Identification of Ni-(L-His)2 as a substrate for NikABCDE-dependent nickel uptake in Escherichia coli†
Peter T.
Chivers‡
*,
Erin L.
Benanti§
,
Vanessa
Heil-Chapdelaine
,
Jeffrey S.
Iwig§
and
Jessica L.
Rowe
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110, USA. E-mail: pchivers@oberlin.edu
First published on 30th July 2012
Abstract
Nickel is an important cofactor for several microbial enzymes. The ATP-dependent NikABCDE transporter is one of several types of uptake pathways known to be important for nickel acquisition in microbes. The Escherichia coli NikA periplasmic binding protein is structurally homologous to the di- and oligopeptide binding proteins, DppA and OppA. This structural similarity raises interesting questions regarding the evolutionary relationships between the recognition of nickel ions and short peptides. We find that in defined minimal growth medium NikABCDE transports nickel ions in the presence of exogenously added L-histidine (L-His), but not D-histidine. Both nickel uptake in cells and nickel binding to purified NikA showed an L-His concentration dependence consistent with recognition of a Ni-(L-His)2 complex. This discovery reveals parallels to the transport of other metal complexes, notably iron, and suggests the structural diversity of nickel transporters may arise from the need to recognize extracellular nickel complexed with different organic ligands, whether they be exogenously or endogenously produced. Further, these results suggest that experiments examining the physiology and ecology of nickel-requiring microbes should account for the possibility that the growth medium may not support nickel uptake.
Introduction
Transition metals are essential cofactors for protein structure and function. They must be acquired from the growth environment, and they are often required at intracellular concentrations much higher than their extracellular levels.1 Metal-acquisition under these limiting concentrations depends upon energy-dependent membrane transporter proteins, of which there are numerous structural classes.2 The ATP-dependent transporters (ABC family) are hetero-oligomeric complexes that use cytoplasmic ATP hydrolysis to power solute transport.3 In bacteria and archaea, ABC transporters use a binding protein to recognize and deliver the metal to the multisubunit transmembrane transporter complex, which is energized by two cytoplasmic ATPase domains. The binding proteins, which can either be soluble or membrane attached depending on the cellular architecture of the microbe, fall into several different structural classes that are associated with the transport of chemically distinct solutes.4 This structural variation provides a solution to the problem of metal speciation in growth environments containing organic ligands, potential chelators of metal ions. For example, the FhuD of E. coli recognizes the ferric–hydroxamate complex,5 while the Zn-periplasmic binding protein ZnuA coordinates the metal ion directly.6E. coli express the NikABCDE ABC-transporter for nickel uptake under anaerobic growth conditions.7 The NikA protein has both sequence and structural homology with the di- and oligopeptide (DppA–OppA) binding proteins.4,7,8 This homology raises the question of how nickel is recognized by NikA. The possibility that NikA does not directly recognize nickel ions but rather a Ni–ligand complex is suggested by NikA co-crystal structures containing metal–ligand complexes.9,10 These structures contain either nickel or iron bound to a polycarboxylic acid molecule. However, these compounds have not been tested in nickel-uptake assays. Additionally, these structures do not exhibit the fully closed conformation observed in periplasmic binding proteins bound to their cognate solute molecule, including the closely related DppA and OppA proteins.11,12
Here, we show that L-histidine (L-His) is sufficient to enable NikABCDE dependent Ni(II)-uptake in E. coli grown in defined minimal medium. Purified NikA binds Ni(II) with high affinity in the presence of L-histidine, as Ni-(L-His)2. These effects were not observed with D-His, indicating the presence of a chiral binding site. These observations suggest that nickel-speciation in different growth environments may affect the mechanism of nickel transport. Additionally, these results suggest that the importance of nickel-dependent microbial physiology, such as hydrogenase activity, may be underestimated in laboratory settings where microbes are often grown under conditions that may not favor Ni-uptake.13
Experimental
Strains, plasmids, and chemicals
Wild-type, ΔnikA,14 ΔnikDE, ΔnikR,14 ΔhisPMQJ, ΔtonB E. coli strains are isogenic versions of E. coli K-12 RZ4500 (ΔlacZ).15 The nikDE and hisPMQJ deletions were created using the Wanner method.16 The ΔtonB strain was created by P1 transduction17 of E. coli KP1344,18 a generous gift from Kathleen Postle (Pennsylvania State University).L-Amino acids were from Fisher Biosciences and used at the concentrations described for LeMaster minimal medium.19D-Histidine, L-histidinol, and L-histidine methyl ester dihydrochloride were purchased from Acros Organics (New Jersey, USA). Methyl-amino L-histidine, deamino L-histidine, L-histidine amide, and Nδ- and Nε-methylhistidines were purchased from Bachem (Torrance, CA). 63NiCl2 (9.87 mCi mg−1; 88 mM) was from PerkinElmer (Boston, MA). 2,5 3H-L-histidine (50 Ci mM−1; 20 μM) was from American Radiolabelled Chemicals (St. Louis, MO). All reagents were used without further purification. Amino acid stock solutions were made in ddH2O and stored at −20 °C.
