Metallomics: whence and whither
Whence: the inorganic side of life
Discoveries in the 19th century demonstrating that plants and yeast depend on iron, manganese, copper, and zinc for growth initiated further studies on defining which chemical elements are essential for life. The area of investigation became known as trace element research and reached its peak with the realization that about one third of the elements – excluding radioactive elements and lanthanides – are essential for life (Fig. 1).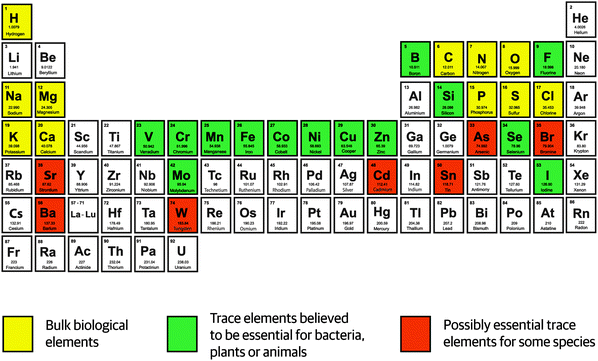 | ||
| Fig. 1 Elements essential for life in the periodic table of the elements.1 | ||
However, several issues remain unresolved, in particular whether or not we indeed have a complete list of the essential elements. Some elements seem to be essential for only some forms of life, and for others there is on-going debate as their molecular functions remain unknown. Much additional work is required to define the exact amounts which optimize biological function and the maximum amounts tolerable to avoid toxicity, both issues with tremendous implications for supporting health and preventing disease.
The difference between an essential and a non-essential element is best described in terms of different dose–response curves (Fig. 2). Characteristic for an essential element is the induction of deficiency symptoms when it becomes limiting. In this case, the response is known as Bertrand's rule.2 An additional property is that elements can positively affect biological function in the absence of any signs of overt deficiency, though it may not always be straightforward to tease apart such a pharmacological effect from an essential physiological function, which depends on the definition of the deficiency syndrome.
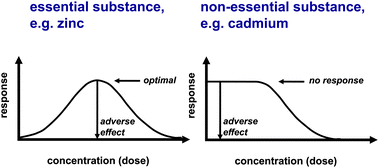 | ||
| Fig. 2 Dose response to an essential (left) and a non-essential (right) element. Potential pharmacological responses are not shown. | ||
In most textbooks, biochemistry is taught almost exclusively as organic chemistry, without much reference to inorganic chemistry. In biology, a separation between the two does not exist. Thus, associating inorganic with the inanimate and organic with the animate world only, as the names imply, is not correct. Another semantic issue, the distinction between macroelements and microelements, also has been a cause of considerable confusion as it is often understood to imply that a substance occurring at a small quantity is relatively less important than a substance occurring at a large quantity. This is clearly not the case as the survival of humans depends on a few micrograms of cobalt in the form of vitamin B12, needed for only two enzymes. Referring to zinc as a trace element illustrates this dilemma. The cellular concentrations of zinc are almost as high as those of ATP, it has roles in about 3000 proteins, and it is much more widely used than any of the vitamins that are generally treated in much greater depth in textbooks of biochemistry.
When structures of metalloproteins became available in the second half of the 20th century, chemists marveled at the coordination environments of their metals. In some cases, they presented structures hitherto unknown in chemistry. To allude to their “unique” characteristics, the metal ions were said to be in an entatic state (from the Greek word entasis, meaning slightly stretched), indicating that macromolecular structure induces a specific reactivity of the metal ion. Structures remain at the core of what fascinates chemists and attracts them to the field. For metal ions involved in catalysis, biological function follows from molecular structure, but deducing functions from the structures of other metalloproteins has not been straightforward. The reason is that biological function is context-specific and therefore interpretation requires knowledge of the underlying biology. Thus, the function of proteins in sensing, storing, and transporting metal ions is revealing yet additional principles of how structure is used for specific biological functions.
The area of research became known as bioinorganic chemistry. The traditional training of investigators in mostly single disciplines, either chemistry or biology, affected the development of the field. Some investigators focused on purely chemical approaches and investigated the coordination chemistry of metal ions with biological ligands. At the extreme, it can mean studying a chemistry that never takes place in biology. In biomimetic chemistry, characterization of synthetic structural and functional analogues of metal sites made valuable inroads into some chemical principles of structure and bonding. Other investigators followed mostly biological approaches without much attention to chemical principles. Thus, depending on the emphasis of the area of investigation, additional names were introduced: inorganic biochemistry, biological inorganic chemistry, inorganic chemical biology, metals in biology, biometals, and other permutations. An unintended consequence of this development was a fragmentation of the field and the establishment of different conferences, journals, and societies. Here, metallomics and Metallomics, the field’s associated journal, has an opportunity to bring together research from these different disciplines.
