DOI:
10.1039/C2AN35744E
(Tutorial Review)
Analyst, 2013,
138, 72-86
Graphene and its derivatives for cell biotechnology
Received
4th June 2012
, Accepted 24th September 2012
First published on 26th September 2012
Abstract
Every few years, a novel material with salient and often unique properties emerges and attracts both academic and industrial interest from the scientific community. The latest blockbuster is graphene, an increasingly important nanomaterial with atomically thin sheets of carbon, which has become a shining star and has shown great promise in the field of material science and nanotechnology. In recent years, it has changed from being the exclusive domain of physicists to the new passion of chemists and biologists. Graphene and its derivatives are now at the forefront of nearly every rapidly developing field of science and engineering, including biochemistry, biomedicine and certain cutting-edge interdisciplines that have intense popularity. The aim of this review is, firstly, to provide readers with a comprehensive, systematic and in-depth prospective of graphene's band structure and properties, and secondly, to concentrate on the recent progress in producing graphene-based nanomaterials, including mechanical exfoliation, chemical vapor deposition, plasma enhanced chemical vapor deposition, chemical reduction of graphene oxide, total organic synthesis, electrochemical synthesis and other fabrication strategies widely accepted by research scientists. At the same time, important definitions related to graphene are also introduced. The focus of this Tutorial Review is to emphasize the current situation and significance of using this new kind of two-dimensional material in the hot and emerging fields that are closely related to human life quality, for instance, cell biochemistry, bioimaging along with other frontier areas. Finally, the latest developments and possible impact that affect the heart of the whole scientific community have been discussed. In addition, the future trends along with potential challenges of this rapidly rising layered carbon have been pointed out in this paper.
 Mei Yang | Mei Yang was born in Sichuan, China, and studied chemistry at China West Normal University. As an undergraduate, she conducted research on the development of new biosensors and their applications in biomolecules analysis such as amino acid, protein and nucleic acid, etc. After receiving her B.S. in chemistry in 2010, she joined the lab of Professor Yixiang Duan in the Research Center of Analytical Instrumentation at Sichuan University, China. Her research interests are in the area of bioanalytical chemistry. Current work focuses on the green and morphology controlled nanomaterials synthesis, the functionalization of inorganic nanoparticles (mainly graphene and quantum dots) with inorganic or organic ligands and integration of these novel materials with various photoelectric analysis techniques. Her other scientific interests include the applications of GC, HPLC and LC-MS on medical diagnostics and pharmaceutical analysis. |
 Jun Yao | Jun Yao studied Chemistry at China West Normal University, where he obtained his B.S. degree in Chemistry in 2006. He worked in the field of bioanalytical chemistry, receiving his M.S. degree from the same university in 2009. Following graduate studies, he became a university teacher of chemistry and pharmacy at Chongqing Three Gorges Medical College, China. Subsequently, he joined the Analytical and Testing Center of Sichuan University as a PhD candidate to research biochemical or medical analysis based on nanomaterials as well as spectroscopy and electrics related technology, especially carbon nanomaterials and fluorescence technique. His research interests revolve around graphene-based nanomaterials, quantum dots and multifunctional nanostructures, spanning from synthetic methodology to device fabrication, with the desire of pursuing nanostructures for useful photoelectric applications, mainly focusing on a variety of biochemical and biomedical applications of graphene and other inorganic nanomaterials such as biosensor, bioimaging, drug delivery, medical diagnostics and biotherapy. |
 Yixiang Duan | Yixiang Duan received his B.S. degree from Fudan University and M.S. degree in analytical chemistry from Changchun Institute of Applied Chemistry, Chinese Academy of Science in 1988 and PhD degree in analytical chemistry jointly from Jilin University, China, and Indiana University, USA in 1994. Then he did his postdoctoral research at Los Alamos National Laboratory. From 1997 to 2010, he was a Principal Investigator and Staff Scientist with the Chemical Diagnostics and Engineering Group, Los Alamos National Laboratory. He is currently a National Special Professor and Director of Research Centre of Analytical Instrumentation, Sichuan University, China. His current research interests include nanomaterial-based photoelectric biosensors, molecular spectrometry, non-invasive medical diagnostics, novel analytical instrumentation, as well as various applications of chemistry to nanomaterials, nanomedicine, and biological science, especially fluorescence instrumentation, fluorescence imaging and development of novel strategies and methods for utilizing graphene in bioanalysis and biomedicine. His other research interests are in the areas of spectrometry, chromatography, mass spectrometry and analytical instrumentation. |
1. Introduction
Nanoscience is always inspiring the imagination of biologists and biotechnologists.1 Recent decades have witnessed the magical glamour of nano- and bio-technology, two important hot topics of interdisciplinary research. They have undergone a rapid growth all over the world and have turned out to be the source of new research directions.1–5 In these cutting-edge fields, the emergence of a broad array of new-found nanomaterials will gradually affect our life. Because of their unique structures, components and properties, these materials have merged in many daily-used products and have provided appealing opportunities for biotechnological development.6–11
Graphene is one of the most fascinating nanomaterials with important applications.12–18 Its discovery has spawned huge interest in the fields of chemistry, physics, biology, medicine, nanoscience and material science.19–27 In the meantime, a lot of papers are published every day according to the bibliometrics predictions.28 There is no doubt that researches on graphene have gone forward at a truly relentless pace and this gathered momentum will keep rapidly increasing over the next few years.29,30 Till now, the blue ribbon for graphene is the Nobel Prize in physics in 2010 that was awarded to Geim and Novoselov, the first group to isolate single-layer graphene from graphite.19,31–33 Since the discovery of graphene, considerable efforts have been dedicated to its structure, fabrication, functionalization and application.13,34–42 A number of researches have suggested that graphene is a single-atom-thick sheet with carbon atoms of sp2 bonding and forming a hexagonal 2D lattice.43–45 It is the thinnest material ever measured,34,46–49 yet is harder than diamond, and possesses a large specific surface area, outstanding thermal conductivity, and favorable electric conductivity, along with eminent quantum Hall effect.13,50–54 Due to these unique physicochemical properties,55–60 graphene has been considered as a promising candidate in many applications and technological aspects such as nanoelectronics,34,61,62 polymer composites,51,53,63–67 sensors,68–86 batteries,87–94 fuel cells and super capacitors.95,96 In this paper, after a brief presentation of its structures, unique properties and preparations, together with the content what is discussed in the vast majority of research articles and reviews, we put most of our energy on its applications for biotechnology that are seldom involved in other peer publications.
2. Basic structure and properties of graphene
Most of the amazing properties of graphene derive from its peculiar lattice structure. Thus, it is imperative to understand the structure of graphene before introducing its properties.97 Graphene comprises a single layer of sp2-hybridized carbon atoms arranged densely in a planar hexagonal honeycomb crystal structure (Fig. 1).98–106 In this configuration, one s-orbital and two p-orbitals of each carbon atom hybridized to form three in-plane σ bonds and have a strong wave function overlap with their three nearest neighbor atoms to give rise to a very strong covalent bond, which results in the mechanical stability of the carbon sheet. While, the unaffected pz orbital, oriented perpendicular to the plane of the 2D sheet, can bind covalently with the adjacent carbon atoms, leading to the formation of a delocalized π bond.
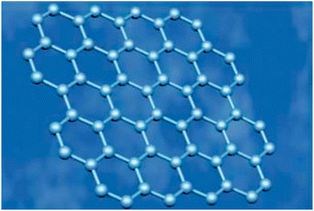 |
| | Fig. 1 Photograph of planar hexagonal honeycomb crystal structure of graphene. Reproduced with permission from ref. 106. Copyright 2006, Nature Publishing Group. | |
Unsurprisingly, the essential 2D structure is responsible for the prominent properties and other excellent characteristics of graphene.13,56,107–109 Here, we will only introduce graphene's characters that are most relevant to cell biotechnology. Because of the largely dislocated π-electrons, graphene possesses remarkably high electron mobility and size-dependent electrical properties, which make it a promising material in high-performance biosensors as well as an efficient fluorescence nanoquencher in the optical-based detection of biomolecules.110–112 Its large specific surface area with a value up to 2600 m2 g−1 can provide an ultrahigh loading capacity for biomolecules and drugs, leading to increased sensitivity.79,97,113,114 Experimental results reveal that graphene has a Young's modulus of 1060 GPa and a breaking strength of ∼40 N m−1.115 Therefore, it has been recognized as an ideal scaffold for cell culture growth.