Nickel transport and lacZ reporter assays
Overnight nickel accumulation (14–16 h, 37 °C) was measured as described previously.20L-Histidine or other molecules were added from concentrated stock solutions to the final concentrations listed. The ice-cold cell resuspension and wash solution in these experiments was growth medium containing 1 mM D-histidine. 63Ni accumulation was determined by scintillation counting for 10 min (0 to 1.3 MeV window). 63Ni per cell was calculated by converting cpm to dpm assuming a 50% counting efficiency. OD600 values were used to calculate the number of cells (cfu) based on a standard curve (cfu = 0.714 × OD600) generated for the spectrophotometer used in this study.63Ni uptake rates were measured using cells grown anaerobically in the absence of added nickel or L-histidine (L-His) (filled 15 mL or 50 mL polypropylene tubes, 37 °C, 14–16 h; 0.3 ≤ OD600 ≤ 0.8). 63Ni-uptake was initiated by addition of cells (1.6 mL) to microfuge tubes containing 63NiCl2, and L-His or other compounds already added from stock solutions to yield the desired final concentration. Tubes were placed in a 37 °C water bath for 2–3 min. 63Ni uptake was quenched by placing the tubes in an ice-water bath (2–5 min). Each tube was then centrifuged, and the cells washed and counted as described above.
LacZ reporter assays were performed as described previously.14 Cells were grown under the same conditions described for the nickel transport assays.
NikA expression and purification
E. coli nikA was PCR amplified from genomic DNA and cloned into pET-28a (Novagen, Madison, WI) using the NcoI and XhoI restriction sites. The first 22 codons, which encode a signal sequence for export to the periplasm, were not included. The plasmid (pNIKA) was sequenced (Seqwright, Houston, TX) to verify the correct gene sequence. NikA was expressed in E. coli DL4119 (DE3) using the Studier autoinduction protocol.21 The protein was purified using a modified version of a previously published procedure.8 Here, the EDTA and β-mercaptoethanol were omitted from the purification buffers to avoid co-purification of metal–ligand complexes, which has been previously observed.9 All other buffer conditions and column chromatography procedures were the same.Nickel binding assays
Nickel-binding reactions were carried out in 20 mM Hepes (pH 7.5), 200 mM NaCl. NikA was incubated with NiCl2 and L-His for at least 15 min (22 °C) prior to measurement. For fluorescence quenching (Cary Eclipse Spectrophotometer; Varian, Palo Alto, CA), 120 μL reactions were excited at 280 nm (5 nm slit width). Filter binding studies used 100 μL reaction volumes, which were pipetted onto a 2.5 cm PVDF membrane under suction. The membrane was washed with 2 × 500 μL of binding buffer without nickel or L-histidine then placed in a scintillation vial with 1 mL of scintillation fluid for 63Ni counting, as described above. Control samples containing 63Ni and L-His but not NikA were used for background subtraction to account for any non-specific binding of the Ni-L-His complex to the membrane.Results and discussion
NikABCDE-dependent Ni-uptake requires L-Histidine
E. coli grown anaerobically in defined M63 minimal medium accumulated less 63Ni compared to cells grown in rich, undefined LB medium (Fig. S1, ESI†). The differential 63Ni-accumulation between the two growth conditions suggested some component of LB caused an increase in nickel uptake. The addition of tryptone, yeast extract, or amino acids to the M63 minimal medium resulted in increased 63Ni-accumulation (Fig. S1, ESI†). Of the amino acids tested, only L-histidine (L-His) was capable of increasing Ni-accumulation, and this effect was not inhibited by the other amino acids (Fig. S1, ESI†). These data suggested that the component of LB medium aiding nickel uptake was L-His, although the contribution of other components, such as L-His-containing peptides, could not be excluded.Nickel accumulation after overnight growth cannot discriminate between the direct use of L-His in Ni-uptake or its modification by the cell before use. To distinguish between these possibilities, initial rates of 63Ni uptake were measured in cells grown anaerobically without added nickel or L-His. The effect of L-His addition on 63Ni uptake was rapid (Fig. 1A), supporting a role for unmodified L-His in transport. The rapid uptake observed with L-His also rules out indirect effects on NikABCDE expression levels in the timeframe of the accumulation assay. L-His-assisted 63Ni uptake required the presence of both NikA (periplasmic binding protein) and the NikDE (cytoplasmic ATPase domains) subunits (Fig. 1B). The absence of nickel accumulation in the ΔnikDE strain confirmed that the 63Ni detected in this assay came from intracellular nickel ions. Any periplasmic NikA:Ni-(L-His)n complex, if present, must be too labile to be observed under these conditions. Additionally, some E. coli ABC transporters have been shown to exhibit interchangeability between periplasmic and membrane protein components.22,23 These data indicate that NikABCDE is the only ABC-transporter involved in L-His dependent Ni-accumulation. Deletion of the tonB gene, which encodes an protein that interacts with outer membrane receptor proteins required for the uptake of iron–ligand complexes and vitamin B12, had no effect on 63Ni uptake (Fig. S2, ESI†). Thus, the low molecular weight Ni-(L-His)n complex probably traverses the outer membrane through non-specific porin proteins.24 The addition of D-His had no effect on 63Ni uptake (Fig. 1B), supporting a structural role for L-His rather than a non-specific effect on Ni-solubility in the phosphate-rich (∼75 mM) M63 minimal medium.
