 Open Access Article
Open Access ArticleThe interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus†
Fabio
Gsaller‡
a,
Martin
Eisendle‡
a,
Beatrix Elisabeth
Lechner
a,
Markus
Schrettl
a,
Herbert
Lindner
b,
Daniela
Müller
a,
Stephan
Geley
c and
Hubertus
Haas
*a
aDivision of Molecular Biology, Biocenter, Innsbruck Medical University, Fritz-Pregl-Str. 3, A-6020 Innsbruck, Austria. E-mail: hubertus.haas@i-med.ac.at; Fax: +43 (0)512-9003-73100; Tel: +43 (0)512-9003-70205
bDivision of Clinical Biochemistry, Biocenter, Innsbruck Medical University, Fritz-Pregl-Str. 3, A-6020 Innsbruck, Austria
cDivision of Molecular Pathophysiology, Biocenter, Innsbruck Medical University, Fritz-Pregl-Str. 3, A-6020 Innsbruck, Austria
First published on 2nd November 2012
Abstract
Iron is an essential element for all eukaryotes but its excess has deleterious effects. Aspergillus fumigatus produces extracellular siderophores for iron uptake and the intracellular siderophore ferricrocin (FC) for distribution and storage of iron. Iron excess has previously been shown to increase the content of ferric FC and the expression of the putative vacuolar iron importer CccA (AFUA_4G12530), indicating a role of both the vacuole and FC in iron detoxification. In this study, we show that CccA-deficiency decreases iron resistance in particular in combination with derepressed iron uptake, while overproduction of CccA increases iron resistance. Green fluorescence protein-tagging confirmed localization of CccA in the vacuolar membrane. In contrast to CccA-deficiency, inactivation of FC biosynthesis did not affect iron resistance, which indicates that vacuolar rather than FC-mediated iron storage is the major iron detoxifying mechanism. After uptake, extracellular siderophore backbones are hydrolyzed and recycled. Lack of FC, CccA, and in particular lack of both increased the cellular content of iron chelated by siderophore breakdown products. These data indicate that the transfer of iron from extracellular siderophores to the metabolism, FC or the vacuole precedes recycling of siderophore breakdown products. Furthermore, this study indicates that CccA does not play an exclusive role in vacuolar iron storage for nutritional reuse.
Introduction
Due to its crucial role in a wide variety of cellular oxidation–reduction reactions, iron is an essential element for all eukaryotes. Nevertheless, iron excess can cause oxidative stress via the Haber–Weiss/Fenton chemistry.1 Therefore, cellular iron homeostasis has to maintain a delicate balance between sufficient iron supply and prevention of iron toxicity. As microorganisms lack iron excretory mechanisms, the maintenance of iron homeostasis results from a tight regulation of iron acquisition and iron storage.Aspergillus fumigatus is a ubiquitous saprophytic mold, but at the same time the most common cause of airborne fungal infections in immunocompromised patients.2 During both saprophytic growth and infection, this opportunistic pathogen has to cope with rapidly changing environments, including high variations of iron availability. For the cellular import, A. fumigatus employs low-affinity iron acquisition and two high-affinity iron uptake systems, reductive iron assimilation and siderophore (low molecular mass, ferric iron chelators)-assisted iron uptake.3 In order to mobilize extracellular iron A. fumigatus excretes two different siderophores, fusarinine C (FSC) and triacetylfusarinine C (TAFC).4,5 After the cellular uptake of the ferri-forms of FSC and TAFC via specific siderophore-iron transporters, these siderophores are degraded for iron release.6,7 Extracellular siderophores are found in most fungal and bacterial species as well as in some plants.6 In contrast to plants and bacteria, most fungi also possess intracellular siderophores. A. fumigatus produces two intracellular siderophores, ferricrocin (FC) for hyphal iron storage and distribution, and hydroxyferricrocin (HFC) for conidial iron storage.4,5 In recent years, individual components of the A. fumigatus siderophore and reductive iron assimilatory systems have been characterized at the molecular level,6e.g. the ornithine hydroxylase SidA that is essential for biosynthesis of both intra- and extracellular siderophores, the non-ribosomal peptide synthetase SidC that is indispensable for biosynthesis of intra- but not extracellular siderophores, and the iron permease FtrA that is essential for reductive iron assimilation. Due to its role in intracellular iron storage and distribution, FC plays a crucial role in germination and conidiation.4 Moreover, FC has also been shown to be essential for sexual development in Aspergillus nidulans.8 Biosynthesis of extra- and intracellular siderophores proved to be crucial for full virulence of A. fumigatus in various aspergillosis models,3,4,9,10 and for intracellular survival after phagocytosis by murine alveolar macrophages.11,12
Inactivation of the transcription factor SreA, which represses excessive iron uptake via the downregulation of siderophore-mediated and reductive iron assimilation, results in toxic accumulation of iron under iron-replete conditions and subsequent growth retardation.6,13 Excessive iron uptake has been shown to elevate the cellular FC content, which indicates an additional role of FC in iron detoxification. Furthermore, iron excess leads to an upregulation of the transcript level of AFUA_4G12530, termed CccA,13 which displays significant similarity to Saccharomyces cerevisiae Ccc1 and Arabidopsis thaliana Vit1. Ccc1 mediates vacuolar import of iron and manganese to prevent toxic levels of these metals in the cytosol.14,15 Vit1 has also been shown to transport iron into the vacuole.16 Both S. cerevisiae and A. thaliana possess vacuolar iron export systems for the reuse of vacuolar-stored iron.17–20
Apart from S. cerevisiae and A. thaliana, which both lack siderophores, the role of vacuolar iron storage has not been functionally characterized in any other species. In the present study we characterized the role of A. fumigatus CccA in vacuolar iron storage and analyzed the interplay between siderophore-mediated and vacuolar iron storage.