3. Fabrication and functionalization of graphene
So far, inspired by pioneering work, tremendous efforts have been devoted to exploring synthetic approaches for graphene and its derivatives.116–121 Broadly speaking, the preparations of graphene can be arranged into four main methods as follows. The first is a physical approach, known as ‘mechanical exfoliation’, where mechanical or chemical energy are essentially used to break the weak van der Waals forces and separate out individual graphene sheets (Fig. 2).122–125 It is promising to produce graphene on a large scale and some of the recent modifications have been employed to produce large-size pieces. But there is still a long way to apply this process into an industrial scale production level due to its low yield. The second is chemical vapor deposition (CVD),126–131 first reported for graphene in 2006, a typical method that eradicates residual metallic impurities and has been used to grow large areas of single- or few-layer graphene on a wide range of metal substrates, e.g. Ni, Ru, and Cu, without changing its properties.132–146 The third strategy is plasma enhanced CVD (PECVD). It has demonstrated the versatility of synthesizing graphene on any substrate and is capable of growing single-layer graphene with high throughput at shorter reaction times and lower deposition temperatures compared to CVD.147 The fourth is the chemical reduction of graphene oxide (GO), both an economical and practical method to acquire graphene.118,148–151 Graphene synthesized by this approach possesses many structural defects, which is advantageous for biosensing applications. Additionally, there are other innovative categories including total organic synthesis, electrochemical synthesis, thermal decomposition of SiC, and unzipping of carbon nanotubes (CNTs).22,55,118,122,125,152–161Fig. 3 shows the different existing ways to unzip CNTs to yield graphene nanoribbons.162 The above-mentioned details revealed that remarkable progress has been made on manufacturing graphene; however, opportunities and challenges still co-exist in developing new synthetic routes for preparing graphene with controlled size, shape, tailored surfaces and defined mesoscopic morphology. For better utilization of graphene in biology and medicine, appropriate functionalization of pristine graphene and the immobilization of biomaterials on it are necessary and play important roles in biocompatibility because the functional groups can create defects on the graphene surface and reduce the strong hydrophobic interaction of graphene with cells and tissues.156 Biomolecules such as protein, DNA and small molecules have been reported to functionalize graphene. The biological activity of graphene-based materials can be associated with their capacities to interfere with molecular processes and functions.163
 |
| | Fig. 2 Mechanical exfoliation of graphene using scotch tape from highly oriented pyrolytic graphite (HOPG). Reproduced with permission from ref. 124. Copyright 2011, Elsevier. | |
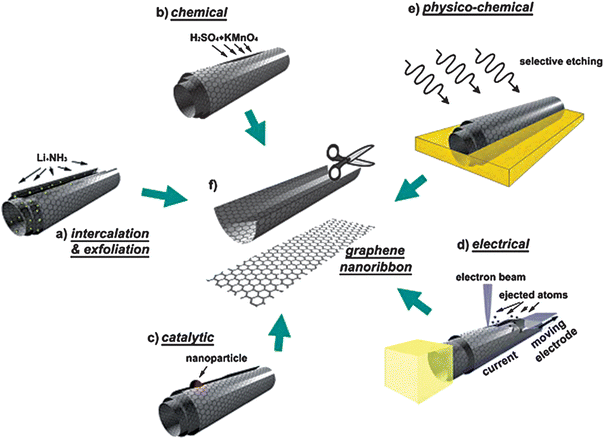 |
| | Fig. 3 Schematics and representative images of several methods for unzipping CNTs into graphene nanoribbons. Reproduced with permission from ref. 162. Copyright 2010, Elsevier. | |
4. Applications in cell biotechnology
4.1 Graphene-based scaffold for cell culture
As we know, the backbone of biomolecules and biological structures is carbon, so it is no doubt to integrate biological systems with nanocarbons. Plenty of evidence has demonstrated that CNTs are able to support the growth and adhesion of live cells;164,165 however, there are also researches certified that this one-dimensional nanocarbon has certain toxic effects on living cells.166–173 Compared with CNTs, the 2D carbon nanomaterials, graphene and its derivatives, being only one atom thick, have a larger surface area and better biocompatibility, introducing the least amount of artificial material possible. Therefore, they have recently captured great interests from scientists as cell culture substrates.174–178 The highest Young's modulus of graphene allows it to be converted to the desired shape.179–181
Graphene and its derivatives have been used as coating materials of substrates to culture human mesenchymal stem cells (hMSCs), a kind of significant stem cell for a myriad of ground-breaking therapies in regenerative medicine.182–190 The survival, proliferation and differentiation of hMSCs are extremely susceptible to numerous environmental factors such as impact from nearby cells, interaction with soluble growth factors and the influence of extracellular matrices. So it is of the utmost urgency to control these factors.191–193 Using graphene-based materials in this field is an elegant solution to these questions as well as a new ray of hope for groups of researches. Firstly, graphene is covered onto the substrates, for example, glass slides, Si/SiO2, polyethylene terephthalate (PET) and polydimethylsiloxane (PDMS), and then hMSCs are seeded on the test chips followed by culturing in normal stem cell medium. Examining the viability and morphology of the hMSCs showed that there was no significant difference between graphene-coated and uncoated substrates. However, these substrates without or with a graphene covering had a different influence on the conversion of hMSCs into specific cell types. In the absence of graphene, hMSCs on the soft substrates, PDMS and PET, could convert into neuronal cells and muscle cells respectively, whereas on stiff substrates such as glass slides and Si/SiO2, these stem cells did not differentiate. Once these substrates had been covered with graphene, the hMSCs were able to convert into osteoblasts. These results proved that graphene played an important role in the differentiation of hMSCs into the bone cells. Besides, the study of cultivating induced pluripotent stem cells (iPSCs) has further proved that graphene and its derivatives could support the cell culture and that their surfaces led to distinct cell proliferation and differentiation characteristics. Other cells like NIH-3T3 fibroblasts,194 L929 cells,195 MC3T3-E1 cells,196 A549197 and mouse hippocampal cells176 can also grow on the graphene-based substrates. A focal adhesion study, proliferation assay, and cell shape analysis together with many other investigations revealed that the cells grow well on the graphene-coated substrates with improved cellular adhesion, proliferation and differentiation, which suggested that these nanomaterials held great potential as surface-coating materials for cell culture. Fig. 4 shows the cell adhesion patterns on different substrates.194 From Fig. 4 we can see that there are large dot-shaped plaques of vinculin in the peripheral regions of the cells cultured on glass, while cells grown on the coated substrates exhibit less vinculin-containing focal contacts at the periphery, which indicate the superior biocompatibility of graphene and its derivatives-coated substrates.
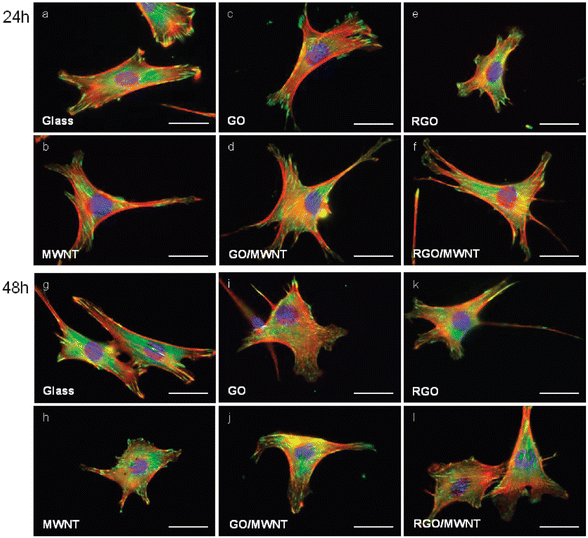 |
| | Fig. 4 Illustration of NIH-3T3 cells incubated on different substrates for 24 and 48 h. Actin, vinculin and the DAPI-stained nucleus are represent in red, green and blue respectively. Reproduced with permission from ref. 194. Copyright 2010, American Chemical Society. | |
Cellular adhesion is a vital prerequisite to subsequent cell functions, such as proliferation, synthesis of proteins and the formation of mineral deposits. It usually depends on the properties of substrates including surface topography, chemical composition, dimensions and wettability.198–203 Cell proliferation is regulated by the induction and inhibition of positive and negative cell cycle regulators. Most reports have shown that graphene-based materials were biocompatible by facilitating the cell adhesion and proliferation of L-929 cells,204 osteoblasts,205 kidney cells,206 and embryonic cells.206 Graphene has a different effect on the differentiation of stem cells into specific tissues. Lee et al. demonstrated that graphene was likely to accelerate the differentiation of hMSCs into the osteoblast lineage; however, they inhibit their differentiation into adipocytes. This was mainly because of different binding interactions between graphene and the growth factors.177 In addition to the literature reporting the good biocompatibility of graphene, there are papers revealing that graphene exhibits concentration-dependent toxicity to cells.207–210 For example, Fan et al. demonstrated that GO nanosheets with a lower dose of 20 μg mL−1 did not show toxicity to A549, and the dose of 85 μg mL−1 had increased cytotoxicity.69 Similar results were also obtained by Cui and colleagues reported that a GO concentration of less than 20 μg mL−1 did not cause appreciable toxicity and GO at a higher concentration (>50 μg mL−1) led to obvious cytotoxicity.211
4.2 Graphene for cell labeling and real-time monitoring
Cellular labeling is a technology that connects special reagents, such as organic dyes or inorganic NPs, to the target cells to trace their functions and behaviours.212–216 It is the foundation of cell imaging, drug delivery and many other cell-related applications. Aptamers are single-stranded nucleic acids selected by a molecular selection process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX).217,218 They can specifically bind to given target molecules with numerous merits, such as high affinity, specificity, recognition capability, small size and little immunogenicity,219–221 and thus have often been used for labeling and differentiating between different cell types.222 However, when delivered into cells the oligonucleotides are prone to be cleaved by enzymes. This problem can be solved smoothly by attaching aptamers to graphene-based structures like graphene oxide nanosheets (GO-nS) because the DNA is able to be effectively constrained on the surface of the graphene via non-covalent binding and the strong interactions between them will generate a steric hindrance effect.223,224 It is this effect that prevents DNAse from attaching to the DNA to initiate enzymatic digestion. Moreover, compared with adsorbing onto CNT where DNA is hypothesized to wrap around it,225 DNA linked to graphene can be more readily released from the surface in the presence of its complementary sequence. Therefore, graphene-based structures can figure as an efficient DNA protector as well as delivering oligonucleotides into cells for real-time monitoring.226,227 In the above example, graphene is just playing a role of vehicle and preserver for the fluorescent probe. Actually, graphene can be applied as a robust biological tag for cell labeling as well because of its distinctive qualities. Not long ago, Zhang and colleagues228 successfully synthesized a kind of graphene-based material, graphene quantum dots (GQDs), with bright yellow photoluminescence by an electrochemical method. After incubation with target cells, these uniform sized GQDs were effortlessly internalized in the stem cells for their unusual penetrating power (Fig. 5). Evidence also demonstrated that the presence of the GQDs did not affect the special characteristics of the cells, which open up a significant opportunity for their biomedical purposes.
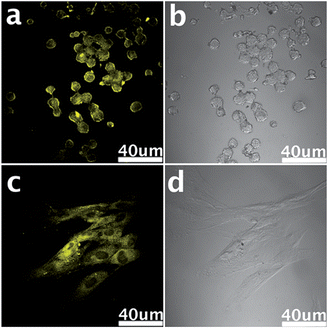 |
| | Fig. 5 Living neurosphere cells (a and b) and pancreas progenitor cells (c and d) labeled by GQDs. Reproduced with permission from ref. 228. Copyright 2012, Royal Society of Chemistry. | |
4.3 Cell analysis based on microfluidic device
Mounting evidence indicates that some cells (e.g., cancer stem cells and circulating cancer cells) in blood or tissues indicate the onset of human diseases and effect of treatment.229,230 But there usually are a small proportion of these cells. As a result, it is a complicated operation procedure needing rigorous conditions to survey these cells for disease diagnosis at an early stage that is ultrasensitive yet simple. Several kinds of promising tricks for cell testing include microfluidic cell separation,231 flow cytometry analysis,232 histological examination,233–236 nanoparticle-based biosensing237 and endogenous nucleic acid analysis.238 Here we emphatically introduce two of them: microfluidic cell separation and flow cytometry analysis. Owing to their large specific surface area and need for less time to separate the target molecule and cell, microfluidic devices have been in widespread use.239 Recently, a microfluidic device with GO for the selective and sensitive detection of circulating tumor cells (CTCs) aimed at the early diagnosis of cancer has been reported (Fig. 6).240 The overall size of the microfluidic device is just 35 mm × 10 mm × 3 mm. It comprises a PDMS layer with a microchannel and a silicon substrate with gold posts. GO functionalized with PEG was attached to the gold nanoposts and then the epithelial cell adhesion molecule (EpCAM) was immobilized on GO. When CTCs go through the microchannel, it would be captured by the nano microfluidic device, consequently, sensitive and specific detection of CTCs was achieved. Flow cytometry is a clinical diagnosis technique for determining thousands of blood-borne cells in a few seconds.241 For its distinct electronic nature that would be easily affected by electrostatic forces from long-range charge scatters, graphene has been integrated with microfluidic flow cytometry for the ‘flow-catch-release’ assay malaria-infected red blood cells at the single-cell level. According to the conductance change and dwell times, specific microscopic information related to the development of disease is capable to be achieved and a dynamic disease diagnostic patterns formed with the help of graphene. In addition, several other features of graphene, for instance optical transparency, easiness of integrating into microfluidic flow cytometry, ability to interface with cell-recognition proteins, endow it immense potential in clinical medicine.
