Co-delivery of genes and drugs with nanostructured calcium carbonate for cancer therapy
Si
Chen
,
Dong
Zhao
,
Feng
Li
,
Ren-Xi
Zhuo
and
Si-Xue
Cheng
*
Key Laboratory of Biomedical Polymers of Ministry of Education, Department of Chemistry, Wuhan University, Wuhan 430072, P. R. China. E-mail: chengsixue@hotmail.com; chengsixue@whu.edu.cn; Fax: +8627 6875 4509
First published on 4th January 2012
Abstract
By using a CaCO3 co-precipitation technique, p53 expression plasmids and doxorubicin hydrochloride (DOX) were encapsulated in nano-sized CaCO3/DNA/DOX co-precipitates for co-delivery of genes and drugs. Under a certain Ca2+/CO32− ratio in the co-precipitation, both plasmid DNA and drugs could be loaded in the CaCO3/DNA/DOX nanoparticles with high encapsulation efficiency. The in vitrocell inhibition of the CaCO3/DNA/DOX nanoparticles was evaluated in HeLa cells by a MTT assay. The results showed the simultaneous treatment by gene and drug could induce cell apoptosis and completely inhibit the cell proliferation. The CaCO3/DNA/DOX nanoparticles exhibited a high cell inhibition rate of about 75%, indicating that the CaCO3/DNA/DOX nanoparticles could effectively mediate gene transfection and deliver the drug to the cells. Compared with the gene delivery system (CaCO3/DNA nanoparticles) or the free drug DOX, the co-delivery system (CaCO3/DNA/DOX nanoparticles) exhibits enhanced cell inhibition rate. The calcium carbonate based approach has great potential in the preparation of gene and drug co-delivery systems, and the CaCO3/DNA/DOX nanoparticles have promising applications in cancer treatment.
Introduction
Gene therapy is a promising approach for the treatment of genetic disorder diseases such as cancers.1,2 To achieve maximum therapeutic efficiency, the strategy of combined drug and gene therapy for tumor treatments has attracted more and more research interests in recent years with the purpose to address the problem of multidrug resistance (MDR) and to achieve a synergic curing effect.3–5 It is well known that long-term chemotherapy causes the development of resistant cell phenotypes, resulting in the loss of sensitivity of cancer cells to anticancer agents. To overcome MDR at a genetic level, the combination of gene therapy and chemotherapy has a unique advantage.3–5 When a chemical therapeutic is administrated, certain genes can be delivered to cancer cells simultaneously with the aim to keep targeted cells sensitive to drugs during the entire treatment period. For example, drug resistance caused by malfunction of genes owing to chromosomal alternations could be overcome by correcting the malfunctioned gene through anticancer gene delivery.3 Tumor suppressor gene p53 plays an important role in cell cycle control and induces apoptosis of cells under DNA damage. p53 mutations occur in almost half of all soft tissue sarcomas, which also contribute to drug resistance.6–10 In addition to inhibiting p53 mutations, the reintroduction of p53 into tumor cells may also enhance the chemosensitivity of tumor cells to chemotherapeutic agents through the inhibition of MDR-1-encoded P-gp expression, which is often closely related to mutant p53.5,11 In addition, wild type p53 protein has a positive response to a variety of stress signals including DNA damage caused by antitumor drugs, like doxorubicin.5,11,12 Thus, the combination of p53 gene therapy and chemotherapy may increase the therapeutic efficacy in cancer treatments.The reported co-delivery systems for p53 gene and doxorubicin include cationic micelles assembled by oligopeptide amphiphile, and cationic polymer prodrug (β-cyclodextrin-polyethylenimine-Dox)/p53 nanocomplexes. Compared with treatments by genes and drugs separately, the co-delivery systems resulted in synergy in cytotoxic effect, i.e. an increased gene expression level as well as an enhanced cell inhibition effect.4,5
Doxorubicin is an anthracycline antibiotic that works by intercalating DNA and it is widely used in cancer chemotherapy in the treatment of a wide range of cancers.13,14 To reduce its side effects including congestive heart failure and dilated cardiomyopathy, a lot of effort has been devoted to develop its delivery systems to directly deliver doxorubicin to cancer cells.15–17