Results
A. fumigatus encodes two homologs of S. cerevisiae Ccc1
BLASTP homology searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi, http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=fungi) and phylogenetic analysis identified homologs of the S. cerevisiae vacuolar iron importer Ccc1 in all fungal genomes available with the exception of Microsporidia, Pleosporales and the Exobasidiomycete Malassezia globosa, i.e., in 68 of 76 fungal species (Fig. 1 and data not shown). Interestingly, Ccc1 homologs are not only found in fungi and plants but also in bacteria such as Pseudomonas aeruginosa and Streptococcus sanguinis. In contrast to most other fungal species, Trichocomaceae (including Neosartorya, Aspergillus, and Penicillium species) and Hypocreales (including Gibberella, Nectria, and Metarhizium species) encode two paralogs (Fig. 1). An amino acid sequence alignment of the Ccc1 homologs from A. fumigatus, Penicillium chrysogenum, S. cerevisiae and A. thaliana is shown in Fig. S1 (ESI†). All these proteins are about 300 amino acids in length and display 12% identity (56 amino acid residues) in the 228 amino acid core region. | ||
| Fig. 1 Phylogenetic analysis (rooted neighbour joining tree) of CccA homologs from selected fungi, bacteria and plants. The Trichocomaceae and Hypocreales paralogs are marked in brackets. Species abbreviations: Af, A. fumigatus; An, A. nidulans; At, A. thaliana; Ca, Candida albicans; Cn, Cryptococcus neoformans; Gz, Gibberella zeae; Ma, Metarhizium anisopliae; Nc, Neurospora crassa; Nf, Neosartorya fischeri; Nh, Nectria haematococca; Pa, P. aeruginosa; Pc, Penicillium chrysogenum; Ss, S. sanguinis; Sc, S. cerevisiae; Sm, Sordaria macrospora; Sp, Schizosaccharomyces pombe; Um, Ustilago maydis. | ||
Expression of CccA is induced by iron
In S. cerevisiae, Ccc1 expression is transcriptionally induced by iron.21 As shown previously13 and confirmed in this study, the transcript level of AFUA_4G12530, termed CccA, is upregulated in the A. fumigatus wild type strain ATCC46645 (wt) during a shift from iron starvation to iron sufficiency (Fig. 2). Furthermore, derepression of iron uptake under iron-replete conditions due to deficiency in the iron uptake repressor SreA increased the cccA steady-state transcript level. As a control for the cellular iron status, Northern analysis was also performed with the siderophore transporter-encoding mirB gene, the expression of which is in contrast to cccA repressed by iron.13 These data indicate the function of CccA in the iron metabolism.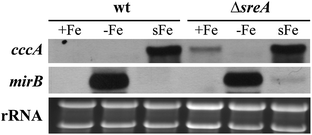 | ||
| Fig. 2 Expression of cccA is induced by iron excess. For Northern analysis of cccA, A. fumigatus wt and ΔsreA strains were grown for 24 h in liquid flask cultures under iron-depleted (−Fe), iron-replete conditions (+Fe; 30 μM FeSO4) or shifted from −Fe to 0.1 mM FeSO4 for 1 h (sFe) to force excessive iron uptake. Ethidium bromide-stained rRNA is shown as a control for loading and quality of RNA. As a control for the cellular iron status, Northern analysis was also performed with the siderophore transporter-encoding mirB gene, the expression of which is repressed by iron.13 | ||
As observed previously,13 transcripts of cccA paralog AFUA_3G09970, termed cccB, could not be detected in Northern analysis, neither in the wt nor in the SreA-deficient strain (data not shown), which suggests a minor role of this gene in iron metabolism. As indicated by phylogenetic analysis (Fig. 1), BLAST searches revealed that the CccA and CccB orthologs are more closely related to each other than to the other homologs (Table S3, ESI†). Interestingly, the CccB orthologs from A. fumigatus and P. chrysogenum display an extended C-terminus, an insertion of two amino acid residues and a deletion of one amino acid in the core region (Fig. S1, ESI†).