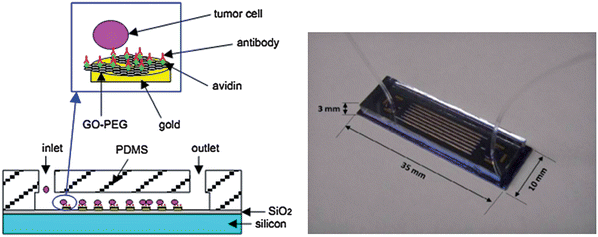 |
| | Fig. 6 Picture of the integrated nano microfluidic device for capturing CTCs on the functionalized GO/gold posts (left) and photograph of the microfluidic device (right). Reproduced with permission from ref. 240. Copyright 2011, Chemical and Biological Microsystems Society. | |
4.4 Cell imaging
Although numerous experimental data employed to structure the mathematical models of molecular networks come from routine analysis, an increasing amount of evidence demonstrates that the determination if unable to provide both the complex and the individual information in living cells. The emergence of cell imaging is capable of resolving this dilemma. Cell imaging provides an opportunity to easily glimpse into the world of biology in a natural setting, which is often difficult or even impossible with other strategies. Among numerous imaging techniques, fluorescence imaging is the most popular and important method for cell biological research. The graphene-based materials, for instance GO and reduced graphene oxide (RGO), have turned out to be fluorescent and thus have been utilized for fluorescence imaging.26,242–245 In a recent study, Wang and co-workers prepared graphene oxide nanoparticles (GONs) approximately sized at around 30 nm. They then functionalized them with transferrin (Trf) molecules and poly(ethylene glycol) (PEG) for targeting cancer cells and prolonging the blood circulation of nanoparticles respectively. After labeling these composites on cancer cells and irradiating them with an ultrafast pulsed laser, strong photoluminescence of GONs can be observed. As shown in Fig. 7, signal disturbance from cell auto-fluorescence and a molecular dye, fluorescein isothiocyanate (FITC), is possible to be avoided effectively with a low laser power of 7 mW.246 When the laser power was reduced to 4 mW, intensive microbubble and instant cell damage occurred, which showed that this system was not only suitable for target cell imaging but also for possible cell therapy. In addition, graphene and its derivatives can also be applied to cellular magnetic resonance imaging (MRI). Due to its capability of producing excellent quality and high-resolution images, allowing a fast scan time as well as no need of radiochemicals, MRI has been proved as the most important modality in both clinical and research settings.247–257 Therefore, integrating this advanced technique into cell (especially tumor cell) imaging will greatly boost the development of diagnostics. In MRI, superparamagnetic Fe3O4 NPs are widely used. They are usually decorated with suitable functionalities to obtain better biocompatibility and physiological stability along with an enhanced MRI contrast effect.258 Thanks to the specific structure and characters of GO, Fe3O4 NPs can anchor onto the surface of the GO sheet to prevent them from aggregating, which will result in a considerable relaxation rate.259–263 Compared with isolated Fe3O4 NPs, the Fe3O4–GO composites show significantly enhanced cellular MRI.
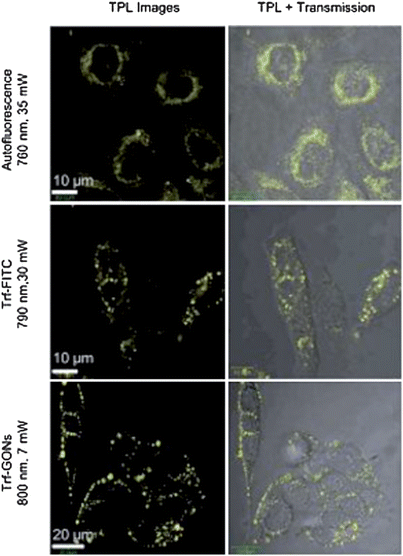 |
| | Fig. 7 Comparison of the in vitro two-photon luminescence of GONs with cell auto-fluorescence and the molecular dye FITC. Reproduced with permission from ref. 246. Copyright 2012, Wiley. | |
4.5 Gene delivery
Gene therapy is a novel and promising approach for the treatment of many acquired and inherited life-threatening diseases, including AIDS, cystic fibrosis, cancer, Parkinson's disease, etc.264–266 An efficacious treatment result requires that a nucleic acid drug can be successfully delivered to the interior of the target cell.267 So the development of a safe gene delivery vector with the ability to protect DNA from enzymatic cleavage and facilitate cellular uptake of DNA with high efficiency is a significant factor.268 It proved that graphene could bind to single-stranded DNA effectively through the π–π interaction and was able to protect oligonucleotides from nuclease degradation. Zhang et al. reported a gene delivery system based on GO functionalized with branched polyethylenimine (PEI-GO).269 The complexation of PEI-GO with plasmid DNA (pDNA) was allowed via an electrostatic interaction arising from the cationic PEI. Intracellular tracking of Cy3-labelled pGL-3 demonstrated that PEI-GO was able to effectively deliver pDNA into cells (Fig. 8). In addition, Kim and colleagues fabricated a hybrid gene carrier by conjugating low-molecular weight branched polyethylenimine (BPEI) to GO. Results confirmed that BPEI−GO possessed high gene delivery efficiency and exhibited high cell viability.270 In another investigation, GO was covalently functionalized with chitosan (CS) by an amide linkage.271 At the same time, pDNA could be condensed on the GO-CS sheet to form stable nanoparticle, which exhibited reasonable transfection efficiency in HeLa cells at certain nitrogen/phosphate (N/P) ratios.
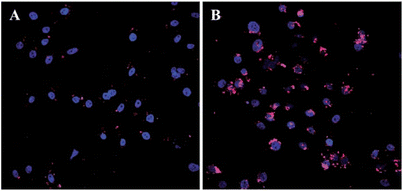 |
| | Fig. 8 Intracellular tracking of Cy3-labelled pGL-3 (red) in HeLa cells. A and B are for 4 h and 24 h post-transfection respectively. Reproduced with permission from ref. 269. Copyright 2011, Royal Society of Chemistry. | |
4.6 Determining dynamic secretion of living cells
Cell secretion is a fundamental and ubiquitous cell function involved in the regulation of various physiological processes, such as the digestion of food, neurotransmission, hormonal control of cell- and reproductive-cycles, and many other life processes.272 Investigation of cell secretions is of great value in diagnosing, monitoring, assessing and treating related disease. Routine assays using ensemble biochemical measurements from a large cell population with a low temporal resolution are unable to resolve the fast kinetics of cell secretion and detect trace amounts of released molecules.273 Fortunately, the emergence and utilization of nanomaterials provide new opportunities for the analysis of this cellular event and provide spatiotemporal information on fundamental researches and drug discovery. Nanomaterials such as nanoparticles, carbon nanotubes and more recently graphene have been reported to determine cell secretions. For example, Chen and co-workers employed single-walled carbon nanotube (SWNT) network-based field-effect transistors (SWNT-net FET) for the label-free detection of ATP release from living astrocytes.274 Subsequently, they coupled SWNT-net FET with neuroendocrine PC12 cells to detect their secretion of hormone catecholamines upon triggering.275 Compared to CNTs, graphene is more advantageous for detecting cell secretions because of its easy functionalization, large sensing area and suitability for interfacing with cell membranes. Zhang et al. synthesized large-area ultrathin RGO films to fabricate nanoelectronic FETs and then applied these RGO-based devices to the label-free monitored dynamic secretion of catecholamine molecules from PC12 cells.149 As illustrated in Fig. 9, PC12 cells were directly cultured on top of the FET devices and membrane depolarization resulting from a physiological stimulation opened voltage-gated Ca2+ ion channels. Once catecholamines were released from the cells, they quickly diffused onto the RGO films and interacted with them by π–π stacking, which would lead to current spikes. As a result, cell secretions were detected by monitoring the change in current. Although it is at an early stage, graphene for analysing the dynamic secretion of biomolecules is promising.
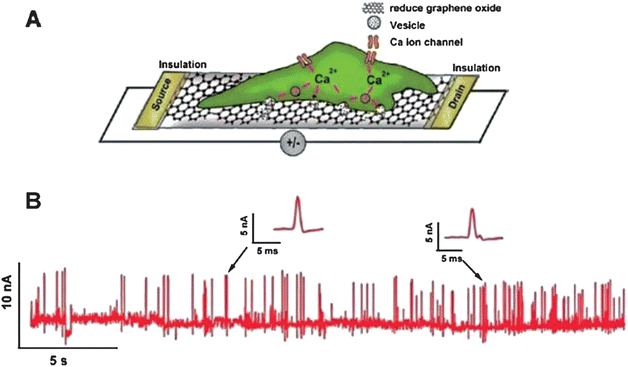 |
| | Fig. 9 (A) Schematic of the interface between a PC12 cell and an RGO FET. (B) Real-time response of RGO to the vesicular secretion of catecholamines from PC12 cells stimulated by high K+ solution. Reproduced with permission from ref. 149. Copyright 2010, American Chemical Society. | |
4.7 Probing the activity of enzymes
Enzymes are a kind of biocatalyst generated from the living cells of the biosome and, in fact, a majority of enzymes are proteins.276–281 They can catalyze all kinds of biochemical reactions, promoting metabolic progress with high efficiency. Accordingly, it is of vital significance for diagnosis and treatment to monitor the activity of enzymes. As its synthesis is facile, can hold surface functionalities, has a large surface area to mass ratio and a fluctuant surface, graphene can act as a potential artificial receptor and inhibit the activity of α-Chymotrypsin (ChT), a proteolytic enzyme capable of decomposing the denatured protein quickly.282–284 Results showed that graphene was strongly and compatibly attached by ChT through reversible interactions between the carboxylate groups of functionalized GO and the cationic surface residues of ChT (Fig. 10),285,286 hence there were no changes of the native conformation of ChT. Another strategy of the fluorescence resonance energy transfer (FRET)-based platform to assess enzymatic activity is a very popular means too. In this method, a pair of energy transfer acceptor and donor are obligatory. Under ordinary circumstances, the acceptor is graphene whereas the donor may be a quantum dot (QD) or fluorescent dye. According to the quenching and recovery of fluorescence, enzymatic activity is able to be obtained.287–291 Taking the assay of protease activity for example, in this measurement, as demonstrated in Fig. 11, a protease substrate peptide with one end attached to biotin and another end bonded with cysteine was employed as the link bridge between GO and QDs.292,293 Due to the ultra-efficient quenching capability of GO, the fluorescence of the QDs could be completely quenched through the FRET procedure. In the presence of target protease, the designated peptide was cleaved into two parts followed by the release of QDs from the surface of GO and the protease activity was detected based on the recovered fluorescence initially quenched. This system is useful for diagnosing protease-related disorders and screening potential drugs.
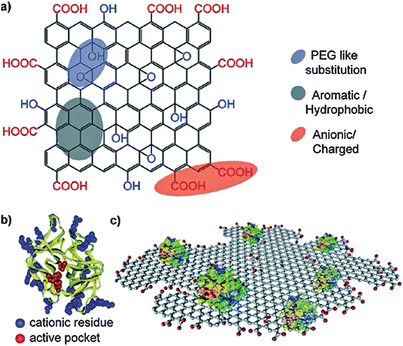 |
| | Fig. 10 The structures of (a) functionalized GO, and (b) α-chymotrypsin. (c) Schematic of reversible interactions between carboxylate groups of functionalized GO and the cationic surface residues of ChT. Reproduced with permission from ref. 286. Copyright 2011, American Chemical Society. | |
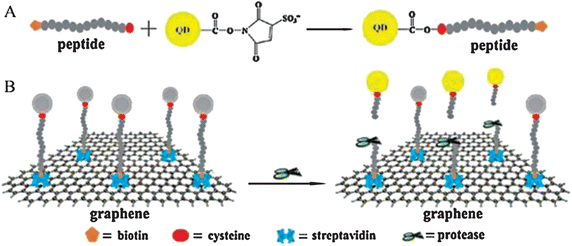 |
| | Fig. 11 (A) Schematic representation of constructing the peptide conjugated QDs. (B) Detecting protease activity through energy transfer by using graphene oxide. Reproduced with permission from ref. 292. Copyright 2011, Elsevier. | |
5. Conclusions and outlook
In summary, the recent advances of graphene applied for biotechnology, in particular, cell culture, real-time monitoring, living cell detection, imaging, gene delivery, investigation of cell secretions and surveying enzymatic activity, are revealed to readers. It is well known that there have been just 8 years in total since the discovery of graphene in 2004 and successful transfer from its original application field, physics, to the realm with better practical significance including biomedicine, biochemistry, materiology and leading-edge disciplines rising in recent times, has been realized with great achievements.294 Certainly, it is no exaggeration to say that the introduction of graphene into these areas brings infinite vitality to scientific researches and enormously enlivens the scientist's enthusiasm for study. Nevertheless, merging graphene into the aforementioned domains is still in its infancy and is conducted only at the laboratory level; consequently, a long journey with numerous challenges lies ahead to summon scientists to apply graphene in reality (e.g. hospitals).53,69,156,206,224,295–298 Therefore, future works are mainly concentrated on the following aspects, for example, tailoring the synthesis of nanohybrid materials for meeting specific applications and needs;224,295,296,299–305 investigating and clarifying the long-term exposure effects of this nanomaterial; combining graphene with other nanomaterials to form multifunctional materials accompanied by better performance; and integrating it into various advanced instruments and techniques such as MS, LC, MRI, photodynamic therapy (PDT), etc. Although these goals cannot be easily obtained, there is every reason to believe that with the joint efforts of biologists, chemists, physicists and medics, all of the difficulties and obstacles will be triumphed over as a better tomorrow follows.