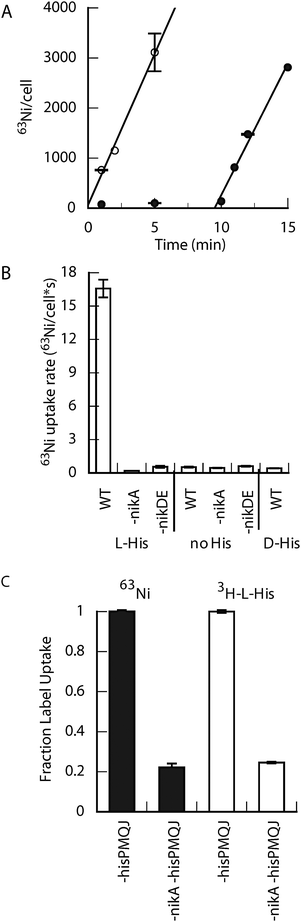 | ||
| Fig. 1 The effect of L-histidine on Ni-uptake in E. coli. (A) The initial rate of 63Ni uptake. L-His (400 μM final) was added at t = 0 or 10 min to cells already containing 5 nM 63NiCl2. The initial rate is ∼10 Ni per cell per s. (B) The effect of different nik operon mutants on 63Ni uptake. Different strains were incubated with 20 nM 63NiCl2 (2.5 min) without or with L- or D-His (400 μM final). (C) 3H-L-His uptake in wild type and ΔnikA cells. All strains in this panel lack the hisPMQJ operon (denoted-hisPMQJ), which encodes the high-affinity L-His transporter.22 Final concentrations were 63NiCl2 or NiCl2 (250 nM) and 3H-L-His or L-His (1 μM). Cells were grown at 37 °C for 14–16 h. | ||
L-His is co-transported with the nickel ion
To further probe the mechanism of L-His-dependent Ni-uptake, the uptake of 3H-L-His was compared between wild type and ΔnikA strains. These strains were deleted for the L-His ABC transporter (hisPMJQ) to eliminate the high capacity for L-His uptake by E. coli,22 which would obscure the comparatively lower amounts of L-His required by the NikABCDE pathway. Additionally, 100 μM of both L-Tyr and L-Trp were present to saturate the aromatic amino acid uptake pathways that can also be utilized by L-His.22L-Histidinol was added at 100 μM to supplement the histidine biosynthetic pathway. The deletion of nikA decreased the uptake of both 63Ni and 3H-L-His by 4-fold (Fig. 1C), consistent with the transport of a Ni-L-His complex.L-His is required at biologically relevant concentrations
Ni(II) forms 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexes with L-His, with stability constants (K1, 105.88, K2, 104.88) that have been determined under conditions similar to those used here.25 The dependence of 63Ni uptake on L-His concentration can therefore resolve the stoichiometry of the complex that is recognized by the NikABCDE pathway. The 63Ni uptake rate was saturable with respect to both Ni and L-His concentration (Fig. 2A and B), consistent with the presence of a binding site that recognizes a Ni-(L-His)n complex. The concentration ranges over which Ni uptake was observed (KL-His 25 μM; KNi 33 nM) were consistent with levels of L-His (10−5 to 10−4 M) in the lower intestine26 and dietary nickel.27 Significantly, the L-His concentration range over which Ni-uptake was observed correlated with the abundance of Ni-(L-His)2, suggesting that NikA binds to this species (Fig. 2C).