Efficient and safe gene transfer vectors are of critical importance for the applications of gene therapy. Compared with viral vectors presenting safety problems, non-viral vectors have attracted much attention because of their low immune response and safety.18 However, the biocompatibility of the widely investigated non-viral vectors based on cationic polymers and cationic liposomes is still not satisfactory. Among different non-viral gene delivery methods, the technique of co-precipitation of Ca2+ with DNA in the presence of inorganic anions present in bodies is an attractive option because of its good biocompatibility and biodegradability.19–22 Compared with the calcium phosphate based approach to prepare Ca–P/DNA co-precipitates which has been extensively studied, the preparation procedure of calcium carbonate based approach to prepare CaCO3/DNA co-precipitates is more simple and no buffer solution is needed to adjust the pH value.23 In our previous study, we optimized the Ca2+/CO32− ratio to obtain nano-sized CaCO3/DNA co-precipitates which could achieve efficient gene transfection with expression levels higher than that of the commercially available transfection reagent Lipofectamine 2000 in the presence of serum.23
In the current study, we prepared CaCO3/DNA/DOX nanoparticles for co-delivery of a p53 gene and DOX. By using the CaCO3 co-precipitation technique, both plasmid DNA and drug could be loaded in the CaCO3/DNA/DOX nanoparticles with high encapsulation efficiency. The in vitrocell inhibition study showed the co-delivery of p53 gene and the drug could induce a higher cell inhibition and promote tumor cell apoptosis more effectively. Our study indicates that the calcium carbonate based approach has great potential in the preparation of gene and drug co-delivery systems, and the CaCO3/DNA/DOX nanoparticles have promising applications in cancer treatments.
Experimental
Materials
Anhydrous calcium chloride and anhydrous sodium carbonate of analytical grade were supplied by Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China) and used as received. Doxorubicin hydrochloride (DOX) was provided by Zhejiang Hisun Pharmaceutical Co. Ltd. (China).Human cervical carcinoma cell line HeLa was obtained from China Center for Typical Culture Collection (Wuhan, China). The medium for cell culture was Dulbecco's Modified Eagle's Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS), 2 mg ml−1NaHCO3, and 100 U/ml penicillin/streptomycin. Cells were incubated at 37 °C in humidified air/5% CO2.
The p53 expression plasmid containing wild type human p53 cDNA (pDsRed2-N1-p53) was amplified in Escherichia coli and extracted and purified by QIAfilter Plasmid Mega Kit (QIAGEN). Plasmid DNA was suspended in de-ionized water and stored at −20 °C.
Preparation of CaCO3/DNA and CaCO3/DNA/DOX nanoparticles
10 μg of plasmid DNA was diluted with de-ionized water to a volume of 125 μl, and then 120 μl of aqueous solution containing 60 μmol of CaCl2 was added and mixed gently at 25 °C. The mixture was added drop-wise to 255 μl of Na2CO3 solution containing 1.6 μmol of Na2CO3 to obtain 500 μl of CaCO3/DNA nanoparticles containing solution. The solution was incubated for 5 min at 25 °C before transfection.10 μg of plasmid DNA was diluted with de-ionized water to a volume of 125 μl, and then 120 μl of aqueous solution containing 60 μmol of CaCl2 and 1 μg of doxorubicin hydrochloride was added and mixed gently at 25 °C. The mixture was added drop-wise to 255 μl of Na2CO3 solution containing 1.6 μmol of Na2CO3 to obtain 500 μl of CaCO3/DNA/DOX nanoparticles containing solution. The solution was incubated for 5 min at 25 °C before transfection.
Determination of loading content and encapsulation efficiency of DNA and DOX
500 μl of nanoparticles containing solution was centrifuged at 4 °C for 1 h at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. After centrifugation, the amount of non-precipitated free DNA remaining in the supernatant of solution was determined by the Quant-iT™ PicoGreen® dsDNA Assay Kit (Molecular Probes) according to the manufacturer's protocol using a spectrofluorophotometer (RF-5301 PC, Shimadzu). The amount of free DOX remaining in the supernatant of solution was determined by the absorbance at 485 nm in a UV-vis spectrophotometer (PerkinElmer Lambda Bio 40). Data were given as average values based on 3 independent measurements. The loading content and encapsulation efficiency were calculated as follows.
