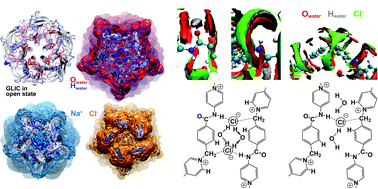
We combine the statistical–mechanical 1D/3D-RISM-KH molecular theory of solvation with DFT electronic structure calculations and MD and coarse-grained DPD simulations, and apply them to study association phenomena in synthetic organic soft matter and biomolecular systems. This combined approach from electronic to molecular structure and coarse-graining allows us to address complex processes occurring in solution on very long timescales, such as protein–ligand binding, distributions of electrolyte solution species in ion channels, and oligomeric polyelectrolyte gelation. The statistics of rare events involving supramolecular solute and solvent is accounted for analytically within the 3D-RISM-KH molecular theory of solvation. The hybrid MD/3D-RISM-KH method for molecular dynamics of the biomolecule in the potential of mean force obtained with the molecular theory of solvation has been implemented in the Amber package. The 3D molecular theory of solvation also replaces MM/GB(PB)SA post-processing using empirical treatment of non-polar contributions with MM/3D-RISM-KH evaluation of the solvation thermodynamics. The 3D-RISM-KH theory accurately yields the solvation structure for biomolecular systems as large as the GroEL chaperonin complex, GLIC ion channel, and 3D maps of ligand binding affinity of antiprion compound to mouse PrPC prion protein at once without phenomenological approximations. We apply the DFT/1D-3D-RISM-KH/DPD multiscale approach to investigate the structural and dynamical properties and gelation ability of oligomeric polyelectrolyte gelators, towards understanding of the gelation mechanism.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...