CccA but not siderophore biosynthesis is important for iron resistance
In order to functionally characterize CccA, the cccA coding region was replaced by the pyrithiamine resistance gene ptrA in the A. fumigatus wt, yielding strain ΔcccA, as described in Materials and methods and Fig. S1 (ESI†). Additionally, CccA-deficiency was combined with deficiency in siderophore biosynthesis and reductive iron assimilation (ΔcccAΔsidAΔftrA), FC biosynthesis (ΔcccAΔsidC), the iron uptake repressor SreA (ΔcccAΔsreA), as well as SreA and siderophore biosynthesis (ΔcccAΔsidAΔsreA). Radial growth of wt and mutant strains was compared on plates containing different iron concentrations (Fig. 3). As shown previously,3,4 deficiency in siderophore biosynthesis (ΔsidA) blocked growth in the presence of the iron chelator bathophenanthroline disulfonic acid (BPS). FC-deficiency (compare wt/ΔsidC) decreased growth in the presence of BPS, and deficiency in both siderophore-mediated and reductive iron assimilation (compare wt/ΔsidAΔftrA) blocked growth up to an iron concentration of 30 μM. Genetically unidentified low-affinity iron uptake enabled partial growth of ΔsidAΔftrA at an iron concentration of 1 mM and wt-like growth at ≥3 mM. SreA-deficiency decreased growth under conditions of high iron availability, which was best visible at 5 mM, due to toxicity of excessive iron uptake (compare wt/ΔsreA).13 In all SreA-functional genetic backgrounds, CccA-deficiency decreased growth under high iron conditions, which was best seen at 5 mM iron (compare wt/ΔcccA, ΔsidCΔ/ΔcccAΔsidC, ΔsidAΔftrA/ΔcccAΔsidAΔftrA). Combined deficiency in both SreA and CccA reduced growth already at iron concentrations ≥30 μM (compare ΔsreA/ΔcccAΔsreA, ΔsidAΔsreA/ΔcccAΔsidAΔsreA). Taken together, CccA-deficiency led to decreased iron resistance, especially in strains with derepressed iron uptake due to lack of SreA. These data indicate a crucial role of CccA in iron detoxification. Consistently, CccA-deficiency did not affect growth in any genetic background under iron limitation (Fig. 3). | ||
| Fig. 3 CccA-deficiency decreases iron resistance. 104 conidia of the respective fungal strain were point-inoculated on AMM plates containing the indicated iron concentration and incubated for 48 h at 37 °C. The BPS medium contained 0.2 mM of the ferrous iron chelator bathophenanthroline disulfonic acid, which inhibits reductive iron assimilation. CEA10 genetic background strains are marked with *, the other strains have an ATCC46645 background. The fungal strains are listed and described in Table 1. | ||
In further agreement with the function of CccA in iron detoxification, expression of cccA under the control of the P. chrysogenum xylP promoter, which is xylose-inducible,22,23 increased iron resistance in wt (strain wtcccAoe) and ΔcccA (strain ΔcccAcccAoe) strains under inducing but not repressing conditions (Fig. 4A).
 | ||
| Fig. 4 Xylose-induced expression of cccA increases iron resistance in wt and ΔcccA backgrounds (A) and GFP-tagged CccA localizes to the vacuolar membrane (B). (A) 104 conidia of the respective fungal strain were point-inoculated on AMM plates containing FeSO4 at a final concentration of 10 mM and 30 mM, respectively, with or without 0.5% xylose and incubated for 48 h at 37 °C. (B) The GFP fluorescence of ΔcccAcccAgfp was visualized using confocal microscopy. Cellular membranes were stained by preincubation with 8 μM FM4-64 for 1 h. | ||
In Ustilago maydis, simultaneous deficiency in the SreA ortholog Urbs1 and siderophore biosynthesis appears to be lethal, indicating an essential role of intracellular siderophores in iron detoxification.24 In A. fumigatus, inactivation of neither the entire siderophore production nor FC biosynthesis decreased iron resistance in any background tested (compare wt/ΔsidA, wt/ΔsidC). Moreover, the simultaneous deficiency in SreA and siderophore biosynthesis was not lethal and did not result in increased iron sensitivity (ΔsreAΔsidA). Even simultaneous deficiency in SreA, siderophore biosynthesis and CccA was not lethal (ΔcccAΔsreAΔsidA). These data demonstrate that in A. fumigatus siderophore biosynthesis is not essential for iron detoxification. Notably, the mutants described here have two different genetic backgrounds (ATCC46645 and CEA10) but in the comparisons above only mutants with the same genetic background have been included. Moreover, iron metabolism and siderophore metabolism are highly similar in these two genetic backgrounds.3,25
It is generally believed that iron leads to damage through the generation of oxygen radicals.1 To investigate the basis of iron susceptibility caused by CccA-deficiency we analyzed the impact of CccA-deficiency in wt and ΔsidC genetic backgrounds on resistance to oxidative stress caused by 2 mM hydrogen peroxide under iron starvation, iron sufficiency (30 μM) and iron excess (3 mM) in radial growth assays. Remarkably, CccA-deficiency did not decrease resistance to hydrogen peroxide under any of the tested conditions indicating that iron toxicity may not be solely due to oxidative damage (data not shown). In addition, neither hydrogen peroxide-caused oxidative stress nor inactivation of various components of the oxidative damage response including the master regulator of oxidative stress response, Yap1, affected the iron susceptibility of the S. cerevisiae Δccc1 mutant.26 Moreover, iron toxicity mediated by Ccc1-deficiency occurs in this system under both aerobic and anaerobic conditions.26
CccA is located in the vacuolar membrane