000 rpm. After centrifugation, the amount of non-precipitated free DNA remaining in the supernatant of solution was determined by the Quant-iT™ PicoGreen® dsDNA Assay Kit (Molecular Probes) according to the manufacturer's protocol using a spectrofluorophotometer (RF-5301 PC, Shimadzu). The amount of free DOX remaining in the supernatant of solution was determined by the absorbance at 485 nm in a UV-vis spectrophotometer (PerkinElmer Lambda Bio 40). Data were given as average values based on 3 independent measurements. The loading content and encapsulation efficiency were calculated as follows.
| Loading content = (WT − WF)/WNP × 100% | (1) |
| Encapsulation efficiency = (WT − WF)/WT × 100% | (2) |
Measurement of in vitrodrug release from CaCO3/DNA/DOX nanoparticles
The CaCO3/DNA/DOX nanoparticle containing solution (4 ml with 80 μg DNA and 8 μg DOX) was put in a dialysis bag and immersed in 30 mL of phosphate buffer (PBS) (pH 7.4) and shaken in a water bath at 37 °C. At predetermined intervals, 4 ml of solution outside the dialysis bag was taken out and replaced by 4 ml of fresh PBS. The drug concentration was determined by the absorbance at 485 nm in a UV-vis spectrophotometer (PerkinElmer Lambda Bio 40).Particle size measurements
The size and size distribution of nanoparticles were measured by a Zetasizer (Nano ZS, Malvern Instruments) at 25 °C. Prior to measurements, 900 μl de-ionized water was added to 100 μl of the freshly prepared nanoparticles containing solution for dilution.Evaluation of in vitrocell inhibition
HeLa cells in 1 ml of complete medium (DMEM containing 10% FBS) were seeded directly in the well of a 24-well plate (5 × 104cells per well) and incubated at 37 °C for 24 h. Then the freshly prepared CaCO3/DNA/DOX nanoparticle containing solution (100 μl with 2 μg DNA and 0.2 μg DOX) was added to each well, and the cells were incubated at 37 °C for predetermined time. Then the cell inhibition rate was determined by an MTT assay. The details are as follows. After the medium containing the co-precipitates was removed, 1 ml of complete medium and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (60 μl, 5 mg ml−1) were added to each well, followed by incubation at 37 °C for 4 h. Then the supernatant was carefully removed, and 1 ml DMSO was added to each well to dissolve the formazan crystals produced by viable cells. The absorbance of the solution was measured using a microplate reader (Bio-Rad 550) at 570 nm to determine the OD value. The data were given as mean ± standard deviation (SD) based on 3 independent measurements. The cell inhibition rate was calculated as follows.
| Cell inhibition rate = (1-ODtreated/ODcontrol) × 100% | (3) |
For comparison, HeLa cells were also treated by CaCO3/DNA nanoparticles in the absence of DOX. The other conditions were the same as that of the treatment of CaCO3/DNA/DOX nanoparticles.
For comparison, HeLa cells were also treated by CaCO3/DNA nanoparticles in the presence of free DOX. The details are as follows. HeLa cells in 1 ml of complete medium (DMEM containing 10% FBS) were seeded directly in the well of a 24-well plate (5 × 104cells per well) and incubated at 37 °C for 24 h. Then the freshly prepared CaCO3/DNA nanoparticle containing solution (100 μl with 2 μg DNA) was added to each well, and then free DOX (2 μl of with 0.2 μg DOX) solution was added to each well. After the cells were incubated at 37 °C for a predetermined time, the cell viability was determined by an MTT assay.
The statistical significance between two sets of data was calculated using Student's t-test. A P value < 0.05 was considered statistically significant.