2 complexes with L-His, with stability constants (K1, 105.88, K2, 104.88) that have been determined under conditions similar to those used here.25 The dependence of 63Ni uptake on L-His concentration can therefore resolve the stoichiometry of the complex that is recognized by the NikABCDE pathway. The 63Ni uptake rate was saturable with respect to both Ni and L-His concentration (Fig. 2A and B), consistent with the presence of a binding site that recognizes a Ni-(L-His)n complex. The concentration ranges over which Ni uptake was observed (KL-His 25 μM; KNi 33 nM) were consistent with levels of L-His (10−5 to 10−4 M) in the lower intestine26 and dietary nickel.27 Significantly, the L-His concentration range over which Ni-uptake was observed correlated with the abundance of Ni-(L-His)2, suggesting that NikA binds to this species (Fig. 2C).
![l-His dependent Ni-uptake is saturable as a function of Ni-(l-His2) concentration. (A) and (B) l-His was present at 10, 100, or 1000 μM and 63NiCl2 was added at 5, 15, 45, 135, or 405 nM. The resulting plots of initial rate vs. [l-His] or [NiCl2] were fit to the Michaelis–Menton equation. (C) The speciation of Ni with l-His based on the affinity constants (Ni-His, 105.88; Ni-His2, 104.59) determined in phosphate buffer by ITC25 is shown with the grey lines. The plotted points (filled squares) correspond to initial uptake rates for 15 nM 63NiCl2 and l-His concentrations from 2 μM to 1 mM.](/image/article/2012/MT/c2mt20139a/c2mt20139a-f2.gif) | ||
| Fig. 2 L-His dependent Ni-uptake is saturable as a function of Ni-(L-His2) concentration. (A) and (B) L-His was present at 10, 100, or 1000 μM and 63NiCl2 was added at 5, 15, 45, 135, or 405 nM. The resulting plots of initial rate vs. [L-His] or [NiCl2] were fit to the Michaelis–Menton equation. (C) The speciation of Ni with L-His based on the affinity constants (Ni-His, 105.88; Ni-His2, 104.59) determined in phosphate buffer by ITC25 is shown with the grey lines. The plotted points (filled squares) correspond to initial uptake rates for 15 nM 63NiCl2 and L-His concentrations from 2 μM to 1 mM. | ||
Modifications of L-His affect Ni-uptake
The modification of the likely coordinating atoms in L-His provides a way to test the proposed transport ligand. Modifications of the carboxyl moiety of L-histidine (histidinol, L-His-CONH2, and L-His methyl ester) were all deleterious to 63Ni uptake (Fig. 3). Removal of the L-His NH2 group (deamino) also knocked out nickel uptake. Monomethylation of the NH2 group, however, still provided close to normal rates of uptake. Methylation of the imidazolium ring nitrogens (Nδ and Nε) had differing effects. Methylation of Nδ, the ring nitrogen that forms the more stable complex, eliminated Ni uptake, while methylation of Nε only slightly affected uptake rate. These data suggest coordination of Ni via the![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) H2−, C
H2−, C![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) O− and imidazolium Nδ atoms. Unrelated polycarboxylic acid molecules (nitrilotriacetic acid, EDTA, citrate, fumarate, and succinate) were not effective at Ni-uptake (Fig. S3, ESI†).
O− and imidazolium Nδ atoms. Unrelated polycarboxylic acid molecules (nitrilotriacetic acid, EDTA, citrate, fumarate, and succinate) were not effective at Ni-uptake (Fig. S3, ESI†).