To investigate the localization of CccA, we generated an A. fumigatus strain expressing CccA N-terminally tagged with enhanced green fluorescence protein (GFP) under the control of the constitutive cgxA-XAH promoter in ΔcccA (strain ΔcccAcccAgfp) as described in Materials and methods and Fig. S1 (ESI†).27 Epifluorescence microscopy revealed that GFP-CccA co-localizes to the vacuolar membrane with the FM4-64 marker (Fig. 4B) as previously shown for N-terminal GFP fusions of the CccA homologs Ccc1 in S. cerevisiae and Vit 1 in A. thaliana.15,16 The GFP-CccA fusion partially cured the iron sensitivity of ΔcccA (Fig. 4A) indicating at least partial correct biochemical function and localization. Lack of full complementation might be due to decreased transport activity caused by the GFP-fusion or inappropriate expression as a result of the use of a heterologous promoter. The latter can be excluded as Northern analysis indicated an even higher transcript level of GFP-tagged cccA compared to cccA (data not shown). Notably, C-terminal GFP-tagged CccA under the native promoter was not functional (data not shown).Taken together, the localization of CccA in the vacuolar membrane suggests that CccA transports iron into the vacuole as shown previously for its homologs in S. cerevisiae and A. thaliana.15,16
Vacuolar iron storage increases the degradation of siderophores
After cellular uptake, ferric forms of TAFC and FSC are degraded via esterase-mediated hydrolysis, the iron is transferred to the metabolism or FC and the FSC/TAFC degradation products are recycled.7 To analyse a possible interplay between vacuolar iron storage and intracellular siderophores, wt, ΔcccA, ΔsidC, and ΔcccAΔsidC strains were grown in liquid cultures for 20 h under iron starvation to activate siderophore production. Subsequently, 100 μM iron were added to the growth media and the strains were further incubated for 4 h, which resulted in uptake of iron chelated by FSC and TAFC. Mycelia were harvested and cell extracts were analyzed by reversed-phase HPLC and mass spectrometry (Fig. 5). After the iron shift, the cell extracts of all strains were orange-coloured (Fig. 5A), which is typical for abundant iron resources chelated by siderophores or siderophore breakdown products.7 Interestingly, the orange coloration of the cell extract was more intense in the case of ΔcccA compared to wt and ΔcccAΔsidC compared to ΔsidC. The latter indicated that the orange coloration was not exclusively caused by ferric FC as ΔsidC strains lack FC. Reversed-phase HPLC (Fig. 5) combined with mass spectrometry identified desferri-FC as a main compound in the wt cell extract during iron starvation. After the addition of iron, the desferri-FC was converted to FC and various FSC/TAFC degradation products accumulated. Compared to wt, CccA-deficiency did not affect the desferri-FC content during iron starvation but resulted in increased accumulation of FSC/TAFC degradation products, in particular N5-anhydromevalonyl-N5-hydroxyornithine (fusarinine). SidC-deficiency blocked FC production and increased accumulation of FSC/TAFC degradation products. The highest levels of FSC/TAFC degradation products were observed in ΔcccAΔsidC, which lacks both vacuolar iron storage and FC-mediated intracellular iron chelation. The orange coloration of the cell extract and the detection of ferric FSC/TAFC breakdown products (Fig. 5) underline that the intracellular siderophore breakdown products indeed chelate iron in vivo. FSC and TAFC are cyclic (N5-anhydromevalonyl-N5-hydroxyornithine)3 and (N2-acetyl-N5-anhydromevalonyl-N5-hydroxyornithine)3, respectively, connected by head to tail ester bonds.28 The detection of N5-anhydromevalonyl-N5-hydroxyornithine-N2-acetyl-N5-anhydromevalonyl-N5-hydroxyornithine demonstrates that TAFC is degraded by hydrolysis and deacetylation and therefore, the detected N5-anhydromevalonyl-N5-hydroxyornithine is most likely not only derived from degradation of FSC but also TAFC.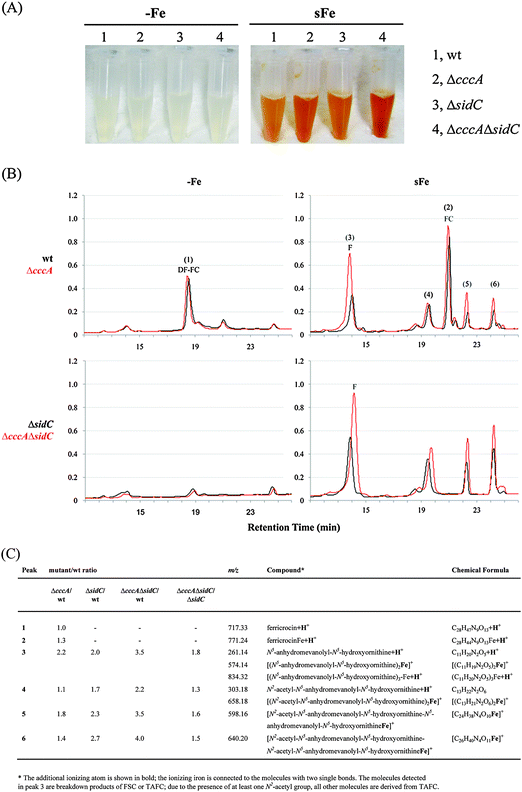 | ||
| Fig. 5 Deficiency in FC, CccA, and in particular both increases the cellular content in FSC/TAFC breakdown products. A. fumigatus wt, ΔcccA, ΔsidC, and ΔcccAΔsidC were grown in liquid cultures for 20 h at 37 °C under iron starvation (−Fe). Subsequently, FeSO4 was added to a final concentration of 0.1 mM and cultures were incubated for another 4 h (sFe). (A) Pictures of cell extracts (50 mg dry weight in 1 ml) before and after the iron shift. The orange coloring indicates the presence of siderophores and/or their degradation products.7 (B) Representative reversed-phase HPLC analysis (absorption at 210 nm in milliabsorption units) of the intracellular siderophore content comparing ΔcccA (in red) and wt (in black) as well as ΔcccAΔsidC (in red) and ΔsidC (in black) during −Fe and sFe, respectively. (C) The detected compounds were analyzed by mass spectrometry and the abundance of the sFe compounds was normalized to that of the wt. The data represent the mean from three experiments; the SD did not exceed 15%. | ||
Taken together, the increased accumulation of FSC/TAFC degradation products due to lack of vacuolar iron storage or intracellular iron chelation by FC and in particular both indicates that the transfer of iron from extracellular siderophores to the metabolism, FC or the vacuole precedes recycling of siderophore breakdown products. In other words, the absence of FC and/or vacuolar iron storage decreases the release of iron from iron-chelating siderophore breakdown products. As chelation decreases iron toxicity, this mechanism might represent another iron detoxification strategy.