Acknowledgements
The authors are grateful to the financial support from National Major Scientific Instruments and Equipments Development Special Funds (no. 2011YQ030113), National Recruitment Program of Global Experts (NRPGE), the Hundred Talents Program of Sichuan Province (HTPSP), and the Startup Funding of Sichuan University for setting up the Research Center of Analytical Instrumentation.
References
- C. M. Niemeyer, Angew. Chem., Int. Ed., 2001, 40, 4128–4158 CrossRef CAS.
- D. Caillerie, A. Mourad and A. Raoult, J. Elasticity, 2006, 84, 33–68 CrossRef.
- H. G. Craighead, Science, 2000, 290, 1532–1535 CrossRef CAS.
- K. L. Ekinci, Small, 2005, 1, 786–797 CrossRef CAS.
- D. A. C. Brownson and C. E. Banks, Analyst, 2010, 135, 2768–2778 RSC.
- C. R. Martin and P. Kohli, Nat. Rev. Drug Discovery, 2003, 2, 29–37 CrossRef CAS.
- T. Sawada and H. Mihara, Mol. BioSyst., 2012, 8, 1264–1274 RSC.
- W. Park, K. Choi, K. Lafdi and C. Yu, J. Heat. Trans.-T. Asme, 2012, 134, 041302 CrossRef.
- W.-S. Kuo, Y.-T. Chang, K.-C. Cho, K.-C. Chiu, C.-H. Lien, C.-S. Yeh and S.-J. Chen, Biomaterials, 2012, 33, 3270–3278 CrossRef CAS.
- H. Dai, E. W. Wong and C. M. Lieber, Science, 1996, 272, 523–526 CAS.
- M. Schrlau and H. Bau, Microfluid. Nanofluid., 2009, 7, 439–450 CrossRef CAS.
- F. Schedin, A. Geim, S. Morozov, E. Hill, P. Blake, M. Katsnelson and K. Novoselov, Nat. Mater., 2007, 6, 652–655 CrossRef CAS.
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef CAS.
- C. Rao, K. Biswas, K. Subrahmanyam and A. Govindaraj, J. Mater. Chem., 2009, 19, 2457–2469 RSC.
- E. C. H. Sykes, Nat. Chem., 2009, 1, 175–176 CrossRef CAS.
- A. Sikora, M. Woszczyna, M. Friedemann, F. J. Ahlers and M. Kalbacc, Micron, 2012, 43, 479–486 CrossRef CAS.
- B. F. Machado and P. Serp, Catal. Sci. Technol., 2012, 2, 54–75 CAS.
- Y. Barlas, K. Yang and A. H. MacDonald, Nanotechnology, 2012, 23, 052001 CrossRef.
- K. Novoselov, A. Geim, S. Morozov, D. Jiang, Y. Zhang, S. Dubonos, I. Grigorieva and A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS.
- S. Park, K.-S. Lee, G. Bozoklu, W. Cai, S. T. Nguyen and R. S. Ruoff, ACS Nano, 2008, 2, 572–578 CrossRef CAS.
- D. Cai, K. Yusoh and M. Song, Nanotechnology, 2009, 20, 085712 CrossRef.
- D. R. Dreyer, S. Park, C. W. Bielawski and R. S. Ruoff, Chem. Soc. Rev., 2010, 39, 228–240 RSC.
- E. R. Margine, M. L. Bocquet and X. Blase, Nano Lett., 2008, 8, 3315–3319 CrossRef CAS.
- D. A. Dikin, S. Stankovich, E. J. Zimney, R. D. Piner, G. H. B. Dommett, G. Evmenenko, S. T. Nguyen and R. S. Ruoff, Nature, 2007, 448, 457–460 CrossRef CAS.
- J. Zhang, H. Yang, G. Shen, P. Cheng, J. Zhang and S. Guo, Chem. Commun., 2010, 46, 1112–1114 RSC.
- F. Xu, H. Shi, X. He, K. Wang, X. Ye, L. Yan and S. Wei, Analyst, 2012, 137, 3989–3994 RSC.
- Y. Liu, D. Yu, C. Zeng, Z. Miao and L. Dai, Langmuir, 2010, 26, 6158–6160 CrossRef CAS.
- P. H. Lv, G. F. Wang, Y. Wan, J. Liu, Q. Liu and F. Ma, Scientometrics, 2011, 88, 399–419 CrossRef CAS.
- M. J. Pitkethly, Mater. Today, 2004, 7, 20–29 CrossRef.
- J. Wu, W. Pisula and K. Müllen, Chem. Rev., 2007, 107, 718–747 CrossRef CAS.
- K. S. Novoselov, Angew. Chem. Int. Ed., 2011, 50, 6986–7002 CrossRef CAS.
- K. S. Novoselov, Int. J. Mod. Phys. B, 2011, 25, 4081–4106 CrossRef CAS.
- A. K. Geim, Int. J. Mod. Phys. B, 2011, 25, 4055–4080 CrossRef CAS.
- J. C. Meyer, A. K. Geim, M. I. Katsnelson, K. S. Novoselov, T. J. Booth and S. Roth, Nature, 2007, 446, 60–63 CrossRef CAS.
- H. Lin, J. Huo, A. Zhang, Y. Liu, Q. Wang, Y. Cai, W. Ying, W. Qin, Y. Zhang and X. Qian, Analyst, 2012, 137, 3620–3623 RSC.
- H. Jang, Y. K. Kim, H. M. Kwon, W. S. Yeo, D. E. Kim and D. H. Min, Angew. Chem., Int. Ed., 2010, 122, 5839–5843 CrossRef.
- J. Balapanuru, J.-X. Yang, S. Xiao, Q. Bao, M. Jahan, L. Polavarapu, J. Wei, Q.-H. Xu and K. P. Loh, Angew. Chem., Int. Ed., 2010, 49, 6549–6553 CrossRef CAS.
- C. Rao, A. Sood, K. Subrahmanyam and A. Govindaraj, Angew. Chem., Int. Ed., 2009, 48, 7752–7777 CrossRef CAS.
- J. Zhu, Nat. Nanotechnol., 2008, 3, 528–529 CrossRef CAS.
- L. Wang, L.-H. Tian, G.-D. Wei, F.-M. Gao, J.-J. Zheng and W.-Y. Yang, J. Inorg. Mater., 2011, 26, 1009–1019 CrossRef CAS.
- T. Gan and S. Hu, Microchim. Acta, 2011, 175, 1–19 CrossRef CAS.
- M. S. Artiles, C. S. Rout and T. S. Fisher, Adv. Drug Delivery Rev., 2011, 63, 1352–1360 CrossRef CAS.
- P. Y. Huang, C. S. Ruiz-Vargas, A. M. van der Zande, W. S. Whitney, M. P. Levendorf, J. W. Kevek, S. Garg, J. S. Alden, C. J. Hustedt, Y. Zhu, J. Park, P. L. McEuen and D. A. Muller, Nature, 2011, 469, 389–392 CrossRef CAS.
- Z. Ni, H. Wang, J. Kasim, H. Fan, T. Yu, Y. Wu, Y. Feng and Z. Shen, Nano Lett., 2007, 7, 2758–2763 CrossRef CAS.
- H. Liu, S. Ryu, Z. Chen, M. L. Steigerwald, C. Nuckolls and L. E. Brus, J. Am. Chem. Soc., 2009, 131, 17099–17101 CrossRef CAS.
- N. G. Shang, P. Papakonstantinou, M. McMullan, M. Chu, A. Stamboulis, A. Potenza, S. S. Dhesi and H. Marchetto, Adv. Funct. Mater., 2008, 18, 3506–3514 CrossRef CAS.
- M. Ishigami, J. Chen, W. Cullen, M. Fuhrer and E. Williams, Nano Lett., 2007, 7, 1643–1648 CrossRef CAS.
- D. Konatham and A. Striolo, Nano Lett., 2008, 8, 4630–4641 CrossRef CAS.
- D. Graf, F. Molitor, K. Ensslin, C. Stampfer, A. Jungen, C. Hierold and L. Wirtz, Solid State Commun., 2007, 143, 44–46 CrossRef CAS.
- H. Xu, H. Dai and G. Chen, Talanta, 2010, 81, 334–338 CrossRef CAS.
- L. Jiang, R. Yuan, Y. Chai, Y. Yuan, L. Bai and Y. Wang, Analyst, 2012, 137, 2415–2420 RSC.
- S. Park and R. S. Ruoff, Nat. Nanotechnol., 2009, 4, 217–224 CrossRef CAS.
- Y. Zhu, S. Murali, W. Cai, X. Li, J. W. Suk, J. R. Potts and R. S. Ruoff, Adv. Mater., 2010, 22, 3906–3924 CrossRef CAS.
- Y. Zhang, Y. W. Tan, H. L. Stormer and P. Kim, Nature, 2005, 438, 201–204 CrossRef CAS.
- L. Jiao, L. Zhang, X. Wang, G. Diankov and H. Dai, Nature, 2009, 458, 877–880 CrossRef CAS.
- M. Liu, L. Wang, J. Deng, Q. Chen, Y. Li, Y. Zhang, H. Li and S. Yao, Analyst, 2012, 137, 4577–4583 RSC.
- S. Stankovich, D. A. Dikin, G. H. B. Dommett, K. M. Kohlhaas, E. J. Zimney, E. A. Stach, R. D. Piner, S. T. Nguyen and R. S. Ruoff, Nature, 2006, 442, 282–286 CrossRef CAS.
- J. S. Bunch, A. M. van der Zande, S. S. Verbridge, I. W. Frank, D. M. Tanenbaum, J. M. Parpia, H. G. Craighead and P. L. McEuen, Science, 2007, 315, 490–493 CrossRef CAS.
- Y. Xu, H. Bai, G. Lu, C. Li and G. Shi, J. Am. Chem. Soc., 2008, 130, 5856–5857 CrossRef CAS.
- X. Dong, Y. Shi, Y. Zhao, D. Chen, J. Ye, Y. Yao, F. Gao, Z. Ni, T. Yu, Z. Shen, Y. Huang, P. Chen and L.-J. Li, Phys. Rev. Lett., 2009, 102, 135501 CrossRef.
- K. Novoselov, A. Geim, S. Morozov, D. Jiang, M. I. K. I. V. Grigorieva, S. Dubonos and A. Firsov, Nature, 2005, 438, 197–200 CrossRef CAS.
