David Keir Building, Stranmillis Road, BT9 5AG, Belfast, UK. E-mail: p.manesiotis@qub.ac.uk; Fax: +44 (0)28 9097 6524; Tel: +44 (0)28 9097 4515
bPharmaceutical and Molecular Biotechnology Research Centre, Waterford Institute of Technology, Waterford, Ireland
First published on 23rd April 2013
Abstract
Molecularly Imprinted Polymers (MIPs) against imiquimod, a highly potent immune response modifier used in the treatment of skin cancer, were synthesised using a template analogue strategy and were compared with imprints of the drug itself. An investigation of the complexation between the functional monomer and the template analogue revealed an association constant of 1376 ± 122 M−1, significantly higher than previously reported values for similar systems. The binding characteristics of the synthesised imprinted polymers were evaluated and extremely strong binding for imiquimod was observed while imprinting factors as high as 17 were calculated. When applied as sorbents in solid-phase extraction of imiquimod from aqueous, urine and blood serum samples, clean extracts and recoveries up to 95% were achieved, and it is concluded that while imiquimod imprints exhibited higher capacity for the drug, template analogue imprints are more selective. The results obtained suggest potential applications of imiquimod imprints as sorbents in rapid extraction and monitoring of undesirable systemic release of the drug.
1 Introduction
Molecularly imprinted polymers are synthetic materials that contain specific binding sites, complementary to a compound of interest in terms of size, shape and functional group orientation.1–3 Such imprinted sites are generated by locking in place solution complexes between the template of interest and a complementary functional monomer, by co-polymerising these complexes with an excess of a cross-linking monomer. Upon completion of the polymerisation reaction, the removal of the template reveals the specific binding sites that are then able to reversibly rebind the template or closely related substances and, depending on the application in hand, extract it from a complicated sample matrix (MI-SPE),4 resolve enantiomeric forms in a racemic mixture of chiral compounds (MI-CSP)5 or signal the binding event acting as a sensing element.6The requirement of a template or a mould around which the molecularly imprinted polymer will be built can in some cases be a limiting factor, as quite often the targeted compound can be prohibitively expensive, incompatible with the polymerisation protocol, e.g. heat or light sensitive, insoluble in polymerisation solvents of choice, dangerous to the researchers' health or simply unavailable. In such cases scientists have employed template substitutes or “dummy” templates, as they are often called, whereby a compound of closely related structure to the substance of interest is imprinted instead of the actual target itself. The resulting polymers have been shown to exhibit excellent recognition properties not only for the substitute template, but also for the actual target, largely depending on the similarity of the chosen template to the targeted structure.7–9 9-Ethyladenine is a typical example of such a template, used by Spivak et al. as an organic solvent soluble analogue of adenine in the study of nucleotide base imprinted polymers.10–12
In the present report, our aim was to develop molecularly imprinted polymers that will enable selective separation and quantitation of imiquimod (IMQ), a highly potent prescribed medication that acts as an immune response modifier. Imiquimod was approved by the FDA for treatment of various types of skin cancer, as well as genital warts, in 1997, and it is formulated as a 5% patient-applied cream under the trade name Aldara™ (3M Healthcare). Although imiquimod is applied topically, a systemic release of the drug is known to occur, with severe side-effects reported in some patient cases.13,14 Thus, a rapid and robust method for isolation and pre-concentration of imiquimod from biological fluids could be an invaluable tool in early detection of systemic release and curtailment of its impact on patient well-being.
Imiquimod is sparingly soluble in organic or aqueous solvents at neutral pH. This poses an immediate limitation to the development of an imprinting protocol, hence a template analogue strategy was originally chosen. However, based on the observation that the drug is more soluble in acidic environments, it was possible to reach the desirable concentration by slight modification of the pre-polymerisation solution composition and thus prepare an imiquimod imprinted polymer, whose performance was subsequently compared to the template analogue based material.