Cell morphology observation
The HeLa cells exposed to different agents at 24 h and 48 h were observed by an inverted microscope (Olympus IX70) equipped with a digital color camera (Roper CoolSnap Color) under magnification of 200.Apoptosis assay
The cell apoptosis was assayed by FITC Annexin V/Dead Cell Apoptosis Kit with FITC annexin V and PI (Invitrogen). HeLa cells exposed to CaCO3/DNA/DOX co-precipitates containing solution for predetermined time were washed with cold PBS and 1× annexin-binding buffer. Then 100 μL of 1× annexin-binding buffer, 5 μl of the annexin V conjugate (Component A) and 1 μl of the PI working solution (100 μg mL−1) were added to the cells. After incubation for 15 min, the cells were washed with 1× annexin-binding buffer and observed using a confocal laser scanning microscope (Nikon C1–si TE2000) with excitation at 488 nm under magnification of 400.Results and discussion
Preparation and characterization of CaCO3/DNA/DOX nanoparticles
According to our previous study, the transfection efficiency of the calcium carbonate based approach is strongly dependent on the Ca2+/CO32− ratio of CaCO3/DNA co-precipitates. When the Ca2+/CO32− ratio is in an appropriate range, effective gene expression could be achieved because DNA could be well encapsulated in the CaCO3/DNA co-precipitates and the size of CaCO3/DNA co-precipitates is suitable for cell internalization. Based on our studies on the gene transfection of a CaCO3/pGL3–Luc system, the Ca2+/CO32− molar ratio of 37.5 is suitable for the gene delivery.23 Thus in this study we used the same Ca2+/CO32− ratio for co-delivery of pDsRed2-N1-p53 and DOX.In the current investigation, both CaCO3/DNA and CaCO3/DNA/DOX nanoparticles were prepared. As shown in Table 1, the DNA encapsulation efficiencies for CaCO3/DNA and CaCO3/DNA/DOX nanoparticles are higher than 90%, indicating the co-precipitation technique could effectively load DNA in the co-precipitates. In addition, the DOX encapsulation efficiency for CaCO3/DNA/DOX nanoparticles is also high, confirming that the co-precipitation technique could effectively encapsulate DNA and DOX simultaneously.
| Sample | Loading content (wt%) | Encapsulation efficiency (%) |
|---|---|---|
| DNA in CaCO3/DNA nanoparticles | 1.30 | 93.3 |
| DNA in CaCO3/DNA/DOX nanoparticles | 1.29 | 92.1 |
| DOX in CaCO3/DNA/DOX nanoparticles | 0.63 | 90.0 |
The DOX release profile from CaCO3/DNA/DOX nanoparticles in PBS medium is shown in Fig. 1. Through being entrapped in the CaCO3/DNA/DOX nanoparticles, the release of DOX could be effectively sustained.
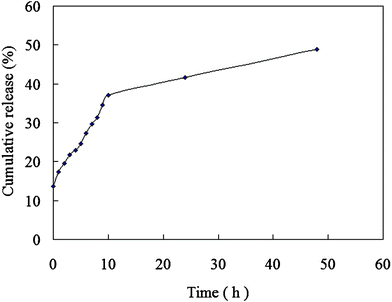 | ||
| Fig. 1 DOX release from CaCO3/DNA/DOX nanoparticles. | ||
The particle sizes and size distributions of CaCO3/DNA and CaCO3/DNA/DOX nanoparticles are presented in Fig. 2. The mean sizes of two types of nanoparticles are smaller than 100 nm. As compared with CaCO3/DNA nanoparticles, CaCO3/DNA/DOX nanoparticles exhibit a slight larger size.
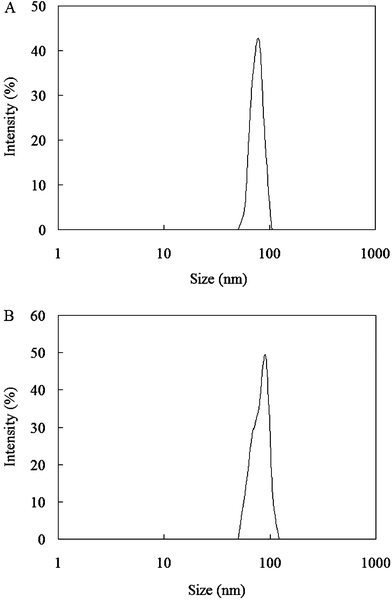 | ||
| Fig. 2 Particle size distributions of (A) CaCO3/DNA nanoparticles and (B) CaCO3/DNA/DOX nanoparticles. | ||
In vitro cell inhibition evaluation
To evaluate the cell proliferation inhibition rate of our gene and drug co-delivery system, HeLa cells were treated by CaCO3/DNA/DOX nanoparticles as compared with CaCO3/DNA nanoparticles, free DOX, and the mixture of CaCO3/DNA nanoparticles and free DOX (CaCO3/DNA + DOX). The results are summarized in Fig. 3. For the cells treated by particular agents, it is obvious that the cell inhibition rates at 48 h are much higher than that at 24 h, and the difference is statistically significant (p < 0.05) for each particular treatment. Comparing the inhibition effects of different agents, we can find that the co-delivery system, CaCO3/DNA/DOX, exhibits the highest cell inhibition rate of about 75%. The mixture