CccA-deficiency does not affect the putative nutritional use of vacuolar iron
Both S. cerevisiae and A. thaliana are able to recycle vacuolar-stored iron and deficiency in the CccA homolog blocks the use of the vacuole as an iron storage compartment in both organisms.15,17–20 The current study indicates that CccA is a major vacuolar iron transporter in A. fumigatus.To study the possible recycling of vacuolar iron in A. fumigatus, we compared in a first approach wt and ΔcccA strains during a shift from iron sufficiency (30 μM TAFC) to iron starvation (Fig. 6). The two strains did not significantly differ in timing and the extent of siderophore synthesis (Fig. 6A) or biomass production (Fig. 6B). As reuse of vacuolar iron is expected to delay iron starvation and consequently siderophore production as a symptom of iron starvation in wt compared to ΔcccA, these data indicate that CccA does not play an exclusive role in the use of the vacuole as a nutritional iron source.
 | ||
| Fig. 6 CccA-deficiency does not affect the potential reuse of vacuolar iron. CccA-deficiency does not influence production of TAFC (A) and biomass (B) of 19 h-liquid iron-replete cultures (AMM + 30 μM TAFC as an iron source) transferred to iron-free AMM. TAFC and biomass production is displayed normalized to wt. (C) CccA-deficiency does not influence radial growth after a shift from iron sufficiency to iron starvation in a ΔsidAΔftrA background. 104 conidia were point-inoculated on plates containing the indicated concentration of TAFC or FC for 18 h separated from the growth media by a dialysis membrane. Subsequently, the mycelia-containing dialysis membranes were transferred onto plates lacking iron and incubated for 5 days. (D) During a shift from iron sufficiency to iron starvation, CccA-deficiency does not influence the expression pattern of mirB and cycA in a ΔsidAΔftrA background, the transcription of which is activated and repressed by iron starvation, respectively. For Northern analysis, RNA was isolated from mycelia of 20 h-liquid cultures containing 15 μM TAFC or FC before (0 h) or after transfer (2–8 h) into iron-lacking media. Ethidium bromide-stained rRNA is shown as a control for loading and quality of RNA. | ||
In a second approach, ΔsidAΔftrA and ΔcccAΔsidAΔftrA strains, which lack both reductive and siderophore-mediated iron assimilation and therefore high-affinity iron uptake, were compared to exclude compensation of possible defects by uptake of extracellular iron (Fig. 6C). The two mutants were iron-saturated by incubation for 16 h at 37 °C on plates containing either TAFC or FC separated from the growth media by dialysis membranes. Subsequently, the mycelia-containing dialysis membranes were transferred onto iron-depleted minimal media, from which they are unable to acquire iron (compare Fig. 3). After incubation for five days, CccA-deficiency did not decrease growth but even supported increased conidiation. Again, these data make an exclusive role of CccA in nutritional vacuolar iron storage unlikely.
In the third approach, ΔsidAΔftrA and ΔcccAΔsidAΔftrA strains were iron-saturated by culturing in liquid media for 18 h with TAFC or FC as an iron source. Subsequently, mycelia were washed and transferred into iron-depleted media and the iron status was monitored in a time course by Northern analysis of mirB and cycA expression (Fig. 6D), which are repressed and induced by iron, respectively.13,29 Interestingly, mirB transcription was induced significantly later and, consistently, cycA expression was repressed later in FC- compared to TAFC-precultured mycelia. These data suggest that FC is a better iron source compared to TAFC due to either better uptake or better intracellular iron supply. Nevertheless, the cycA and mirB expression patterns did not reveal significant differences in the two strains (Fig. 6D), which again makes an exclusive role of CccA in vacuolar iron storage for nutritional use unlikely.