2 Experimental
2.1 Materials and methods
Ethyleneglycol dimethacrylate (EDMA), methacrylic acid (MAA), 1,1′-azobis(cyclohexanecarbonitrile) (ACCN), adenine, thymine, cytosine, uracil, 5-fluorouracil, 1-bromo-2-methylpropane, potassium carbonate, HPLC grade solvents, CDCl3, DMSO-d6, and anhydrous N,N-dimethylformamide (DMF) were purchased from Sigma-Aldrich (Wicklow, Ireland). Imiquimod was a donation from EirGen Pharma (Waterford, Ireland). Water was purified by a SG Ultra Clear TWF UV Plus TM water purification system (Whitewater, Blackrock, Ireland). EDMA was purified by extraction with 10% NaOH, washing with brine, drying over anhydrous magnesium sulphate, and subsequent distillation under reduced pressure. MAA was distilled under reduced pressure.NMR spectra were obtained using a Jeol ECX 400 spectrometer (Tokyo, Japan). HPLC measurements were performed using an Agilent 1200 system equipped with a diode array detector. Surface area analysis was performed using a Micromeritics Gemini VI Nitrogen sorption analyser (Particular Sciences, Dublin, Ireland).
2.2 Template analogue synthesis
The template analogue, 9-isobutyladenine (9IBA), was synthesised using a protocol previously described,15 adapted as follows: 2.6 g (20 mmol) of adenine were suspended in 100 mL of anhydrous DMF and 3.5 g (25 mmol) of K2CO3 were added. The mixture was stirred at room temperature for 24 h in order to form the potassium salt of adenine. Then, 3.4 g (25 mmol) of 1-bromo-2-methylpropane were added to the slurry mixture and allowed to react for a further 72 h. DMF was then removed under reduced pressure and the solid residue was suspended in water. The aqueous layer was extracted with CHCl3 (3 × 50 mL), the organic layers were combined, dried over MgSO4 and finally the solvent was evaporated under reduced pressure to yield 9IBA as a white solid. This was recrystallised from butan-2-one to yield the final product. 1H NMR (DMSO-d6): δ (ppm): 0.80 (d, 6H, –CH(CH3)2), 2.14 (m, 1H, –CH(CH3)2), 3.91 (d, 2H, –NCH2CH(CH3)2), 7.16 (s, br, 2H, –NH2), 8.08 (s, 1H, imidazole CH), 8.09 (s, 1H, pyrimidine CH). 13C NMR (DMSO-d6): δ (ppm): 20.1, 29.0, 50.5, 119.2, 141.7, 150.3, 153.9, 156.5. LC-MS analysis: calculated MW for C9H13N5: 191.12, found m/z: 192.1 (MH+).2.3 Pre-polymerisation studies
Monomer–template complexation was studied prior to polymer synthesis by means of a 1H NMR titration in CDCl3 in order to establish the strength of interactions present in the pre-polymerisation solution. Hence, to a solution of 9IBA (1 mmol L−1) were added increasing amounts of MAA. The complexation-induced shift (CIS) of 9IBA's H2 proton was followed, and a titration curve was then constructed. The raw titration data were fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding isotherm, and the association constant was obtained by nonlinear regression. The stoichiometry of the 9IBA–MAA complex was confirmed using Job's method of continuous variation.16 Hence, equimolar solutions (10 mmol L−1) of 9IBA and MAA were mixed in different ratios and a plot of Δδ against the molar fraction of 9IBA multiplied by the CIS (X9IBA × Δδ) was constructed.
1 binding isotherm, and the association constant was obtained by nonlinear regression. The stoichiometry of the 9IBA–MAA complex was confirmed using Job's method of continuous variation.16 Hence, equimolar solutions (10 mmol L−1) of 9IBA and MAA were mixed in different ratios and a plot of Δδ against the molar fraction of 9IBA multiplied by the CIS (X9IBA × Δδ) was constructed.
2.4 Preparation of imprinted polymers
9IBA imprinted polymers (9IBA-MIP) were synthesised using a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of functional monomer (MAA) to template plus 20 equivalents of the cross-linker (EDMA). Thus, 0.191 g (1 mmol) of 9IBA and 0.342 mL (4 mmol) of MAA were dissolved in 5.6 mL of acetonitrile followed by addition of 4 g (20 mmol) of EDMA in a 20 mL screw-cap vial. The solution was degassed using N2 while cooling in an ice-bath for 5 minutes. Finally, 0.04 g of the free radical initiator ACCN were added and the vial was sealed and placed in a UV reactor at 4 °C for 24 hours. Imiquimod imprinted polymers (IMQ-MIP) were prepared in the same manner however, a total of 7 mmol of MAA were required in order to dissolve the drug in the pre-polymerisation mixture, resulting in a 7
1 ratio of functional monomer (MAA) to template plus 20 equivalents of the cross-linker (EDMA). Thus, 0.191 g (1 mmol) of 9IBA and 0.342 mL (4 mmol) of MAA were dissolved in 5.6 mL of acetonitrile followed by addition of 4 g (20 mmol) of EDMA in a 20 mL screw-cap vial. The solution was degassed using N2 while cooling in an ice-bath for 5 minutes. Finally, 0.04 g of the free radical initiator ACCN were added and the vial was sealed and placed in a UV reactor at 4 °C for 24 hours. Imiquimod imprinted polymers (IMQ-MIP) were prepared in the same manner however, a total of 7 mmol of MAA were required in order to dissolve the drug in the pre-polymerisation mixture, resulting in a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of MAA
1 ratio of MAA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IMQ. In both cases, the corresponding non-imprinted polymers (9IBA-NIP and IMQ-NIP) were synthesised with omission of the template. The resulting monoliths were smashed to coarse particles and washed with methanol for 24 hours using a Soxhlet apparatus. Finally, the polymers were manually ground and sieved collecting the 25–38 μm fraction for packing into HPLC columns and SPE cartridges.