- Z.-S. Wu, S. Pei, W. Ren, D. Tang, L. Gao, B. Liu, F. Li, C. Liu and H.-M. Cheng, Adv. Mater., 2009, 21, 1756–1760 CrossRef CAS.
- W. Yang, K. R. Ratinac, S. P. Ringer, P. Thordarson, J. J. Gooding and F. Braet, Angew. Chem., Int. Ed., 2010, 49, 2114–2138 CrossRef CAS.
- F. Liu, J. Y. Choi and T. S. Seo, Biosens. Bioelectron., 2010, 25, 2361–2365 CrossRef CAS.
- J. H. Jung, D. S. Cheon, F. Liu, K. B. Lee and T. S. Seo, Angew. Chem., Int. Ed., 2010, 49, 5708–5711 CrossRef CAS.
- C. Xu, X. Wang and J. Zhu, J. Phys. Chem. C, 2008, 112, 19841–19845 CAS.
- S. Wang, M. Tambraparni, J. Qiu, J. Tipton and D. Dean, Macromolecules, 2009, 42, 5251–5255 CrossRef CAS.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef CAS.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4, 4317–4323 CrossRef CAS.
- K. Yang, S. Zhang, G. Zhang, X. Sun, S.-T. Lee and Z. Liu, Nano Lett., 2010, 10, 3318–3323 CrossRef CAS.
- L. Zhang, J. Xia, Q. Zhao, L. Liu and Z. Zhang, Small, 2010, 6, 537–544 CrossRef CAS.
- D. Du, L. Wang, Y. Shao, J. Wang, M. H. Engelhard and Y. Lin, Anal. Chem., 2011, 83, 746–752 CrossRef CAS.
- Y. Song, Y. Chen, L. Feng, J. Ren and X. Qu, Chem. Commun., 2011, 47, 4436–4438 RSC.
- L. Feng, Y. Chen, J. Ren and X. Qu, Biomaterials, 2011, 32, 2930–2937 CrossRef CAS.
- X. Sun, Z. Liu, K. Welsher, J. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203–212 CrossRef CAS.
- T. Cohen-Karni, Q. Qing, Q. Li, Y. Fang and C. M. Lieber, Nano Lett., 2010, 10, 1098–1102 CrossRef CAS.
- M. Kalbacova, A. Broz, J. Kong and M. Kalbac, Carbon, 2010, 48, 4323–4329 CrossRef CAS.
- S. Y. Park, J. Park, S. H. Sim, M. G. Sung, K. S. Kim, B. H. Hong and S. Hong, Adv. Mater., 2011, 23, H263–H267 CrossRef CAS.
- Z. Liu, J. T. Robinson, X. Sun and H. Dai, J. Am. Chem. Soc., 2008, 130, 10876–10877 CrossRef CAS.
- B. Standley, W. Bao, H. Zhang, J. Bruck, C. N. Lau and M. Bockrath, Nano Lett., 2008, 8, 3345–3349 CrossRef CAS.
- N. Mohanty and V. Berry, Nano Lett., 2008, 8, 4469–4476 CrossRef CAS.
- S. Alwarappan, A. Erdem, C. Liu and C.-Z. Li, J. Phys. Chem. C, 2009, 113, 8853–8857 CAS.
- S. Alwarappan, C. Liu, A. Kumar and C.-Z. Li, J. Phys. Chem. C, 2010, 114, 12920–12924 CAS.
- S. Alwarappan, S. Boyapalle, A. Kumar, C.-Z. Li and S. Mohapatra, J. Phys. Chem. C, 2012, 116, 6556–6559 CAS.
- S. Alwarappan, R. K. Joshi, M. K. Ram and A. Kumar, Appl. Phys. Lett., 2010, 96, 263702 CrossRef.
- S. Alwarappan, S. R. Singh, S. Pillai, A. Kumar and S. Mohapatra, Anal. Lett., 2012, 45, 746–753 CrossRef CAS.
- X. Wang, L. Zhi and K. Müllen, Nano Lett., 2008, 8, 323–327 CrossRef CAS.
- Y. K. Kim and D. H. Min, Langmuir, 2009, 25, 11302–11306 CrossRef CAS.
- S. Gilje, S. Han, M. Wang, K. L. Wang and R. B. Kaner, Nano Lett., 2007, 7, 3394–3398 CrossRef CAS.
- M. D. Stoller, S. Park, Y. Zhu, J. An and R. S. Ruoff, Nano Lett., 2008, 8, 3498–3502 CrossRef CAS.
- Y. Xu, Z. Liu, X. Zhang, Y. Wang, J. Tian, Y. Huang, Y. Ma, X. Zhang and Y. Chen, Adv. Mater., 2009, 21, 1275–1279 CrossRef CAS.
- W. Hong, Y. Xu, G. Lu, C. Li and G. Shi, Electrochem. Commun., 2008, 10, 1555–1558 CrossRef CAS.
- L. Zhao, Y. Xu, T. Qiu, L. Zhi and G. Shi, Electrochim. Acta, 2009, 55, 491–497 CrossRef CAS.
- F.-R. F. Fan, S. Park, Y. Zhu, R. S. Ruoff and A. J. Bard, J. Am. Chem. Soc., 2009, 131, 937–939 CrossRef CAS.
- M. Pumera, Chem. Rec., 2009, 9, 211–223 CrossRef CAS.
- L. Chang, S. Alwarappan, C. Zhongfang, K. Xiangxing and L. Chen-Zhong, Biosens. Bioelectron., 2010, 25, 1829–1833 CrossRef.
- W. Choi, I. Lahiri, R. Seelaboyina and Y. S. Kang, Crit. Rev. Solid State Mater. Sci., 2010, 35, 52–71 CrossRef CAS.
- A. H. Castro Neto, F. Guinea, N. M. R. Peres, K. S. Novoselov and A. K. Geim, Rev. Mod. Phys., 2009, 81, 109–162 CrossRef CAS.
- L. Yilun, X. Bo, Z. Zhong, Z. Quanshui and X. Zhiping, J. Mech. Phys. Solids, 2012, 60, 591–605 CrossRef.
- C. Baykasoglu and A. Mugan, Comput. Mater. Sci., 2012, 55, 228–236 CrossRef CAS.
- Z. Liuyan, H. Rui, R. Kwang Taeg, T. Schiros, K. Keun Soo, Z. Hui, C. Gutierrez, S. P. Chockalingam, C. J. Arguello, L. Palova, D. Nordlund, M. S. Hybertsen, D. R. Reichman, T. F. Heinz, P. Kim, A. Pinczuk, G. W. Flynn and A. N. Pasupathy, Science, 2011, 333, 999–1003 CrossRef.
- C. Jin Sik, K. Jin-Soo, B. Ik-Su, L. Duk Hyun, L. Mi Jung, P. Bae Ho, L. Changgu, Y. Duhee, C. Hyeonsik, L. Ki Ho, S. Young-Woo, P. Jeong Young and M. Salmeron, Science, 2011, 333, 607–610 CrossRef.
- N. M. Gabor, J. C. W. Song, Q. Ma, N. L. Nair, T. Taychatanapat, K. Watanabe, T. Taniguchi, L. S. Levitov and P. Jarillo-Herrero, Science, 2011, 334, 648–652 CrossRef CAS.
- L. Britnell, R. V. Gorbachev, R. Jalil, B. D. Belle, F. Schedin, A. Mishchenko, T. Georgiou, M. I. Katsnelson, L. Eaves, S. V. Morozov, N. M. R. Peres, J. Leist, A. K. Geim, K. S. Novoselov and L. A. Ponomarenko, Science, 2012, 335, 947–950 CrossRef CAS.
- D. Basko, Science, 2011, 334, 610–611 CrossRef CAS.
- R. Van Noorden, Nature, 2006, 442, 228–229 CrossRef CAS.
- N. Peres, J. Phys.: Condens. Matter, 2009, 21, 323201 CrossRef CAS.
- L. Malard, M. Pimenta, G. Dresselhaus and M. Dresselhaus, Phys. Rep., 2009, 473, 51–87 CrossRef CAS.
- D. S. Hecht, L. Hu and G. Irvin, Adv. Mater., 2011, 23, 1482–1513 CrossRef CAS.
- N. Tombros, C. Jozsa, M. Popinciuc, H. T. Jonkman and B. J. Van Wees, Nature, 2007, 448, 571–574 CrossRef CAS.
- K. Bolotin, K. Sikes, J. Hone, H. Stormer and P. Kim, Phys. Rev. Lett., 2008, 101, 96802 CrossRef CAS.
- K. I. Bolotin, K. J. Sikes, Z. Jiang, M. Klima, G. Fudenberg, J. Hone, P. Kim and H. L. Stormer, Solid State Commun., 2008, 146, 351–355 CrossRef CAS.
- L. Schadler, S. Giannaris and P. Ajayan, Appl. Phys. Lett., 1998, 73, 3842 CrossRef CAS.
- H. K. Chae, D. Y. Siberio-Pérez, J. Kim, Y. B. Go, M. Eddaoudi, A. J. Matzger, M. O'Keeffe and O. M. Yaghi, Nature, 2004, 427, 523–527 CrossRef CAS.
- C. Lee, X. Wei, J. W. Kysar and J. Hone, Science, 2008, 321, 385–388 CrossRef CAS.
- M. J. Allen, V. C. Tung and R. B. Kaner, Chem. Rev., 2010, 110, 132–145 CrossRef CAS.
- H. Gao, L. Song, W. H. Guo, L. Huang, D. Z. Yang, F. C. Wang, Y. L. Zuo, X. L. Fan, Z. Liu, W. Gao, R. Vajtai, K. Hackenberg and P. M. Ajayan, Carbon, 2012, 50, 4476–4482 CrossRef CAS.
- D. Li, M. B. Müller, S. Gilje, R. B. Kaner and G. G. Wallace, Nat. Nanotechnol., 2008, 3, 101–105 CrossRef CAS.
- C. Gómez-Navarro, R. T. Weitz, A. M. Bittner, M. Scolari, A. Mews, M. Burghard and K. Kern, Nano Lett., 2007, 7, 3499–3503 CrossRef.
- X. Li, G. Zhang, X. Bai, X. Sun, X. Wang, E. Wang and H. Dai, Nat. Nanotechnol., 2008, 3, 538–542 CrossRef CAS.
- S. Stankovich, D. A. Dikin, R. D. Piner, K. A. Kohlhaas, A. Kleinhammes, Y. Jia, Y. Wu, S. B. T. Nguyen and R. S. Ruoff, Carbon, 2007, 45, 1558–1565 CrossRef CAS.
- H. C. Schniepp, J. L. Li, M. J. McAllister, H. Sai, M. Herrera-Alonso, D. H. Adamson, R. K. Prud'homme, R. Car, D. A. Saville and I. A. Aksay, J. Phys. Chem. B, 2006, 110, 8535–8539 CrossRef CAS.
- X. Lu, M. Yu, H. Huang and R. S. Ruoff, Nanotechnology, 1999, 10, 269 CrossRef CAS.
- V. Singh, D. Joung, L. Zhai, S. Das, S. I. Khondaker and S. Seal, Prog. Mater. Sci., 2011, 56, 1178–1271 CrossRef CAS.
- U. Stöberl, U. Wurstbauer, W. Wegscheider, D. Weiss and J. Eroms, Appl. Phys. Lett., 2008, 93, 051906 CrossRef.
- M. A. Fanton, J. A. Robinson, C. Puls, Y. Liu, M. J. Hollander, B. E. Weiland, M. LaBella, K. Trumbull, R. Kasarda and C. Howsare, ACS Nano, 2011, 8062–8069 CrossRef CAS.