Taken together, these data indicate that vacuolar iron-import by CccA does not seem to play an exclusive role for nutritional reuse, which contrasts the situation in S. cerevisiae.15 Possibly A. fumigatus lacks recycling of vacuolar iron in general. S. cerevisiae extracts vacuolar iron via vacuolar membrane-localized paralogs of proteins involved in reductive iron assimilation, termed Fet5 and Fth1.17 In agreement with a possible lack of recycling of vacuolar iron, A. fumigatus does not possess paralogs to the proteins involved in reductive iron assimilation.6 However, iron might be exported from the vacuole in A. fumigatus by other mechanisms and the data could be explained by the presence of CccA-independent vacuolar iron importers. One potential alternative vacuolar iron importer could be the CccB. However, expression of the encoding gene could not be detected, even in strains lacking CccA (data not shown).
Materials and methods
Growth conditions and siderophore analysis
Generally, A. fumigatus strains were grown at 37 °C in Aspergillus minimal medium (AMM) according to Pontecorvo et al.30 containing 1% (wt/vol) glucose as a carbon source, 20 mM glutamine as a nitrogen source, and 30 μM FeSO4. For iron-depleted conditions, iron was omitted. Production of conidia was performed on AMM plates containing 30 μM FeSO4; for strains lacking high affinity iron uptake systems (ΔsidAΔftrA, ΔcccAΔsidAΔftrA) 4 mM FeSO4 was supplemented. The xylPP promoter was induced by adding 0.5% xylose to AMM. Siderophore production was analyzed by reversed phase HPLC as described previously.31Mass-spectrometric analysis
Determination of the molecular mass of the samples obtained by RP-HPLC was carried out using a LTQ Velos ion trap mass spectrometer (Thermo Fisher Scientific) equipped with an electrospray source (ESI-MS, electrospray ionization mass spectrometry). Samples were dissolved in 50% aqueous methanol containing 0.1% formic acid, and infused directly into the ion source using the syringe pump. The electrospray voltage was set at 4.0 kV and the heated capillary was held at 270 °C.cccA-disruption (ΔcccA), cccA-overexpression (cccAoe) and gfp-cccA fusion (cccAgfp)
All strains used in this study are listed and described in Table 1. For generating the cccA-deletion strain, the bipartite marker technique was used.32 Therefore, A. fumigatus was co-transformed with two DNA fragments, each containing overlapping but incomplete fragments of the pyrithiamine resistance-conferring ptrA gene fused to 0.9-kb 5′- and 1.2-kb c-flanking regions of cccA, respectively. The cccA 5′-flanking region was PCR-amplified from genomic DNA using primers oAfcccA-1 and oAfcccA-4r. For amplification of the 3′-flanking region primers oAfcccA-3 and oAfcccA-2r were employed. Subsequent to gel-purification, these fragments were digested with StuI (5′-flanking region) and HindIII (3′-flanking region), respectively. The ptrA selection marker was released from the plasmid pSK275 by digestion with SmaI and HindIII and ligated with the 5′- and 3′-flanking region, respectively. The transformation construct A (2266 bp, fusion of the cccA 5′-flanking region and the prtA split marker) was amplified from the ligation product using primers oAfcccA-5 and optrA-2r. For amplification of the transformation construct B (2267 bp, fusion of the cccA 3′-flanking region and the supplementary ptrA split marker) primers occcA-6r and optrA-1 were employed. For transformation of A. fumigatus strains, both constructs A and B were used simultaneously. In the ΔcccA mutant, the region −150 to 1155 bp relative to the cccA sequence was replaced by the ptrA resistance cassette.| Strains | Genotype | Ref. |
|---|---|---|
| wt ATCC46645 | American type culture collection | |
| wt CEA10 | 37 | |
| ΔcccA | ATCC46645; ΔcccA::ptrA | This study |
| ΔsreA | ATCC46645; ΔsreA::hph | 13 |
| ΔsidA | ATCC46645; ΔsidA::hph | 3 |
| ΔsidC | ATCC46645; ΔsidC::hph | 4 |
| ΔcccAΔsreA | ATCC46645; ΔcccA::ptrA; ΔsreA::hph | This study |
| ΔcccAΔsidC | ATCC46645; ΔcccA::ptrA; ΔsidC::hph | This study |
| ΔsidAΔftrA | CEA17; ΔsidA::pyrG; ΔftrA::hph | 3 |
| ΔcccAΔsidAΔftrA | CEA17; ΔsidA::pyrG; ΔftrA::hph; ΔcccA::ptrA | This study |
| ΔsidAΔsreA | CEA17; ΔsidA::pyrG; ΔsreA::hph | This study |
| ΔcccAΔsidAΔsreA | CEA17; ΔcccA::ptrA ΔsidA::pyrG; ΔsreA::hph | This study |
| cccA oe | ATCC46645; pxylPPcccA; ptrA | This study |
| ΔcccAcccAoe | ATCC46645; ΔcccA::ptrA; pxylPPcccA; hph | This study |
| ΔcccAcccAgfp | ATCC46645; ΔcccA::ptrA; pgfpcccA; hph | This study |
For the reconstitution of ΔcccA and overexpression of cccA in wt and ΔcccA, strains were transformed with the cccA coding sequence plus 110 bp of its 3′ untranslated region under the control of the xylose-inducible promoter of the P. chrysogenum endoxylanase xylP gene.22 Therefore, a 1.6 kb fragment of the xylP promoter (xylPP) was amplified using pXyluidA-P as a template with synthetic oligos XylP-NotI and XylP-NcoI, thus introducing a NcoI site into the xylP start codon and a NotI site at 1650 bp.22 The fragment was then digested and ligated to the NcoI/NotI-cleaved pGEM5zf(+) vector (Promega) resulting in pxylPP. Next, a 2.2 kb fragment of cccA was amplified using XylPcccA1 and oAfcccA-6r, introducing a PciI site into the cccA start codon. The fragment was then cleaved and ligated to NcoI-cleaved pxylPP, giving rise to pxylPPcccA.