IMQ. In both cases, the corresponding non-imprinted polymers (9IBA-NIP and IMQ-NIP) were synthesised with omission of the template. The resulting monoliths were smashed to coarse particles and washed with methanol for 24 hours using a Soxhlet apparatus. Finally, the polymers were manually ground and sieved collecting the 25–38 μm fraction for packing into HPLC columns and SPE cartridges.
2.5 Chromatographic evaluation
Imprinted or non-imprinted particles of 25–38 μm were slurry packed in 50 mm × 4.6 mm i.d. HPLC columns using a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol
1 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture. The columns were then connected to a HPLC instrument and equilibrated with CH3CN–1% CH3COOH until a stable baseline was obtained. Analyses were performed at 1 mL min−1 by injecting 5 μL of a 1 mmol L−1 solution of each analyte and recording its elution profile at 240 or 260 nm. All injections were repeated at least three times, alternating between the different analytes to avoid column overloading.
water mixture. The columns were then connected to a HPLC instrument and equilibrated with CH3CN–1% CH3COOH until a stable baseline was obtained. Analyses were performed at 1 mL min−1 by injecting 5 μL of a 1 mmol L−1 solution of each analyte and recording its elution profile at 240 or 260 nm. All injections were repeated at least three times, alternating between the different analytes to avoid column overloading.
Staircase frontal chromatography was performed on 9IBA-MIP and 9IBA-NIP using solutions of 9IBA in CH3CN–1% CH3COOH as the mobile phase and mixing them in a step-wise fashion of 10% increments with the pure mobile phase to produce a staircase frontal chromatogram with a total of 30 steps in the concentration range 10−6 to 10−3 mol L−1. From each step the corresponding amount of bound analyte was calculated and the collected results were plotted against the corresponding concentration of 9IBA to produce binding isotherms that were fitted using the appropriate binding model.
2.6 Solid phase extractions
25 mg of MIP and NIP particles were dry packed in 3 mL SPE cartridges using 20 μm porous PTFE frits. Equilibration of the columns, loading and washing were performed using 1 mL aliquot of the corresponding solutions and elution of the retained analytes was achieved with 1 mL of CH3OH–5% CH3COOH. Full vacuum was applied between each step for 3 minutes in order to dry the stationary phases. The collected fractions were analysed by HPLC using a 150 mm × 3.9 mm, 5 μm C18 column and isocratic elution using a mixture of CH3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) aqueous CH3COONa (10 mM, pH = 4) 40
aqueous CH3COONa (10 mM, pH = 4) 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 with the flow-rate set at 1 mL min−1. Under these conditions imiquimod eluted at 3.9 minutes. Blood serum samples were prepared by allowing the collected blood to clot at room temperature for approximately 45 minutes, followed by centrifugation at 3000 rpm for 15 minutes, and careful collection of the clear top layer. Serum samples were stored at −80 °C. SPE analysis of serum samples was performed on spiked samples (10 μg mL−1) after dilution with ultra-pure water. Urine samples were filtered, spiked and analysed following dilution with ultra-pure water. The protocol for biological sample analysis was approved by the Institutional Ethics Committee and written consent was obtained.
60 with the flow-rate set at 1 mL min−1. Under these conditions imiquimod eluted at 3.9 minutes. Blood serum samples were prepared by allowing the collected blood to clot at room temperature for approximately 45 minutes, followed by centrifugation at 3000 rpm for 15 minutes, and careful collection of the clear top layer. Serum samples were stored at −80 °C. SPE analysis of serum samples was performed on spiked samples (10 μg mL−1) after dilution with ultra-pure water. Urine samples were filtered, spiked and analysed following dilution with ultra-pure water. The protocol for biological sample analysis was approved by the Institutional Ethics Committee and written consent was obtained.