of CaCO3/DNA nanoparticles and free DOX also has a good inhibition effect, with a slightly lower cell inhibition rate. The CaCO3/DNA/DOX nanoparticles could ensure that both drug and gene can be delivered to the same cancer cells. The mixture system may deliver the gene, drug, or both to particular cells, thus the corresponding synergistic effect could not be maximized.5 However, it should be noted that the difference in in vitrocell inhibition rates between the mixture (CaCO3/DNA + DOX) and the CaCO3/DNA/DOX nanoparticular co-delivery system is small. As we know, doxorubicin hydrochloride is a water soluble anticancer drug, and it can be directly uptaken by the cells, resulting in effective cell inhibition. The cell inhibition efficiencies of the free drug and the drug delivery systems are dependent on many factors such as the cell uptake and the drug release rate of the drug delivery systems, and cell types.17 Based on previous research carried out by other researchers, the nano-sized DOX delivery systems may even exhibit a lower cytotoxicity compared with the free DOX at the same drug concentration.16 The possible reason for the lower cytotoxicity of drug-loaded nanoparticles may be due to the fact that the majority of the drug entrapped in the nanoparticles has not been released out. It also should be noted that these in vitro results were obtained under ideal conditions, i.e. the samples were added in the wells of the cell culture plates and the nanoparticles and the water soluble free DOX can directly access the cells. In real in vivo conditions, the concentration of free DOX in the targeted sites will become much lower after systematic injection and may cause side effects to untargeted organs. That is the reason we need to apply the drug delivery technique and encapsulate the drug in the nanoparticles.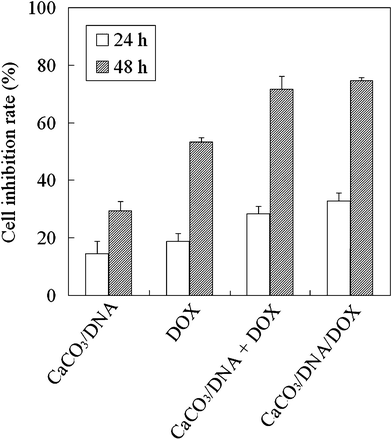 | ||
| Fig. 3 Cell inhibition rate of HeLa cells after being treated by different agents for 24 h and 48 h. (The comparison between the CaCO3/DNA/DOX and CaCO3/DNA shows p < 0.05, and the comparison between the CaCO3/DNA/DOX and DOX shows p < 0.05.) | ||
In this study, the cumulative DOX release from CaCO3/DNA/DOX nanoparticles is less than 50% at 48 h (Fig. 1). Nevertheless the cell inhibition rates of CaCO3/DNA/DOX nanoparticles are slightly higher than that of the mixture of CaCO3/DNA nanoparticle and free DOX at 24 h and 48 h, implying that the dominant release mechanism is the cell uptake. After being uptaken by the cells, the DOX loaded in the nanoparticles could be released, leading to effective cell inhibition.
According to our previous study, CaCO3/DNA nanoparticles can effectively enter the cells by cell uptake. The detailed mechanism of cell uptake of CaCO3 based nanoparticles has been reported in our previous article.23 In our previous study, we investigated the internalization route of CaCO3/DNA co-precipitates. Chloroquine was used as a lysosomotrophic agent as it could buffer endosomes so that it could give an overall measure of the contribution of endocytosis. Wortmannin was used as an inhibitor of both macropinocytosis and clathrin-mediated endocytosis as it could act as phosphoinositide 3-kinase inhibitor, and cytochalasin D was used as an inhibitor of macropinocytosis as it could prohibit actin polymerization. The effects of treatments of these agents on the transfection mediated by CaCO3/DNA co-precipitates were determined. We found that CaCO3/DNA co-precipitates were internalized viaendocytosis since chloroquine with a proper concentration could improve the gene expression level significantly and both treatments by wortmannin and cytochalasin D resulted in reduced gene expression levels. In addition, macropinocytosis was the main route of internalization because the treatment of cytochalasin D could greatly decrease the gene expression.