- A. Reina, X. Jia, J. Ho, D. Nezich, H. Son, V. Bulovic, M. S. Dresselhaus and J. Kong, Nano Lett., 2009, 9, 3087–3087 CrossRef CAS.
- D. Wei, Y. Liu, Y. Wang, H. Zhang, L. Huang and G. Yu, Nano Lett., 2009, 9, 1752–1758 CrossRef CAS.
- X. Li, C. W. Magnuson, A. Venugopal, J. An, J. W. Suk, B. Han, M. Borysiak, W. Cai, A. Velamakanni and Y. Zhu, Nano Lett., 2010, 4328–4334 CrossRef CAS.
- H. Seul Ki, S. Seung Min, S. Onejae and C. Byung Jin, J. Electrochem. Soc., 2012, 159, K107–K109 CrossRef.
- N. Lam Van and K. Eui-Tae, J. Electrochem. Soc., 2012, 159, K93–K96 CrossRef.
- K. S. Kim, Y. Zhao, H. Jang, S. Y. Lee, J. M. Kim, K. S. Kim, J.-H. Ahn, P. Kim, J.-Y. Choi and B. H. Hong, Nature, 2009, 457, 706–710 CrossRef CAS.
- X. Li, W. Cai, J. An, S. Kim, J. Nah, D. Yang, R. Piner, A. Velamakanni, I. Jung, E. Tutuc, S. K. Banerjee, L. Colombo and R. S. Ruoff, Science, 2009, 324, 1312–1314 CrossRef CAS.
- P. W. Sutter, J.-I. Flege and E. A. Sutter, Nat. Mater., 2008, 7, 406–411 CrossRef CAS.
- N. Xiao, X. Dong, L. Song, D. Liu, Y. Tay, S. Wu, L.-J. Li, Y. Zhao, T. Yu, H. Zhang, W. Huang, H. H. Hng, P. M. Ajayan and Q. Yan, ACS Nano, 2011, 5, 2749–2755 CrossRef CAS.
- X. Dong, B. Li, A. Wei, X. Cao, M. B. Chan-Park, H. Zhang, L.-J. Li, W. Huang and P. Chen, Carbon, 2011, 49, 2944–2949 CrossRef CAS.
- M. Eizenberg and J. Blakely, Surf. Sci., 1979, 82, 228–236 CrossRef CAS.
- T. Aizawa, R. Souda, S. Otani, Y. Ishizawa and C. Oshima, Phys. Rev. Lett., 1990, 64, 768–771 CrossRef CAS.
- P. R. Somani, S. P. Somani and M. Umeno, Chem. Phys. Lett., 2006, 430, 56–59 CrossRef CAS.
- C. Rao, A. Sood, R. Voggu and K. Subrahmanyam, J. Phys. Chem. Lett., 2010, 1, 572–580 CrossRef CAS.
- A. Obraztsov, E. Obraztsova, A. Tyurnina and A. Zolotukhin, Carbon, 2007, 45, 2017–2021 CrossRef CAS.
- H. Ueta, M. Saida, C. Nakai, Y. Yamada, M. Sasaki and S. Yamamoto, Surf. Sci., 2004, 560, 183–190 CrossRef CAS.
- J. Coraux, A. T. N'Diaye, C. Busse and T. Michely, Nano Lett., 2008, 8, 565–570 CrossRef CAS.
- A. Vázquez de Parga, F. Calleja, B. Borca, M. Passeggi, Jr, J. Hinarejos, F. Guinea and R. Miranda, Phys. Rev. Lett., 2008, 100, 56807 CrossRef.
- A. Starodubov, M. Medvetskii, A. Shikin and V. Adamchuk, Phys. Solid State, 2004, 46, 1340–1348 CrossRef CAS.
- S. Bae, H. Kim, Y. Lee, X. Xu, J.-S. Park, Y. Zheng, J. Balakrishnan, T. Lei, H. R. Kim, Y. I. Song, Y.-J. Kim, K. S. Kim, B. Ozyilmaz, J.-H. Ahn, B. H. Hong and S. Iijima, Nat. Nanotechnol., 2010, 5, 574–578 CrossRef CAS.
- Y. Wang, X. Xu, J. Lu, M. Lin, Q. Bao, B. Özyilmaz and K. P. Loh, ACS Nano, 2010, 4, 6146–6152 CrossRef CAS.
- X. Qi, K.-Y. Pu, H. Li, X. Zhou, S. Wu, Q.-L. Fan, B. Liu, F. Boey, W. Huang and H. Zhang, Angew. Chem., Int. Ed., 2010, 49, 9426–9429 CrossRef CAS.
- Q. He, H. G. Sudibya, Z. Yin, S. Wu, H. Li, F. Boey, W. Huang, P. Chen and H. Zhang, ACS Nano, 2010, 4, 3201–3208 CrossRef CAS.
- X. Qi, K.-Y. Pu, X. Zhou, H. Li, B. Liu, F. Boey, W. Huang and H. Zhang, Small, 2010, 6, 663–669 CrossRef CAS.
- X. Zhou, X. Huang, X. Qi, S. Wu, C. Xue, F. Y. C. Boey, Q. Yan, P. Chen and H. Zhang, J. Phys. Chem. C, 2009, 113, 10842–10846 CAS.
- D. V. Kosynkin, A. L. Higginbotham, A. Sinitskii, J. R. Lomeda, A. Dimiev, B. K. Price and J. M. Tour, Nature, 2009, 458, 872–876 CrossRef CAS.
- C. Berger, Z. Song, X. Li, X. Wu, N. Brown, C. Naud, D. Mayou, T. Li, J. Hass and A. N. Marchenkov, Science, 2006, 312, 1191 CrossRef CAS.
- A. G. Cano-Márquez, F. J. Rodríguez-Macías, J. Campos-Delgado, C. G. Espinosa-González, F. Tristán-López, D. Ramírez-González, D. A. Cullen, D. J. Smith, M. Terrones and Y. I. Vega-Cantú, Nano Lett., 2009, 9, 1527–1533 CrossRef.
- Z.-S. Wu, W. Ren, L. Gao, B. Liu, C. Jiang and H.-M. Cheng, Carbon, 2009, 47, 493–499 CrossRef CAS.
- Y. Shao, J. Wang, H. Wu, J. Liu, I. A. Aksay and Y. Lin, Electroanalysis, 2010, 22, 1027–1036 CrossRef CAS.
- A. Kongkanand, R. M. Dominguez and P. V. Kamat, Nano Lett., 2007, 7, 676–680 CrossRef CAS.
- P. Brown, K. Takechi and P. V. Kamat, J. Phys. Chem. C, 2008, 112, 4776–4782 CAS.
- G. Williams, B. Seger and P. V. Kamat, ACS Nano, 2008, 2, 1487–1491 CrossRef CAS.
- N. Yang, J. Zhai, D. Wang, Y. Chen and L. Jiang, ACS Nano, 2010, 4, 887–894 CrossRef CAS.
- V. C. Tung, M. J. Allen, Y. Yang and R. B. Kaner, Nat. Nanotechnol., 2009, 4, 25–29 CrossRef CAS.
- M. Terrones, A. R. Botello-Mendez, J. Campos-Delgado, F. Lopez-Urias, Y. I. Vega-Cantu, F. J. Rodriguez-Macias, A. L. Elias, E. Munoz-Sandoval, A. G. Cano-Marquez, J.-C. Charlier and H. Terrones, Nano Today, 2010, 5, 351–372 CrossRef.
- C. Ménard-Moyon, K. Kostarelos, M. Prato and A. Bianco, Chem. Biol., 2010, 17, 107–115 CrossRef.
- S. Garibaldi, C. Brunelli, V. Bavastrello, G. Ghigliotti and C. Nicolini, Nanotechnology, 2006, 17, 391 CrossRef CAS.
- J. Meng, L. Song, H. Kong, G. Zhu, C. Wang, L. Xu, S. Xie and H. Xu, J. Biomed. Mater. Res., Part A, 2006, 79A, 298–306 CrossRef CAS.
- X. Zhao and R. Liu, Environ. Int., 2012, 40, 244–255 CrossRef CAS.
- S. Jain, V. S. Thakare, M. Das, C. Godugu, A. K. Jain, R. Mathur, K. Chuttani and A. K. Mishra, Chem. Res. Toxicol., 2011, 24, 2028–2039 CrossRef CAS.
- D. Gutierrez-Praena, S. Pichardo, E. Sanchez, A. Grilo, A. Maria Camean and A. Jos, Toxicol. in Vitro, 2011, 25, 1883–1888 CrossRef CAS.
- I. Fenoglio, E. Aldieri, E. Gazzano, F. Cesano, M. Colonna, D. Scarano, G. Mazzucco, A. Attanasio, Y. Yakoub, D. Lison and B. Fubini, Chem. Res. Toxicol., 2012, 25, 74–82 CrossRef CAS.
- M. L. Di Giorgio, S. Di Bucchianico, A. M. Ragnelli, P. Aimola, S. Santucci and A. Poma, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2011, 722, 20–31 CrossRef CAS.
- J. Wang, P. Sun, Y. Bao, J. Liu and L. An, Toxicol. in Vitro, 2011, 25, 242–250 CrossRef CAS.
- A. K. Patlolla, S. M. Hussain, J. J. Schlager, S. Patlolla and P. B. Tchounwou, Environ. Toxicol., 2010, 25, 608–621 CrossRef CAS.
-
J. Kayat, V. Gajbhiye, R. K. Tekade and N. K. Jain, Nanomedicine: Nanotechnology, Biology and Medicine, 2011, vol. 7, pp. 40–49 Search PubMed.
- O. N. Ruiz, K. A. S. Fernando, B. Wang, N. A. Brown, P. G. Luo, N. D. McNamara, M. Vangsness, Y.-P. Sun and C. E. Bunker, ACS Nano, 2011, 5, 8100–8107 CrossRef CAS.
- T. R. Nayak, H. Andersen, V. S. Makam, C. Khaw, S. Bae, X. Xu, P.-L. R. Ee, J.-H. Ahn, B. H. Hong, G. Pastorin and B. Oezyilmaz, ACS Nano, 2011, 5, 4670–4678 CrossRef CAS.
- N. Li, X. Zhang, Q. Song, R. Su, Q. Zhang, T. Kong, L. Liu, G. Jin, M. Tang and G. Cheng, Biomaterials, 2011, 32, 9374–9382 CrossRef CAS.
- W. C. Lee, C. H. Y. X. Lim, H. Shi, L. A. L. Tang, Y. Wang, C. T. Lim and K. P. Loh, ACS Nano, 2011, 5, 7334–7341 CrossRef CAS.
- G. Y. Chen, D. W. P. Pang, S. M. Hwang, H. Y. Tuan and Y. C. Hu, Biomaterials, 2012, 33, 418–427 CrossRef CAS.
- I. W. Frank, D. M. Tanenbaum, A. M. Van der Zande and P. L. McEuen, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.–Process., Meas., Phenom., 2007, 25, 2558–2561 CrossRef CAS.
- J.-U. Lee, D. Yoon and H. Cheong, Nano Lett., 2012, 12, 4444–4448 CrossRef CAS.
- Y. Lee, S. Bae, H. Jang, S. Jang, S.-E. Zhu, S. H. Sim, Y. I. Song, B. H. Hong and J.-H. Ahn, Nano Lett., 2010, 10, 490–493 CrossRef CAS.
- B. Xu, G. Song, Y. Ju, X. Li, Y. Song and S. Watanabe, J. Cell. Physiol., 2012, 227, 2722–2729 CrossRef CAS.