3 Results and discussion
3.1 Choice of the template analogue
Given the limited solubility of imiquimod in the organic solvents commonly used in molecular imprinting it was originally decided to follow a template analogue approach. Upon examination of the drug's structure, it is noticeable that its core ring structure bears significant similarities to that of adenine. It was thus proposed that substitution of the N9-position of the DNA base with an isobutyl group could serve a dual purpose: enhance the solubility of the template analogue and, more importantly, make the latter structurally similar to the targeted drug (Scheme 1). Thus, 9-isobutyladenine (9IBA) was chosen as a template analogue and synthesised in a 2-step reaction in moderate to high yields.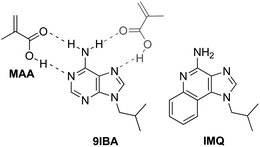 | ||
| Scheme 1 Proposed primary (black) and secondary (grey) mode of interaction between the functional monomer (MAA) and 9IBA, and the chemical structure of imiquimod (IMQ). | ||
3.2 Pre-polymerisation studies
The interaction between nucleic acid bases and carboxylic acids has been previously investigated. In an early report by Lancelot,17 butyric acid was used as an analogue of amino acid side chain acidic functionalities and the strength as well as its mode of interaction with the DNA bases was extensively studied using 1H-NMR titrations in CDCl3. The findings suggested an association constant between butyric acid and 9-ethyladenine of 160 M−1 at 303 K, while it was proposed that 3 different complexes contributed to the total association event. More recently, Janke et al.18 studied the complexation of adenosine with acetic acid using low temperature 1H NMR as well as 2D NMR and found an association constant of 296 M−1 at 293 K between the two counterparts.The system studied here is not very dissimilar to the ones reported previously however it was decided to calculate the binding constant between the specific compounds involved. Interestingly, our findings suggest a much stronger association between 9IBA and methacrylic acid, and a Ka = 1376 ± 122 M−1 was calculated when 9IBA's H2 was monitored (Fig. 1), almost an order of magnitude higher than the values previously reported. Job plot experiments verified a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex stoichiometry, warranting the use of a 1
1 complex stoichiometry, warranting the use of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model to fit the obtained experimental results (Fig. 1, inset).
1 binding model to fit the obtained experimental results (Fig. 1, inset).
 | ||
| Fig. 1 1H NMR titration binding isotherm of 9IBA with MAA: experimental data (◇) and the corresponding fitted curve. Inset: Job plot for the 9IBA–MAA complex. | ||
This significantly higher Ka value can be attributed to the fact that methacrylic acid has different pKa value than the carboxylic acids used in previous studies and more importantly, to the concentration of host (9IBA) used in this study (1 mmol L−1) compared to 20–35 mmol L−1 used in previous studies. The lower concentration used here should eliminate the influence of self-association of both guest and/or host molecules occurring at higher concentrations, but also probe stronger complexation events, occurring at low concentrations. In any case, a Ka of this magnitude is in agreement with the findings of fundamental studies on MIPs, whereby exceptionally strong binding against adenine derivatives using MAA as the functional monomer has been reported.10,11
3.3 Chromatographic evaluation of imprinted polymers
Nitrogen sorption analysis of the synthesised polymer particles revealed that all tested materials have a microporous character with very similar specific surface areas and pore dimensions (Table 1). Furthermore, both imprinted polymers have 10–15% higher specific surface areas than the corresponding non-imprinted polymers, demonstrating the influence of the template in the formation of the porous network. These particles were packed in 50 mm × 4.6 mm HPLC columns and connected to a HPLC instrument for chromatographic evaluation. This was performed by consecutive injections of the template (9IBA), the analyte of interest (IMQ) and the DNA and RNA bases. 5-Fluorouracil (5-FU) was also included in the injection series.| 9IBA-MIP | 9IBA-NIP | IMQ-MIP | IMQ-NIP | |
|---|---|---|---|---|
| Surface area (m2 g−1) | 241.5 | 210.2 | 229.8 | 210.7 |
| Pore volume (cm3 g−1) | 0.44 | 0.56 | 0.51 | 0.49 |
| Pore diameter (Å) | 82.0 | 99.5 | 102.7 | 128.9 |
In agreement with the findings of Shea et al.,19 MAA-based MIPs against adenine derivatives exhibit extremely long retention times for their template when tested using the porogen as a mobile phase. Thus, when 9IBA was injected on the 9IBA-MIP column prepared here, no elution was observed after 1.5 hours of analysis using acetonitrile as a mobile phase. Similarly, IMQ, adenine and cytosine were also retained for >1.5 h while thymine, uracil and 5-FU all eluted at or near the solvent front on both columns (data not shown). The latter observation is attributed to the lack of complementarity between the imide functionality of the pyrimidine bases and the donor–acceptor hydrogen bond array of methacrylic acid.