In order to determine the subcellular localization of CccA, the plasmid pgfpcccA was generated. Therefore the cccA gene was amplified using the oligonucleotides oAfcccA-BglII.f and oAfcccA-NotI.r. After digestion, the fragment was subcloned into the BglII/NotI digested plasmid pCAME-703-AohapX-full replacing AohapX.27
For selection, A. fumigatus strains harbouring pxylPPcccA and pgfpcccA A. fumigatus wt were cotransformed with the plasmid pSK275 carrying the pyrithiamine resistance gene and mutant strains with pAN7-1 carrying the hygromycin resistance gene, respectively. Generally, 0.1 μg ml−1 pyrithiamine (Sigma) or 0.2 mg ml−1 hygromycin B (Sigma) was used for selection. Screening of transformants was performed by Southern blot analysis (Fig. S1, ESI†). The hybridization probes for Southern blot analysis of ΔcccA, ΔcccAcccAgfp and ΔcccAcccAoe strains were generated by PCR using the primers listed in Table S1 (ESI†). For extraction of genomic DNA, mycelia were homogenized and DNA was isolated according to Sambrook et al.33 For general DNA propagations Escherichia coli DH5α strain was used as the host.
Northern analysis
RNA isolation was performed using TRI Reagent® (Sigma). For Northern analysis, 10 μg of total RNA was electrophoresed on 1.2% agarose–2.2 M formaldehyde gels and blotted onto Hybond N membranes (Amersham). The hybridization probes used in this study were generated by PCR. Hybridization probes and primers used are listed in Table S1 (ESI†).Fluorescence microscopy
Confocal microscopy images were taken on a confocal laser scanning microscope (Axiovert 100 LSM510 [Carl Zeiss] or SP5 [Leica]) equipped with a 63× oil immersion objective (numerical aperture, 1.40). Images were processed using LSM Image Browser (version 4.2) software, ImageJ (version 1.44o), and Adobe Photoshop CS3.Phylogenetic analysis
A BLASTP search was carried out with the PSI-BLAST algorithm with a NCBI protein database containing 76 different fungal species to determine CccA homologs in different fungi.34 A multiple sequence alignment was done with ClustalW35 using selected sequences with E values higher than 2 × 10–4 in the PSI-BLAST search. Based on this alignment, a rooted tree was generated (Fig. 1). The alignment was generated using ClustalW and BOXSHADE 3.31.36Abbreviations
| BPS | Bathophenanthroline disulfonic acid |
| GFP | Enhanced green fluorescence protein |
| FC | Ferricrocin |
| FSC | Fusarinine C |
| TAFC | Triacetylfusarinine C |
| wt | Wild type |
Acknowledgements
This work was supported by the Austrian Science Foundation grant FWF P21643-B11 (to HH) the “Medizinische Forschungsförderung Innsbruck” (MFI No. 4319 to ME) and the European Science Foundation via its Research Networking Programme Fuminomics.References
- B. Halliwell and J. M. Gutteridge, Biochem. J., 1984, 219, 1–14 CAS.
- F. Tekaia and J. P. Latge, Curr. Opin. Microbiol., 2005, 8, 385–392 CrossRef CAS.
- M. Schrettl, E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. N. Arst Jr, K. Haynes and H. Haas, J. Exp. Med., 2004, 200, 1213–1219 CrossRef CAS.
- M. Schrettl, E. Bignell, C. Kragl, Y. Sabiha, O. Loss, M. Eisendle, A. Wallner, H. N. Arst, K. Haynes and H. Haas, PLoS Pathog., 2007, 3, e128 Search PubMed.
- A. Wallner, M. Blatzer, M. Schrettl, B. Sarg, H. Lindner and H. Haas, Appl. Environ. Microbiol., 2009, 75, 4194–4196 CrossRef CAS.
- H. Haas, M. Eisendle and G. B. Turgeon, Annu. Rev. Phytopathol., 2008, 46, 149–187 CrossRef CAS.
- C. Kragl, M. Schrettl, B. Abt, B. Sarg, H. H. Lindner and H. Haas, Eukaryotic Cell, 2007, 6, 1278–1285 CrossRef CAS.
- M. Eisendle, M. Schrettl, C. Kragl, D. Muller, P. Illmer and H. Haas, Eukaryotic Cell, 2006, 5, 1596–1603 CrossRef CAS.