Compared with the treatments by gene and drug simultaneously, the treatments by CaCO3/DNA and free DOX separately lead to much weaker cell inhibition. To directly show the HeLa cell numbers after being treated by different agents, the OD values after treatments for 24 h and 48 h are listed in Table 2. Clearly, the simultaneous treatments by gene and drug can completely inhibit the proliferation of HeLa cells since the OD values at 48 h are lower than that at 24 h. And the application of gene or drug alone can not stop HeLa cells from proliferation since the OD values increase with increasing time.
| Agent | OD (24 h) | OD (48 h) |
|---|---|---|
| Control | 1.78 | 3.58 |
| CaCO3/DNA | 1.53 | 2.33 |
| DOX | 1.45 | 1.54 |
| CaCO3/DNA + DOX | 1.28 | 0.94 |
| CaCO3/DNA/DOX | 1.20 | 0.84 |
Cell morphology observation
The cell morphology of HeLa cells exposed to different agents was observed by an inverted microscope (Fig. 4). The cells without any treatment show a stretched cell morphology and are well attached on the cell culture plate. (Fig. 4a). After exposure to CaCO3/DNA nanoparticles, the growth of HeLa cells slows down (Fig. 4b). After treatment for 48 h, some cells become round and some cells are suspended in the culture medium, and a few cells show blebs. The cell morphology clearly indicates the inhibition effect due to the expression of the p53 gene.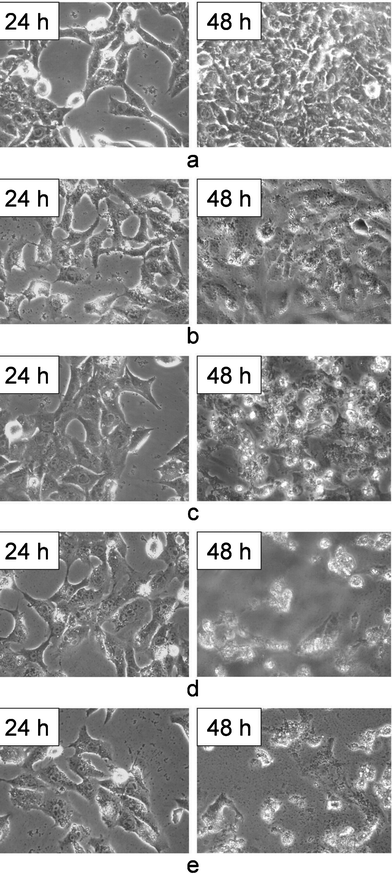 | ||
| Fig. 4 Morphology of HeLa cells observed by an inverted microscope after being treated by different agents for 24 h and 48 h. (a) control, (b) CaCO3/DNA, (c) free DOX, (d) CaCO3/DNA and free DOX (e) CaCO3/DNA/DOX. The images were obtained under magnification of 200. | ||
After exposure to free DOX, the number of HeLa cells obviously decreases as compared with the control sample. After 48 h, many cells become rounded or suspended in the culture medium (Fig. 4c).
Compared with the treatments of the p53 gene and DOX separately, the co-treatments of p53 gene and DOX exhibit much stronger cell inhibition effects. After co-incubation with the mixture of CaCO3/DNA nanoparticles and free DOX for 48 h, cell shrinkage could be clearly observed. Most cells become rounded or suspended in the culture media, together with cell fragments produced. Blebbing is visible for many cells (Fig. 4d). The cells treated by CaCO3/DNA/DOX nanoparticular co-delivery system show a similar morphology and significantly reduced cell number (Fig. 4e).
These morphology observation results are inconsistent with the cell inhibition rates determined by the MTT assay.
Apoptosis observation
In this investigation, the mechanism of the cell inhibition effect of CaCO3/DNA/DOX nanoparticles was studied using a FITC Annexin V/Dead Cell Apoptosis Kit. As we know, cell apoptosis is the regulated programmed process of cell death which is distinguished from necrosis, or accidental cell death, by characteristic morphological and biochemical changes, including compaction and fragmentation of the nuclear chromatin, shrinkage of the cytoplasm, and loss of membrane asymmetry. In normal live cells, phosphatidylserine (PS) is located on the cytoplasmic surface of the cell membrane. While in apoptotic cells, PS is translocated from the inner to the outer leaflet of the cell membrane. The human anticoagulant, annexin V, is a Ca2+–dependent phospholipid-binding protein that has a high affinity for PS. FITC Annexin V can identify apoptotic cells by binding to PS exposed on the outer leaflet. The red-fluorescent propidium iodide nucleic acid binding dye (PI) is impermeant to live cells and apoptotic cells, but stains the nuclei of dead cells with red fluorescence through binding tightly to the nucleic acids in the cell. After staining a cell population with FITC annexin V and PI, apoptotic cells show green fluorescence, dead cells show red and green fluorescence, and live cells show little or no fluorescence under a confocal laser scanning microscope.To evaluate the cell apoptosis after treatment by CaCO3/DNA/DOX nanoparticles, we stained HeLa cells that were exposed to CaCO3/DNA/DOX with FITC annexin V and PI. As shown in Fig. 5, after exposure to CaCO3/DNA/DOX for 24 h, no obvious red fluorescence could be detected, implying that almost no dead cells exist. The green fluorescence indicates apoptosis occurs for some cells. After 48 h, obvious morphology changes of the HeLa cells could be observed, and the majority of the cells shrink and become rounded. The strong red and green fluorescence could be detected for most cells, indicating that cell death is induced by apoptosis.