In order to facilitate elution of the injected analytes from the imprinted and non-imprinted columns, 1% acetic acid was added to the mobile phase, aiming to disrupt hydrogen bonding of the analytes to the stationary phase and reduce elution times. Indeed, under the new analysis conditions 9IBA eluted in 28 minutes while IMQ was retained for 63 minutes (Table 2). An imprinting factor (IF = kMIP/kNIP) of 17 was calculated for IMQ, more than 2.5 times higher than the IF calculated for the template itself (IF = 6.4), supporting the design and choice of 9IBA as a template analogue. Adenine and cytosine did not elute from the imprinted column after 1.5 h however their retention times on the non-imprinted column were significantly reduced to 10 and 6.2 minutes respectively (Fig. 2). This observation clearly demonstrates the shape, size and functional group complementarity of the synthesised imprinted polymer: adenine, slightly smaller and lacking the non-functional bulky side chain of 9IBA, is tightly bound by the binding cavities while cytosine, much smaller but with a very similar functional group orientation to 9IBA, is also strongly bound. Thymine, uracil and 5-FU eluted again at or near the solvent front on both columns.
| Analyte | t R (min) | k | IF | t R (min) | k | IF | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 9IBA–MIP | 9IBA–NIP | 9IBA–MIP | 9IBA–NIP | IMQ–MIP | IMQ–NIP | IMQ–MIP | IMQ–NIP | |||
| 5-Fluorouracil | 1.6 | 1.0 | 1.3 | 0.4 | 3.1 | 1.3 | 1.1 | 0.8 | 0.4 | 1.9 |
| Uracil | 3.8 | 1.3 | 4.3 | 0.8 | 5.5 | 3.9 | 1.3 | 4.2 | 0.7 | 5.8 |
| Thymine | 4.3 | 1.4 | 5.1 | 0.9 | 5.9 | 2.7 | 1.7 | 2.7 | 1.2 | 2.1 |
| 9IBA | 28.0 | 5.1 | 38.0 | 5.9 | 6.4 | >90.0 | 10.1 | >125.0 | 12.2 | >9.8 |
| Imiquimod | 63.0 | 4.5 | 87.0 | 5.1 | 17.0 | >90.0 | 16.2 | >125.0 | 20.2 | >5.9 |
| Adenine | >90.0 | 10.0 | >125.0 | 13.0 | >9.7 | >90.0 | 19.7 | >125.0 | 24.8 | >4.8 |
| Cytosine | >90.0 | 6.2 | >125.0 | 7.6 | >16.6 | >90.0 | 11.4 | >125.0 | 14.0 | >8.6 |
 | ||
| Fig. 2 Retention times (tR, min) of imiquimod and related substances on (a) 9IBA and (b) IMQ imprinted polymer packed HPLC columns (50 mm × 4.6 mm i.d.). Conditions: mobile phase: CH3CN–1% CH3COOH, flow-rate: 1 mL min−1, inj. vol.: 5 μL of 1 mmol L−1 solution, DAD detection at 240 or 260 nm. | ||
HPLC evaluation of IMQ-MIP and IMQ-NIP revealed similar binding characteristics, albeit retention times were significantly longer. Thus, apart from adenine and cytosine, IMQ and 9IBA were also not eluted from the imprinted column after 1.5 h of analysis. Notably, retention times of IMQ-NIP were longer compared to those of 9IBA-NIP, possibly due to the higher amount of MAA used, contributing to an increase in non-specific binding. This led to an overall decrease in the corresponding calculated imprinting factors; nonetheless, these values have only qualitative interest as the exact retention time on the imprinted column could not be determined.