- G. Chamilos, E. M. Bignell, M. Schrettl, R. E. Lewis, K. Leventakos, G. S. May, H. Haas and D. P. Kontoyiannis, Med. Mycol., 2010, 48, 506–510 CrossRef CAS.
- J. L. Slater, L. Gregson, D. W. Denning and P. A. Warn, Med. Mycol., 2011, 49(Suppl. 1), S107–S113 CrossRef CAS.
- M. Schrettl, O. Ibrahim-Granet, S. Droin, M. Huerre, J. P. Latge and H. Haas, Microbes Infect., 2010, 12, 1035–1041 CrossRef CAS.
- M. Seifert, M. Nairz, A. Schroll, M. Schrettl, H. Haas and G. Weiss, Immunobiology, 2008, 213, 767–778 CrossRef CAS.
- M. Schrettl, H. S. Kim, M. Eisendle, C. Kragl, W. C. Nierman, T. Heinekamp, I. Jacobsen, E. R. Werner, A. A. Brakhage and H. Haas, Mol. Microbiol., 2008, 70, 27–43 CrossRef CAS.
- P. J. Lapinskas, S. J. Lin and V. C. Culotta, Mol. Microbiol., 1996, 21, 519–528 CrossRef CAS.
- L. Li, O. S. Chen, D. McVey Ward and J. Kaplan, J. Biol. Chem., 2001, 276, 29515–29519 CrossRef CAS.
- S. A. Kim, T. Punshon, A. Lanzirotti, L. Li, J. M. Alonso, J. R. Ecker, J. Kaplan and M. L. Guerinot, Science, 2006, 314, 1295–1298 CrossRef CAS.
- J. L. Urbanowski and R. C. Piper, J. Biol. Chem., 1999, 274, 38061–38070 CrossRef CAS.
- M. E. Portnoy, X. F. Liu and V. C. Culotta, Mol. Cell. Biol., 2000, 20, 7893–7902 CrossRef CAS.
- A. Singh, N. Kaur and D. J. Kosman, J. Biol. Chem., 2007, 282, 28619–28626 CrossRef CAS.
- V. Lanquar, F. Lelievre, S. Bolte, C. Hames, C. Alcon, D. Neumann, G. Vansuyt, C. Curie, A. Schroder, U. Kramer, H. Barbier-Brygoo and S. Thomine, EMBO J., 2005, 24, 4041–4051 CrossRef CAS.
- L. Li, D. Bagley, D. M. Ward and J. Kaplan, Mol. Cell. Biol., 2008, 28, 1326–1337 CrossRef CAS.
- I. Zadra, B. Abt, W. Parson and H. Haas, Appl. Environ. Microbiol., 2000, 66, 4810–4816 CrossRef CAS.
- S. Yasmin, B. Abt, M. Schrettl, T. A. Moussa, E. R. Werner and H. Haas, Fungal Genet. Biol., 2009, 46, 707–713 CrossRef CAS.
- S. A. Leong and G. Winkelmann, Met. Ions Biol. Syst., 1998, 35, 147–186 CAS.
- M. Blatzer, B. M. Barker, S. D. Willger, N. Beckmann, S. J. Blosser, E. J. Cornish, A. Mazurie, N. Grahl, H. Haas and R. A. Cramer, PLoS Genet., 2011, 7, e1002374 CAS.
- H. Lin, L. Li, X. Jia, D. M. Ward and J. Kaplan, J. Biol. Chem., 2011, 286, 3851–3862 CrossRef CAS.
- H. Goda, T. Nagase, S. Tanoue, J. Sugiyama, S. Steidl, A. Tuncher, T. Kobayashi, N. Tsukagoshi, A. A. Brakhage and M. Kato, Arch. Microbiol., 2005, 184, 93–100 CrossRef CAS.
- H. Haas, Appl. Microbiol. Biotechnol., 2003, 62, 316–330 CrossRef CAS.
- M. Schrettl, N. Beckmann, J. Varga, T. Heinekamp, I. D. Jacobsen, C. Jöchl, T. A. Moussa, S. Wang, F. Gsaller, M. Blatzer, E. R. Werner, W. C. Niermann, A. A. Brakhage and H. Haas, PLoS Pathogens, 2010, 6(9), e1001124 Search PubMed.
- G. Pontecorvo, J. A. Roper, L. M. Hemmons, K. D. MacDonald and A. W. J. Bufton, Adv. Genet., 1953, 5, 141–238 CrossRef CAS.
- H. Oberegger, M. Schoeser, I. Zadra, B. Abt and H. Haas, Mol. Microbiol., 2001, 41, 1077–1089 CrossRef CAS.
- M. L. Nielsen, L. Albertsen, G. Lettier, J. B. Nielsen and U. H. Mortensen, Fungal Genet. Biol., 2006, 43, 54–64 CrossRef CAS.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 1989 Search PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS.
- B. Neron, H. Menager, C. Maufrais, N. Joly, J. Maupetit, S. Letort, S. Carrere, P. Tuffery and C. Letondal, Bioinformatics, 2009, 25, 3005–3011 CrossRef CAS.
- C. d'Enfert, Curr. Genet., 1996, 30, 76–82 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2mt20179h |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2012 |