- F. Tan, F. O'Neill, M. Naciri, D. Dowling and M. Al-Rubeai, Acta Biomater., 2012, 8, 1627–1638 CrossRef CAS.
- F. Crobu, V. Latini, M. F. Marongiu, V. Sogos, F. Scintu, S. Porcu, C. Casu, M. Badiali, A. Sanna, M. F. Manchinu and M. S. Ristaldi, Mol. Biol. Rep., 2012, 39, 3995–4007 CrossRef CAS.
- A. Shakhbazau, D. Shcharbin, N. Petyovka, N. Goncharova, I. Seviaryn, S. Kosmacheva, M. Bryszewska and M. Potapnev, J. Pharm. Sci., 2012, 101, 1546–1554 CrossRef CAS.
- E.-K. Kim, S. Lim, J.-M. Park, J. K. Seo, J. H. Kim, K. T. Kim, S. H. Ryu and P.-G. Suh, J. Cell. Physiol., 2012, 227, 1680–1687 CrossRef CAS.
- D. Tibullo, I. Barbagallo, C. Giallongo, P. La Cava, A. Branca, C. Conticello, F. Stagno, A. Chiarenza, G. A. Palumbo and F. D. Raimondo, Hematol. Oncol., 2012, 30, 27–33 CrossRef CAS.
- F. Sato, H. Sato, D. Jin, U. K. Bhawal, Y. Wu, M. Noshiro, T. Kawamoto, K. Fujimoto, H. Seino, S. Morohashi, Y. Kato and H. Kijima, Biochem. Biophys. Res. Commun., 2012, 419, 441–446 CrossRef CAS.
- N. G. Rim, S. J. Kim, Y. M. Shin, I. Jun, D. W. Lim, J. H. Park and H. Shin, Colloids Surf., B, 2012, 91, 189–197 CrossRef CAS.
- B. F. Pierce, E. Pittermann, N. Ma, T. Gebauer, A. T. Neffe, M. Holscher, F. Jung and A. Lendlein, Macromol. Biosci., 2012, 12, 312–321 CrossRef CAS.
- A. Spradling, D. Drummond-Barbosa and T. Kai, Nature, 2001, 414, 98–104 CrossRef CAS.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS.
- D. E. Discher, D. J. Mooney and P. W. Zandstra, Science, 2009, 324, 1673–1677 CrossRef CAS.
- S.-R. Ryoo, Y.-K. Kim, M.-H. Kim and D.-H. Min, ACS Nano, 2010, 4, 6587–6598 CrossRef CAS.
- H. Fan, L. Wang, K. Zhao, N. Li, Z. Shi, Z. Ge and Z. Jin, Biomacromolecules, 2010, 11, 2345–2351 CrossRef CAS.
- D. Depan, B. Girase, J. S. Shah and R. D. K. Misra, Acta Biomater., 2011, 7, 3432–3445 CrossRef CAS.
- Y. Chang, S. T. Yang, J. H. Liu, E. Dong, Y. Wang, A. Cao, Y. Liu and H. Wang, Toxicol. Lett., 2011, 200, 201–210 CrossRef CAS.
- H. J. Kong, J. Liu, K. Riddle, T. Matsumoto, K. Leach and D. J. Mooney, Nat. Mater., 2005, 4, 460–464 CrossRef CAS.
- D. E. Discher, P. Janmey and Y. L. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS.
- J. Huang, S. V. Grater, F. Corbellinl, S. Rinck, E. Bock, R. Kemkemer, H. Kessler, J. Ding and J. P. Spatz, Nano Lett., 2009, 9, 1111–1116 CrossRef CAS.
- S. Oh, K. S. Brammer, Y. S. J. Li, D. Teng, A. J. Engler, S. Chien and S. Jin, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2130–2135 CrossRef CAS.
- V. Brunetti, G. Maiorano, L. Rizzello, B. Sorce, S. Sabella, R. Cingolani and P. P. Pompa, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 6264–6269 CrossRef CAS.
- N. Sniadecki, R. A. Desai, S. A. Ruiz and C. S. Chen, Ann. Biomed. Eng., 2006, 34, 59–74 CrossRef.
- H. Chen, M. B. Mueller, K. J. Gilmore, G. G. Wallace and D. Li, Adv. Mater., 2008, 20, 3557–3561 CrossRef CAS.
- S. Agarwal, X. Zhou, F. Ye, Q. He, G. C. K. Chen, J. Soo, F. Boey, H. Zhang and P. Chen, Langmuir, 2010, 26, 2244–2247 CrossRef CAS.
- S. Park, N. Mohanty, J. W. Suk, A. Nagaraja, J. An, R. D. Piner, W. Cai, D. R. Dreyer, V. Berry and R. S. Ruoff, Adv. Mater., 2010, 22, 1736–1740 CrossRef CAS.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Chem. Res. Toxicol., 2011, 25, 15–34 CrossRef.
- Y. Zhang, S. F. Ali, E. Dervishi, Y. Xu, Z. Li, D. Casciano and A. S. Biris, ACS Nano, 2010, 4, 3181–3186 CrossRef CAS.
- A. Sasidharan, L. S. Panchakarla, P. Chandran, D. Menon, S. Nair, C. N. R. Rao and M. Koyakutty, Nanoscale, 2011, 3, 2461–2464 RSC.
- Y. Zhang, T. R. Nayak, H. Hong and W. Cai, Nanoscale, 2012, 4, 3833–3842 RSC.
- K. Wang, J. Ruan, H. Song, J. Zhang, Y. Wo, S. Guo and D. Cui, Nanoscale Res. Lett., 2011, 6, 8 Search PubMed.
- X. Wang, F. Wei, A. Liu, L. Wang, J.-C. Wang, L. Ren, W. Liu, Q. Tu, L. Li and J. Wang, Biomaterials, 2012, 33, 3719–3732 CrossRef CAS.
- J. Wang, W. Ding, B. Sun, R. Jing, H. Huang, G. Shi and H. Wang, Mol. Cell. Biochem., 2012, 363, 1–10 CrossRef CAS.
- J. Yue, S. Liu, G. Mo, R. Wang and X. Jing, J. Controlled Release, 2011, 152, E258–E260 CrossRef CAS.
- G. Schmidtke-Schrezenmeier, M. Urban, A. Musyanovych, V. Mailaender, M. Rojewski, N. Fekete, C. Menard, E. Deak, K. Tarte, V. Rasche, K. Landfester and H. Schrezenmeier, Cytotherapy, 2011, 13, 962–975 CrossRef CAS.
- H. Freichels, F. Danhier, V. Preat, P. Lecomte and C. Jerome, Int. J. Artif. Organs, 2011, 34, 152–160 CrossRef CAS.
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11838–11843 CrossRef CAS.
- S. Jhaveri, M. Rajendran and A. D. Ellington, Nat. Biotechnol., 2000, 18, 1293–1297 CrossRef CAS.
- S. M. Nimjee, C. P. Rusconi and B. A. Sullenger, Annu. Rev. Med., 2005, 56, 555 CrossRef CAS.
- K. D. Bake and D. R. Walt, Annu. Rev. Anal. Chem., 2008, 1, 515–547 CrossRef CAS.
- B. Soontornworajit and Y. Wang, Anal. Bioanal. Chem., 2011, 399, 1591–1599 CrossRef CAS.
- D. Shangguan, Z. C. Cao, Y. Li and W. Tan, Clin. Chem., 2007, 53, 1153–1155 CAS.
- Y. Wu, J. A. Phillips, H. Liu, R. Yang and W. Tan, ACS Nano, 2008, 2, 2023–2028 CrossRef CAS.
- Z. Tang, H. Wu, J. R. Cort, G. W. Buchko, Y. Zhang, Y. Shao, I. A. Aksay, J. Liu and Y. Lin, Small, 2010, 6, 1205–1209 CrossRef CAS.
- M. Zheng, A. Jagota, E. D. Semke, B. A. Diner, R. S. McLean, S. R. Lustig, R. E. Richardson and N. G. Tassi, Nat. Mater., 2003, 2, 338–342 CrossRef CAS.
- Y. Wang, Z. Li, D. Hu, C.-T. Lin, J. Li and Y. Lin, J. Am. Chem. Soc., 2010, 132, 9274–9276 CrossRef CAS.
- C.-H. Lu, C.-L. Zhu, J. Li, J.-J. Liu, X. Chen and H.-H. Yang, Chem. Commun., 2010, 46, 3116–3118 RSC.
- M. Zhang, L. Bai, W. Shang, W. Xie, H. Ma, Y. Fu, D. Fang, H. Sun, L. Fan, M. Han, C. Liu and S. Yang, J. Mater. Chem., 2012, 22, 7461–7467 RSC.
- M. Cristofanilli, G. T. Budd, M. J. Ellis, A. Stopeck, J. Matera, M. C. Miller, J. M. Reuben, G. V. Doyle, W. J. Allard, L. Terstappen and D. F. Hayes, N. Engl. J. Med., 2004, 351, 781–791 CrossRef CAS.
-
M. Diehn, R. W. Cho and M. F. Clarke, Seminars in Radiation Oncology, 2009, vol. 19, pp. 78–86 Search PubMed.
- J. A. Phillips, Y. Xu, Z. Xia, Z. H. Fan and W. Tan, Anal. Chem., 2009, 81, 1033–1039 CrossRef CAS.
- B. J. Hicke, S. R. Watson, A. Koenig, C. K. Lynott, R. F. Bargatze, Y. F. Chang, S. Ringquist, L. MoonMcDermot, S. Jennings, T. Fitzwater, H. L. Han, N. Varki, I. Albinana, M. C. Willis, A. Varki and D. Parma, J. Clin. Invest., 1996, 98, 2688–2692 CrossRef CAS.
- B. J. Hicke, A. W. Stephens, T. Gould, Y. F. Chang, C. K. Lynott, J. Heil, S. Borkowski, C. S. Hilger, G. Cook, S. Warren and P. G. Schmidt, J. Nucl. Med., 2006, 47, 668–678 CAS.
- D. Shangguan, L. Meng, Z. C. Cao, Z. Xiao, X. Fang, Y. Li, D. Cardona, R. P. Witek, C. Liu and W. Tan, Anal. Chem., 2008, 80, 721–728 CrossRef CAS.
- D. Proske, M. Blank, R. Buhmann and A. Resch, Appl. Microbiol. Biotechnol., 2005, 69, 367–374 CrossRef CAS.
- S. Li, H. Xu, H. Ding, Y. Huang, X. Cao, G. Yang, J. Li, Z. Xie, Y. Meng, X. Li, Q. Zhao, B. Shen and N. Shao, J. Pathol., 2009, 218, 327–336 CrossRef CAS.
- J. E. Smith, C. D. Medley, Z. Tang, D. Shangguan, C. Lofton and W. Tan, Anal. Chem., 2007, 79, 3075–3082 CrossRef CAS.
- A. Ring, I. E. Smith and M. Dowsett, Lancet Oncol., 2004, 5, 79–88 CrossRef.
- I. Oita, H. Halewyck, B. Thys, B. Rombaut, Y. Vander Heyden and D. Mangelings, Anal. Bioanal. Chem., 2010, 398, 239–264 CrossRef CAS.
-
H. J. Yoon, K. Lee, Z. Zhang, T. M. Pham and S. Nagrath, Presented in Part at the 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Seattle, Washington, USA, 2011 Search PubMed.
- M. Brown and C. Wittwer, Clin. Chem., 2000, 46, 1221–1229 CAS.