Overall, the two imprinted polymers exhibit similar behaviour towards the tested analytes with IMQ-MIP being potentially the better sorbent for the drug. This is attributed to the presence of the drug itself in the pre-polymerisation mixture, producing binding sites of superior complementarity to the targeted structure, but also to the higher ratio of MAA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) template, potentially resulting in more points of attachment for the drug, although the majority of them are of a non-specific nature, as demonstrated by the longer retention times exhibited by IMQ-NIP.
template, potentially resulting in more points of attachment for the drug, although the majority of them are of a non-specific nature, as demonstrated by the longer retention times exhibited by IMQ-NIP.
Frontal chromatography was performed using 9IBA as the test analyte on its corresponding imprinted and non-imprinted polymers; the quantity of the analyte required for the analysis and the price as well as toxicity of IMQ prohibited its use in such experiments. In spite of this, when a 9IBA rebinding experiment was attempted in the low concentration range using IMQ-MIP as the stationary phase, elution times of each step were excessive and did not permit accurate collection of experimental points.
The obtained results revealed the superior binding performance of 9IBA-MIP versus its non-imprinted counterpart (Fig. 3). In order to calculate the corresponding binding isotherms and binding capacities, each isotherm was fitted using the most common binding models used to model similar materials, namely the single binding site model (Langmuir), two-binding site model (Bi-Langmuir), the continuous distribution model (Freundlich)20 and the Langmuir–Freundlich model.21 The obtained results are reported in Table 3 together with the corresponding sums of least squares (R2) for each model, as a measure of fit quality.
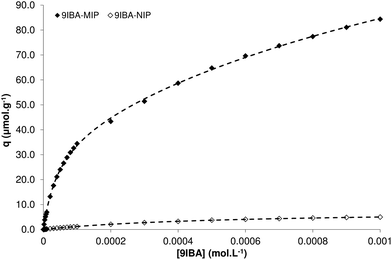 | ||
| Fig. 3 Frontal chromatography binding isotherms of 9IBA on 9IBA-MIP (◆) and 9IBA–NIP (◇) packed HPLC columns and the corresponding curves fitted to the Bi-Langmuir model (dashed lines). | ||
| Binding model | 9IBA–MIP | 9IBA–NIP | ||||
|---|---|---|---|---|---|---|
| K a (L mol−1) | N (μmol g−1) | R 2 | K a (L mol−1) | N (μmol g−1) | R 2 | |
| a Average Ka and N values calculated from affinity distribution curves. b Average Ka calculated using Ka = a1/m. | ||||||
| Langmuir | 6.0 × 103 | 90.8 | 0.9839 | 1.8 × 103 | 7.7 | 0.9998 |
| Bi-Langmuir | 2.3 × 104 | 38.9 | 0.9996 | 8.5 × 105 | 0.04 | 0.9999 |
| 0.5 × 103 | 142.6 | 1.7 × 103 | 7.8 | |||
| Freundlicha | 3.9 × 104 | 66.3 | 0.9919 | 2.0 × 104 | 3.1 | 0.9918 |
| Langmuir–Freundlichb | 1.1 × 103 | 162.9 | 0.9969 | 1.7 × 103 | 8.0 | 0.9998 |
The Langmuir model provides a poor fit in the case of 9IBA-MIP, not unexpectedly, given the well documented heterogeneous character of imprinted polymers. The best fit is achieved when the Bi-Langmuir model is applied, assuming a strong and a weak family of binding sites with affinity constants of 2.3 × 104 L mol−1 and 0.5 × 103 L mol−1 and a calculated population of 38.9 μmol g−1 and 142.6 μmol g−1 respectively. The Freundlich and Langmuir–Freundlich models also offer good fittings of the experimental data, with the former yielding average parameters closer to the “stronger” sites predicted by the Bi-Langmuir model and the latter closer to the “weaker” sites. Although it is conceivable that there are more than just two types of binding sites in the matrix of the imprinted polymer, a combination of the above information provides an accurate representation of the affinity characteristics of the polymer.
In the case of 9IBA-NIP, the Langmuir and Langmuir–Freundlich models yield nearly identical values of average affinity constant and number of binding sites, while the same parameters are calculated for the “weak” sites using the Bi-Langmuir model, which constitute the majority of sites in the polymer. The Freundlich model does not offer as good a fit, and the parameters derived are not comparable with the other models. Overall, it appears that 9IBA-MIP possesses high energy and low energy sites, the latter being larger in population and with an affinity constant comparable to the low energy sites of the non-imprinted counterpart. This indicates that the low energy, or non-specific, binding sites are of similar nature in both polymers.