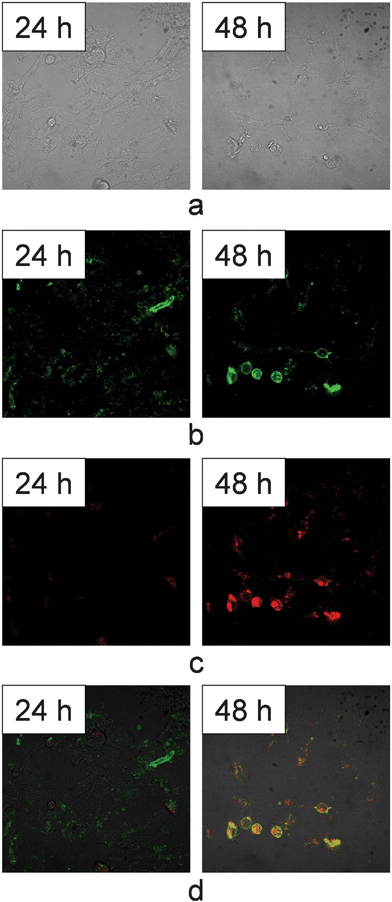 | ||
| Fig. 5 Confocal images of HeLa cells after being treated by CaCO3/DNA/DOX for 24 h and 48 h. (a) DIC images, (b) green fluorescence images (c) red fluorescence images, and (d) overlapped images of red and green fluorescence. The images were obtained under magnification of 400. | ||
Doxorubicin is among the first generation anthracycline antibiotics. According to previous studies, doxorubicin could intercalate into DNA and prevents the DNA double helix from being resealed and thereby stops the process of replication.13,14 For the tumor cells treated by doxorubicin, apoptosis, rather than necrosis, is the major mechanism of cell death.24 p53 gene is a tumor suppressor gene that has a regulatory function associated with DNA damage in cells. p53 can activate DNA repair proteins when DNA is sustained damage, and can induce cell apoptosis if DNA damage is irreparable. The status of the gene has an effect on the growth of cancer cells, apoptosis, and sensitivity to anticancer agents. The introduction of p53 into tumor cells harboring p53 mutations may also enhance the chemosensitivity of tumor cells to chemotherapeutic agents.5,11 In the current study, after the combined application of p53 and doxorubicin, the change in DNA double helix caused by doxorubicin could be identified by p53 as DNA damage, leading to cell apoptosis initiated by p53.
Conclusions
CaCO3/DNA/DOX nanoparticles for co-delivery of p53 expression plasmid and doxorubicin hydrochloride were prepared by CaCO3 co-precipitation technique. Under a proper Ca2+/CO32− ratio, both DNA and drug could be loaded in the CaCO3/DNA/DOX nanoparticles with high encapsulation efficiency and the size of CaCO3/DNA/DOX co-precipitates could be controlled at lower than 100 nm. The in vitrocell inhibition evaluation showed simultaneous treatments by gene and drug could induce an effective cell inhibition and completely inhibited the proliferation of HeLa cells, indicating that the CaCO3/DNA/DOX nanoparticles could effectively mediate gene transfection and deliver the drug to the cells. Compared with the co-delivery of gene and drug, the treatments by CaCO3/DNA and free DOX separately led to much lower cell inhibition rates. The CaCO3/DNA/DOX nanoparticles have promising applications in cancer treatments.Acknowledgements
Financial supports from National Natural Science Foundation of China (21074099) and Ministry of Science and Technology of China (National Basic Research Program of China 2011CB606202) are gratefully acknowledged. This research was also supported by Program for Changjiang Scholars and Innovative Research Team in University (IRT1030).References
- E. D. Dominik and D. M. Nettelbeck, Adv. Drug Delivery Rev., 2009, 61, 554–571 CrossRef.