Whither: Metallomics, an integrated biometal science
The word “metallomics” indicates an approach to studying metals in a system. Relating the functions of molecules to the functions of a system creates enormous opportunities. Conceptually, the approach is different from the typical reductionist approach of chemists focusing on individual molecules. Metallomics is used in juxtaposition to other “omics” sciences addressing the “omes” of other biomolecules (Table 1).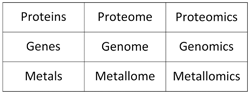
Williams defined the metallome as all the metal species in a biological system,3 and Haraguchi coined the term metallomics in 2002 “to integrate the research fields related to bio-trace metals” and then formally used it in the Journal of Analytical Atomic Spectrometry (JAAS) in 2004.4 Around this time, the area was reviewed by Szpunar.5 She distinguished qualitative metallomics (establishing the identity of metal species), quantitative metallomics (establishing the concentrations of species), and comparative metallomics (establishing the changes of metallomes with time and upon exposure to external stimuli) and emphasized the importance of analytical sciences to the speciation and identification of the function and availability of metals in biological systems. The area was later highlighted in two themed issues of the Journal of Analytical Atomic Spectrometry (JAAS) in 2004 and 2007 on the topic of atomic spectrometry of metallobiomolecules and metallomics.6,7
Metallomics has since been the subject of an IUPAC Technical Report where definitions for the terms “metallome” and “metallomics” are proposed.8 Analytical chemistry has played a crucial role in the emergence of the field, and the Technical Report also contains a critical evaluation of analytical chemistry approaches in the area.
The journal Metallomics was launched with strong participation of analytical chemists, in particular mass spectrometrists, combining element- and molecule-specific mass spectrometry and developing new mass spectrometric techniques. In as much as mass spectrometry has made substantive contributions in other “omics” sciences, it is also one of the key techniques in metallomics.
The name Metallomics serves the journal well as an integration of the biometal sciences. However as a science that strengthens the importance of the inorganic elements for life, it has also become practice to include non-metals as well. A significant number of articles published in Metallomics to date have been focused on metalloid and non-metal trace elements such as arsenic and selenium, adding substantially to the field.
Integrated biometal science is no longer just bridging chemistry and biology within an organism or a cell. It includes other disciplines that look at the influence of the environment on organisms and cells, e.g. how the chemistry in the environment determines speciation and availability of metal ions to biological systems. It also includes systems biology, where at the extreme not a single analysis of a metal or an element is being made, and in bioinformatics it includes approaches, such as high-throughput screening, to study systems. Additional words have already been created to describe parts of the field and it is almost certain that others will follow. Ionome,9 ironome,10 or metalloproteome11,12 may serve as examples.
Important and emerging interfaces covered in Metallomics are:
Medicine
Inorganic chemistry is involved in numerous diseases. Investigations of deficiencies, overloads, or the perturbation of the homeostatic control of essential elements have vast potential to understand etiology and pathology and to apply this knowledge for therapeutics and diagnostics, such as imaging. Metallodrugs is one of the subfields.Nutrition and toxicology
These disciplines look at a spectrum of activities and are complementary: Essential elements are nutrients (purview of nutrition) and have adverse effects (purview of toxicology). Thus, the transition of nutrition focusing on essential elements to toxicology focusing on non-essential elements is “fluid”. With the anthropogenic introduction of many rare elements into the environment to which we are exposed, including new species such as nanoparticles, many basic and applied metallomics issues need to be addressed.Biogeochemistry
Though the “omics” sciences usually refer to biological systems, there are systems and hence “omes” in the inanimate world. How geochemistry and biochemistry interact is a rather specialized area. Elemental cycles and transformations are important for biology. On a scale of time, the issue of chemical evolution is impossible to address without references to inorganic chemistry. With important guiding thoughts having been published by Williams, the area is in its infancy and open (and ripe!) for experimental exploitation.1Bioinformatics
The vast amount of scientific information now available needs to be properly archived, analyzed, curated, and reviewed. The computer became an essential tool for in silico chemistry. We would therefore like to take this opportunity to invite authors in all these areas (and others that we have yet to interact with) to find a new home in the journal Metallomics in order to make their work known to a wider community of interested “biometallogists.” With the inter- and multidisciplinary nature of Metallomics, we hope to continue to provide an exciting new forum that overcomes the split into different disciplines, and that will lead to a wider recognition of the field in the biological sciences.We look forward to working with you in the future.
Wolfgang Maret, Metallomics Editorial Board Chair
May Copsey, Editor, Metallomics
References
- R. J. P. Williams and R. Rickaby, “A chemical account of evolution”, Chem. World, 2012, 6 Search PubMed , http://www.rsc.org/chemistryworld/2012/06/chemical-account-evolution.
- G. Bertrand, “On the role of trace substances in agriculture”, Eighth Int. Congr. Appl. Chem., 1912, 28, 30–40 Search PubMed.
- R. J. P. Williams, Coord. Chem. Rev., 2001, 216–217, 583–595 CrossRef CAS.
- H. Haraguchi, J. Anal. At. Spectrom., 2004, 19, 5–14 RSC.
- J. Szpunar, Anal. Bioanal. Chem., 2004, 378, 54–56 CrossRef CAS.
- J. Anal. At. Spectrom., 2004, 19, 1–200 Search PubMed, Themed issue on Atomic Spectrometry of Metallobiomolecules.
- J. Anal. At. Spectrom., 2007, 22, 101–224 Search PubMed, Themed issue on Metallomics.
- R. Lobinski, J. S. Becker, H. Haraguchi and B. Sarkar, Pure Appl. Chem., 2010, 82, 493–504 CrossRef CAS.
- D. E. Salt, I. Baxter and B. Lahner, Annu. Rev. Plant Biol., 2008, 59, 709–733 CrossRef CAS.
- P. A. Lindahl and G. P. Holmes-Hampton, Curr. Opin. Chem. Biol., 2011, 15, 342–346 CrossRef CAS.
- A. Sanz-Medel, Anal. Bioanal. Chem., 2005, 381, 1–2 CrossRef CAS.
- R. A. Scott, J. E. Shokes, N. J. Cosper, F. E. Jenney and M. W. W. Adams, J. Synchrotron Radiat., 2005, 12, 19–22 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