3.4 Solid phase extractions
The performance of the template analogue and the directly imprinted polymers was finally evaluated as SPE sorbents for the extraction of imiquimod from aqueous samples, urine and blood serum. Following loading of 1 mL of sample, the cartridges were washed with 1 mL H2O in order to remove any hydrophilic matrix substances adsorbed onto the polymer. In early experiments, an attempt was made to elute the cartridges using CH3CN–1% CH3COOH however the average recovery on the imprinted polymers was less than 10% while up to 40% of the drug was recovered on the non-imprinted polymers. This is in agreement with the chromatographic evaluation results, whereby extremely long retention times were recorded for IMQ under similar elution conditions. It was thus decided to use CH3CN–1% CH3COOH in an intermediate “molecular recognition” step, and employ a stronger elution solvent mixture to recover IMQ from the cartridges. Following a series of experiments, CH3OH–5% CH3COOH was selected and the results obtained using the optimised protocol are presented in Fig. 4. It should be noted here that during the initial steps of the optimisation, excessive template bleeding from IMQ-MIP was observed, which resulted in recoveries significantly higher than what was expected for quantitative recovery of the drug. An extensive wash protocol using CH3OH–5% CH3COOH was employed and aliquots were analysed by HPLC until no IMQ could be detected. | ||
| Fig. 4 MI-SPE recoveries (%) of imiquimod on (a) 9IBA and (b) IMQ imprinted polymers and the corresponding imprinting factors (dashed lines). Conditions: loading: 1 mL of sample; 1st wash: 1 mL H2O, 2nd wash: 1 mL CH3CN–1% CH3COOH; elution: 1 mL CH3OH–5% CH3COOH. Blood and urine samples spiked with 10 μg mL−1 imiquimod. | ||
Both imprinted polymers achieved high recoveries of the drug from urine and blood serum samples, albeit IMQ-MIP was more consistent across the different sample matrices and average recoveries of up to 95% were reached. However, similar to the observations during chromatographic evaluation, a significant amount of non-specifically bound IMQ, up to 64.7%, was recovered from IMQ-NIP, resulting in average imprinting factors of 1.6. On the other hand, 9IBA-NIP exhibited substantially lower non-specific binding, less than half of what was measured on IMQ-NIP in some cases, thus imprinting factors for 9IBA polymers were as high as 4.3 in H2O and 3.3 in urine. It is noteworthy that both polymers exhibited their lowest recoveries and highest imprinting factors in the aqueous samples while the opposite was observed as sample complexity increased. The nature and composition of biological samples appear to “push” the hydrophobic drug onto the polymer stationary phases, resulting in higher recoveries in blood serum samples, albeit at the expense of selectivity. Nonetheless, both materials produced clean extracts as seen in Fig. 5, and no interference to the quantitation of IMQ was detected.
 | ||
| Fig. 5 Typical HPLC chromatograms of the different fractions collected by MI-SPE of blood serum samples using (a) 9IBA-MIP and (b) IMQ-MIP. From the bottom, 1st wash (H2O), 2nd wash (CH3CN–1% CH3COOH), eluted sample (CH3OH–5% CH3COOH). | ||
Thus, it is concluded from this study that 9IBA imprinted polymers exhibit higher selectivity for IMQ, while possessing a smaller number of binding sites, resulting in reduced capacity. The opposite is true for IMQ-MIPs that exhibit marginally higher capacity but appear to be less specific towards the drug.
4 Conclusions
9-Isobutyladenine was selected as a template analogue for imiquimod, based on its similar size, shape and functionality and was synthesised in a 2 step reaction in moderate to high yields. Pre-polymerisation studies of the complexes formed between methacrylic acid and the template analogue revealed a binding constant of 1376 ± 122 M−1, higher than those previously reported, but in agreement with the high affinity exhibited by the subsequently synthesised imprinted polymers. Limitations posed by imiquimod's insolubility were overcome by using a higher ratio of functional monomer to template and imiquimod imprinted polymers were synthesised. Chromatographic evaluation of the synthesised polymers revealed longer retention times for IMQ and related substances on IMQ-MIP compared to 9IBA-MIP, however the corresponding non-imprinted polymer exhibited significant non-specific binding, resulting in overall reduced imprinting factors. Analysis of 9IBA-MIP and 9IBA-NIP by frontal chromatography demonstrated the superior binding affinity and capacity of the imprinted polymers, while the “weak” binding sites of MIP and NIP were found to have comparable binding energies. When employed as a SPE sorbent, IMQ-MIP showed the highest recovery for the drug in biological samples, however high levels of non-specific binding were found on the corresponding NIP. 9IBA–MIP was capable of achieving marginally lower recoveries in the same samples however non-specific binding was significantly lower and overall higher imprinting factors were calculated.Based on the above observations on polymer performance, and considering the clear advantages of 9IBA compared to imiquimod in terms of solubility, availability, toxicity and cost, as well as the extensive polymer pre-conditioning required to eliminate template bleeding, 9IBA is proposed as an excellent template analogue and its use is fully justified by the experimental results acquired in this study.