- R. Waehler, S. J. Russell and D. T. Curiel, Nat. Rev. Genet., 2007, 8, 573–587 CrossRef CAS.
- L. Y. Qiu and Y. H. Bae, Biomaterials, 2007, 28, 4132–4142 CrossRef CAS.
- N. Wiradharma, Y. W. Tong and Y. Y. Yang, Biomaterials, 2009, 30, 3100–3109 CrossRef CAS.
- X. Lu, Q. Q. Wang, F. J. Xu, G. P. Tang and W. T. Yang, Biomaterials, 2011, 32, 4849–4856 CrossRef CAS.
- T. J. Wang, M.nS. Huang, C.nY. Hong, V. Tse, G. D. Silverberg and M. Hsiao, Biochem. Biophys. Res. Commun., 2001, 287, 173–180 CrossRef CAS.
- A. Ventura, D. G. Kirsch, M. E. McLaughlin, D. A. Tuveson, J. Grimm, L. Lintault, J. Newman, E. E. Reczek, R. Weissleder and T. Jacks, Nature, 2007, 445, 661–665 CrossRef CAS.
- M. Olivier, D. E. Goldgar, N. Sodha, H. Ohgaki, P. Kleihues, P. Hainaut and R. A. Eeles, Cancer Res., 2003, 63, 6643–6650 CAS.
- M. Idogawa, Y. Sasaki, H. Suzuki, H. Mita, K. Imai, Y. Shinomura and T. Tokino, Clin. Cancer Res., 2009, 15, 3725–3732 CrossRef CAS.
- M. Nakase, M. Inui, K. Okumura, T. Kamei, S. Nakamura and T. Tagawa, Mol. Cancer Ther., 2005, 4, 625–631 CrossRef CAS.
- M. Zhan, D. Yu, A. Lang, L. Li and R. E. Pollock, Cancer, 2001, 92, 1556–1566 CrossRef CAS.
- K. Oshima, Y. Naoi, K. Kishi, Y. Nakamura, T. Iwamoto, K. Shimazu, T. Nakayama, S. J. Kim, Y. Baba, Y. Tamaki and S. Noguchi, Cancer Lett., 2011, 307, 149–157 CrossRef CAS.
- R. L. Momparler, M. Karon, S. E. Siegel and F. Avila, Cancer Res., 1976, 36, 2891–2895 CAS.
- E. Czeczuga-Semeniuk, S. Wołczyński, M. Dąbrowska, J. Dzięcioł and T. Anchim, Folia Histochem. Cyto., 2004, 42, 221–227 CAS.
- N. Cao and S. S. Feng, Biomaterials, 2008, 29, 3856–3865 CrossRef CAS.
- A. Mahmud, X. B. Xiong and A. Lavasanifar, Eur. J. Pharm. Biopharm., 2008, 69, 923–934 CrossRef CAS.
- C. Zheng, J. Xu, X. Yao, J. Xu and L. Qiu, J. Colloid Interface Sci., 2011, 355, 374–382 CrossRef CAS.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS.
- A. Hanifi, M. H. Fathi and H. Mir Mohammad Sadeghi, J. Mater. Sci.: Mater. Med., 2010, 21, 2601–2609 CrossRef CAS.
- B. Sun, K. K. Tran and H. Shen, Biomaterials, 2009, 30, 6386–6393 CrossRef CAS.
- E. H. Chowdhury, A. Maruyama, A. Kano, M. Nagaoka, M. Kotaka, S. Hirose, M. Kunou and T. Akaike, Gene, 2006, 376, 87–94 CrossRef CAS.
- T. Y. Cheang, S. M. Wang, Z. J. Hu, Z. H. Xing, G. Q. Chang, C. Yao, Y. Liu, H. Zhang and A. W. Xu, J. Mater. Chem., 2010, 20, 8050–8055 RSC.
- S. Chen, F. Li, R. X. Zhuo and S. X. Cheng, Mol. BioSyst., 2011, 7, 2841–2847 RSC.
- M. Rossi, E. R. Munarriz, S. Bartesaghi, M. Milanese, D. Dinsdale, M. A. Guerra-Martin, E. T. W. Bampton, P. Glynn, G. Bonanno, R. A. Knight, P. Nicotera and G. Melino, J. Cell Sci., 2009, 122, 3330–3339 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