- J. Shen, Y. Zhu, C. Chen, X. Yang and C. Li, Chem. Commun., 2011, 47, 2580–2582 RSC.
- D. Pan, L. Guo, J. Zhang, C. Xi, Q. Xue, H. Huang, J. Li, Z. Zhang, W. Yu, Z. Chen, Z. Li and M. Wu, J. Mater. Chem., 2012, 22, 3314–3318 RSC.
- C. Galande, A. D. Mohite, A. V. Naumov, W. Gao, L. Ci, A. Ajayan, H. Gao, A. Srivastava, R. B. Weisman and P. M. Ajayan, Sci. Rep., 2011, 1, 85 CrossRef.
- J.-L. Chen and X.-P. Yan, Chem. Commun., 2011, 47, 3135–3137 RSC.
- J.-L. Li, H.-C. Bao, X.-L. Hou, L. Sun, X.-G. Wang and M. Gu, Angew. Chem., Int. Ed., 2012, 51, 1830–1834 CrossRef CAS.
- P. Caravan, J. J. Ellison, T. J. McMurry and R. B. Lauffer, Chem. Rev., 1999, 99, 2293–2352 CrossRef CAS.
-
M. Vogel, D. Schranz and J. Bauer, Cardiology in the Young, 2012, vol. 22, pp. 204–205 Search PubMed.
- I. Rego, M. Severino, C. Micalizzi, M. Faraci, D. Pende, C. Dufour, M. Arico and A. Rossi, Pediatr. Blood Canc., 2012, 58, 810–814 CrossRef.
- S. L. Partington, S. Cheng and J. A. C. Lima, Heart Fail. Clin., 2012, 8, 179–190 CrossRef.
- X. Li, H. Li, G. Liu, Z. Deng, S. Wu, P. Li, Z. Xu, H. Xu and P. K. Chu, Biomaterials, 2012, 33, 3013–3024 CrossRef CAS.
- C. C. Abbott, F. Merideth, D. Ruhl, Z. Yang, V. P. Clark, V. D. Calhoun, F. M. Hanlon and A. R. Mayer, Prog. Neuro-Psychopharmacol. Biol. Psychiatry, 2012, 37, 161–168 CrossRef.
- A. Schmid, B. Rignall, B. J. Pichler and M. Schwarz, Toxicol. Sci., 2012, 126, 52–59 CrossRef CAS.
- L.-M. Wu, J.-R. Xu, M.-J. Liu, X.-F. Zhang, J. Hua, J. Zheng and J.-N. Hu, Acad. Radiol., 2012, 19, 331–340 CrossRef.
- A. Schuster, M. Ishida, G. Morton, B. Bigalke, M. T. Moonim and E. Nagel, Clin. Res. Cardiol., 2012, 101, 237–238 CrossRef.
- D. Luo, Y.-Y. Yao, Y.-F. Li, Z.-L. Sheng, Y. Tang, F. Fang, K. Fang, G.-s. Ma and G.-J. Teng, Cardiovasc. Pathol., 2012, 21, 112–119 CrossRef.
- K. A. Haifley and S. Hecht, J. Am. Vet. Med. Assoc., 2012, 240, 577–579 CrossRef.
- J. Xie, G. Liu, H. S. Eden, H. Ai and X. Chen, Acc. Chem. Res., 2011, 44, 883–892 CrossRef CAS.
- H. Jaganathan, D. L. Hugar and A. Ivanisevic, ACS Appl. Mater. Interfaces, 2011, 3, 1282–1288 CAS.
- H. Ai, C. Flask, B. Weinberg, X. Shuai, M. D. Pagel, D. Farrell, J. Duerk and J. M. Gao, Adv. Mater., 2005, 17, 1949–1952 CrossRef CAS.
- G. Liu, Z. Wang, J. Lu, C. Xia, F. Gao, Q. Gong, B. Song, X. Zhao, X. Shuai, X. Chen, H. Ai and Z. Gu, Biomaterials, 2011, 32, 528–537 CrossRef CAS.
- J. Lu, S. Ma, J. Sun, C. Xia, C. Liu, Z. Wang, X. Zhao, F. Gao, Q. Gong, B. Song, X. Shuai, H. Ai and Z. Gu, Biomaterials, 2009, 30, 2919–2928 CrossRef CAS.
- Z. Wang, G. Liu, J. Sun, B. Wu, Q. Gong, B. Song, H. Ai and Z. Gu, J. Nanosci. Nanotechnol., 2009, 9, 378–385 CrossRef CAS.
- F. McCormick, Nat. Rev. Cancer, 2001, 1, 130–141 CrossRef CAS.
- Z. R. Yang, H. F. Wang, J. Zhao, Y. Y. Peng, J. Wang, B. A. Guinn and L. Q. Huang, Cancer Gene Ther., 2007, 14, 599–615 CrossRef CAS.
- X. Guo and L. Huang, Acc. Chem. Res., 2011, 45, 971–979 CrossRef.
- I. M. Verma and N. Somia, Nature, 1997, 389, 239–242 CrossRef CAS.
- L. Naldini, U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma and D. Trono, Science, 1996, 272, 263–267 CAS.
- B. Chen, M. Liu, L. Zhang, J. Huang, J. Yao and Z. Zhang, J. Mater. Chem., 2011, 21, 7736–7741 RSC.
- H. Kim, R. Namgung, K. Singha, I.-K. Oh and W. J. Kim, Bioconjugate Chem., 2011, 22, 2558–2567 CrossRef CAS.
- H. Bao, Y. Pan, Y. Ping, N. G. Sahoo, T. Wu, L. Li, J. Li and L. H. Gan, Small, 2011, 7, 1569–1578 CrossRef CAS.
- S. Trikha, E. C. Lee and A. M. Jeremic, The Scientific World Journal, 2010, 10, 2054–2069 CrossRef CAS.
- Y. Huang, D. Cai and P. Chen, Anal. Chem., 2011, 83, 4393–4406 CrossRef CAS.
- Y. Huang, H. G. Sudibya, D. Fu, R. Xue, X. Dong, L.-J. Li and P. Chen, Biosens. Bioelectron., 2009, 24, 2716–2720 CrossRef CAS.
- H. G. Sudibya, J. Ma, X. Dong, S. Ng, L.-J. Li, X.-W. Liu and P. Chen, Angew. Chem., Int. Ed., 2009, 48, 2723–2726 CrossRef CAS.
- T. P. Khaket, J. Singh, P. Attri and S. Dhanda, Curr. Pharm. Des., 2012, 18, 220–230 CrossRef CAS.
- N. N. Nalivaeva, C. Beckett, N. D. Belyaev and A. J. Turner, J. Neurochem., 2012, 120, 167–185 CrossRef CAS.
- S.-H. Kim and C. R. Dass, DNA Cell Biol., 2012, 31, 1–7 CrossRef CAS.
- T. C. Chen, T. Sakaki, K. Yamamoto and A. Kittaka, Anticancer Res., 2012, 32, 291–298 CAS.
- A. N. Godet, J. Guergnon, A. Galioot, P.-B. Falanga, J.-H. Colle, X. Cayla and A. Garcia, M S-Med. Sci., 2011, 27, 1106–1111 Search PubMed.
- C. S. Ferguson and R. F. Tyndale, Trends Pharmacol. Sci., 2011, 32, 708–714 CrossRef CAS.
- Y. Yamaguchi, N. Kato, H. Azuma, T. Nagasaki and J. Ohkanda, Bioorg. Med. Chem. Lett., 2012, 22, 2354–2358 CrossRef CAS.
- A. Narai-Kanayama, T. Hanaishi and K. Aso, J. Biotechnol., 2012, 157, 428–436 CrossRef CAS.
- T. Matsuo, C. Imai, T. Yoshida, T. Saito, T. Hayashi and S. Hirota, Chem. Commun., 2012, 48, 1662–1664 RSC.
- Y. Shimohigashi, T. Nose, Y. Yamauchi and I. Maeda, Biopolymers, 1999, 51, 9–17 CrossRef CAS.
- M. De, S. S. Chou and V. P. Dravid, J. Am. Chem. Soc., 2011, 133, 17524–17527 CrossRef CAS.
- M. Oliveira, R. J. S. Torquato, M. F. M. Alves, M. A. Juliano, D. Bromme, N. M. T. Barros and A. K. Carmona, Peptides, 2010, 31, 562–567 CrossRef CAS.
- M. F. Marcondes, R. J. S. Torquato, D. M. Assis, M. A. Juliano, M. A. F. Hayashi and V. Oliveira, Biochem. Biophys. Res. Commun., 2010, 391, 123–128 CrossRef CAS.
- A. K. Carmona, M. A. Juliano and L. Juliano, An. Acad. Bras. Cienc., 2009, 81, 381–392 CrossRef CAS.
- K. Boeneman, J. B. Delehanty, K. Susumu, M. H. Stewart, J. R. Deschamps and I. L. Medintz, Adv. Exp. Med. Biol., 2012, 733, 63–74 CrossRef.
- D. Stockholm, M. Bartoli, G. Sillon, N. Bourg, J. Davoust and I. Richard, J. Mol. Biol., 2005, 346, 215–222 CrossRef CAS.
- J. Li, C.-H. Lu, Q.-H. Yao, X.-L. Zhang, J.-J. Liu, H.-H. Yang and G.-N. Chen, Biosens. Bioelectron., 2011, 26, 3894–3899 CrossRef CAS.
- X. Gu, G. Yang, G. Zhang, D. Zhang and D. Zhu, ACS Appl. Mater. Interfaces, 2011, 3, 1175–1179 CAS.
- J. Yao, Y. Sun, M. Yang and Y. Duan, J. Mater. Chem., 2012, 22, 14313–14329 RSC.
- M. J. Allen, V. C. Tung, L. Gomez, Z. Xu, L.-M. Chen, K. S. Nelson, C. Zhou, R. B. Kaner and Y. Yang, Adv. Mater., 2009, 21, 2098–2102 CrossRef CAS.
- B. Schmaltz, T. Weil and K. Muellen, Adv. Mater., 2009, 21, 1067–1078 CrossRef CAS.
- Y. Wang, Z. Li, J. Wang, J. Li and Y. Lin, Trends Biotechnol., 2011, 29, 205–212 CrossRef CAS.
- T. Premkumar and K. E. Geckeler, Prog. Polym. Sci., 2012, 37, 515–529 CrossRef CAS.
- A. Reina, X. Jia, J. Ho, D. Nezich, H. Son, V. Bulovic, M. S. Dresselhaus and J. Kong, Nano Lett., 2009, 9, 30–35 CrossRef CAS.
- M. Musameh, M. R. Notivoli, M. Hickey, I. L. Kyratzis, Y. Gao, C. Huynh and S. C. Hawkins, Adv. Mater., 2011, 23, 906–910 CrossRef CAS.
- A. K. Geim, Science, 2009, 324, 1530–1534 CrossRef CAS.
- T. Kuilla, S. Bhadra, D. Yao, N. H. Kim, S. Bose and J. H. Lee, Prog. Polym. Sci., 2010, 35, 1350–1375 CrossRef CAS.
- R. Stine, J. T. Robinson, P. E. Sheehan and C. R. Tamanaha, Adv. Mater., 2010, 22, 5297–5300 CrossRef CAS.
- S. Roy and Z. Gao, Nano Today, 2009, 4, 318–334 CrossRef CAS.
- H. Bai, C. Li and G. Shi, Adv. Mater., 2011, 23, 1089–1115 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2013 |
Click here to see how this site uses Cookies. View our privacy policy here. 