Acknowledgements
Financial support from Enterprise Ireland's Applied Research Enhancement Programme (Contract number: RE-2008-11) and donation of imiquimod from EirGen Pharma are gratefully acknowledged.References
- Molecularly imprinted polymers. Man made mimics of antibodies and their applications in analytical chemistry, ed. B. Sellergren, Elsevier Science B.V., Amsterdam, 2001 Search PubMed.
- C. Alexander, H. S. Andersson, L. I. Andersson, R. J. Ansell, N. Kirsch, I. A. Nicholls, J. O'Mahony and M. J. Whitcombe, J. Mol. Recognit., 2006, 19, 106–180 CrossRef CAS.
- P. Manesiotis and G. Theodoridis, in Encyclopedia of Chromatography, ed. J. Cazes, Taylor & Francis, 3rd edn, 2009, pp. 24–30 Search PubMed.
- M. Lasakova and P. Jandera, J. Sep. Sci., 2009, 32, 799–812 CrossRef CAS.
- P. Manesiotis, Q. Osmani and P. McLoughlin, J. Mater. Chem., 2012, 22, 11201–11207 RSC.
- K. Haupt and K. Mosbach, Chem. Rev., 2000, 100, 2495–2504 CrossRef CAS.
- P. Manesiotis, C. Borrelli, C. S. A. Aureliano, C. Svensson and B. Sellergren, J. Mater. Chem., 2009, 19, 6185–6193 RSC.
- P. Manesiotis, A. Hall, J. Courtois, K. Irgum and B. Sellergren, Angew. Chem., Int. Ed., 2005, 44, 3902–3906 CrossRef CAS.
- G. Theodoridis, A. Kantifes, P. Manesiotis, N. Raikos and H. Tsoukali-Papadopoulou, J. Chromatogr., A, 2003, 987, 103–109 CrossRef CAS.
- D. A. Spivak, M. A. Gilmore and K. J. Shea, J. Am. Chem. Soc., 1997, 119, 4388–4393 CrossRef CAS.
- D. A. Spivak and K. J. Shea, Macromolecules, 1998, 31, 2160–2165 CrossRef CAS.
- D. A. Spivak and K. J. Shea, Anal. Chim. Acta, 2001, 435, 65–74 CrossRef CAS.
- C. Hanger, J. Dalrymple and D. Hepburn, N. Z. Med. J., 2005, 118, 1682 Search PubMed.
- S. Adams, L. Kozhaya, F. Martiniuk, T. C. Meng, L. Chiriboga, L. Liebes, T. Hochman, N. Shuman, D. Axelrod, J. Speyer, Y. Novik, A. Tiersten, J. D. Goldberg, S. C. Formenti, N. Bhardwaj, D. Unutmaz and S. Demaria, Clin. Cancer Res., 2012, 18, 6748–6757 CrossRef CAS.
- Synthetic Procedures in Nucleic Acid Chemistry, ed. W. W. Zorbach and R. S. Tipson, Interscience, New York, 1968 Search PubMed.
- K. A. Connors, Binding constants; The measurement of molecular complex stability, John Wiley & Sons, New York, 1987 Search PubMed.
- G. Lancelot, J. Am. Chem. Soc., 1977, 99, 7037–7042 CrossRef CAS.
- E. M. B. Janke, H.-H. Limbach and K. Weisz, J. Am. Chem. Soc., 2003, 126, 2135–2141 CrossRef.
- K. J. Shea, D. A. Spivak and B. Sellergren, J. Am. Chem. Soc., 1993, 115, 3368–3369 CrossRef CAS.
- R. J. Umpleby, S. C. Baxter, M. Bode, J. K. Berch, R. N. Shah and K. D. Shimizu, Anal. Chim. Acta, 2001, 435, 35–42 CrossRef CAS.
- R. J. Umpleby, S. C. Baxter, Y. Z. Chen, R. N. Shah and K. D. Shimizu, Anal. Chem., 2001, 73, 4584–4591 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2013 |