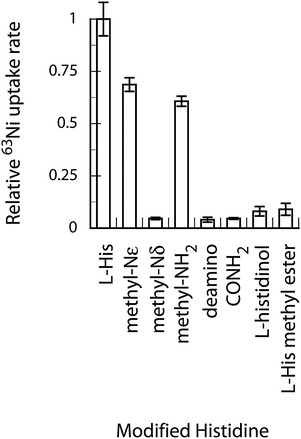 | ||
| Fig. 3 Modifications of L-His affect Ni-uptake rate. Modified L-His molecules were present at 1 mM and 63Ni was added to 10 nM. | ||
Metal selectivity of NikABCDE pathway
L-His dependent nickel uptake via NikABCDE pathway was also selective. Cobalt was the only first-row divalent metal (Fig. 4) to significantly affect Ni-accumulation during overnight growth, and only when present in 100-fold molar excess over 63Ni. The differences in metal–ligand distances and coordination geometries of these metal-histidine complexes (Table S1, ESI†) are consistent with the specific recognition of an octahedral Ni-(L-His)2 complex by NikA. Co(II) forms an octahedral complex28 with slightly longer metal–ligand atom distances compared to Ni(II). In contrast, the Zn(II) and Cu(II) bis-His complexes29,30 have lower coordination numbers and greater differences in bond lengths compared to Ni(II).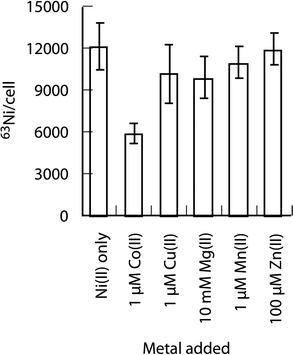 | ||
| Fig. 4 The effect of divalent metals on L-His dependent 63Ni accumulation. Cells were added to tubes containing 10 nM 63NiCl2 + 400 μM L-His plus a competitor metal added at the different concentrations indicated. | ||
The effect of L-His on nickel-dependent gene regulation in E. coli
The effect of L-His regulation of NikABCDE expression by the NikR repressor was examined using a PNikABCDE-lacZ reporter assay.14 Higher levels of intracellular nickel result in greater NikR activity and lower expression from PNikABCDE. Previously, LacZ activity was shown to be independent of the presence of NikABCDE in minimal growth medium lacking L-His.14 In the presence of 400 μM L-His, a 2.5-fold lower basal level of PNik-lacZ expression was observed (wild-type in Fig. 5A and B), which correlated with the increase in intracellular nickel due to the presence of L-His (open squares in Fig. 5A and B). The deletion of nikA resulted in an almost 5-fold decrease in NikR activity in the presence of L-His (Fig. 5A), which contrasts with negligible effect of a nikA deletion in cells grown without L-His.14 The addition of L-His also shifted the nickel-dependent transition required for full repression by NikR (Fig. 5A and B). This effect was independent of NikA, as observed previously,14 and further supports the role of a non-specific nickel uptake pathway in NikR-dependent repression of NikABCDE expression. The NiCl2 concentration at which cell growth was reduced by more than 10% was also shifted, and in a NikA-independent manner.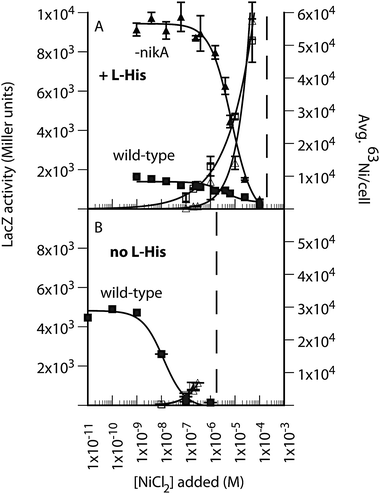 | ||
| Fig. 5 The effect of L-His on nickel-dependent gene regulation in E. coli. (A) 63Ni accumulation (open symbols) and PNik-lacZ expression (closed symbols) in wild-type (squares) and ΔnikA (triangles) E. coli grown in M63 minimal medium (14–16 h) in the presence of 400 μM L-His. (B) The same experiment in the absence of L-His. Left axis, LacZ activity (Miller units), which decreases with increasing added NiCl2. Right axis, average 63Ni per cell, which increases with added NiCl2. The y-axes in Panels A and B are to the same scale for the corresponding measured value. The dashed lines represent the NiCl2 concentration at which overnight growth was reduced by greater than 10% based on final OD600 values. | ||
The non-specific pathway was further probed by examining the concentration dependence of 63Ni-uptake in the absence of L-His (Fig. S4, ESI†). The linear relationship of uptake rate versus nickel concentration suggests that this pathway was either non-saturable, thus non-specific, or a high Kmpathway. The non-specific pathway was also of lower capacity than NikABCDE based on the total nickel accumulated under similar conditions (Fig. 5A and B). Additionally, as expected from Fig. 5, in the absence of nikA the presence of L-His is strongly inhibitory to 63Ni uptake, due to the formation of Ni-(L-His)2 (Fig. S4, ESI†). Half-maximal inhibition was observed at 80 μM L-His.
These data suggest that the transition to complete repression of PNik expression is likely a stress response that can be buffered by stable nickel–ligand complex in the medium, not just those formed by L-His. This interpretation is also consistent with the observation that full repression of PNik by NikR is coincident with induction of de-repression of the RcnR-repressed RcnA nickel efflux pathway,20 which is also shifted in response to the composition of the growth medium. The data also indicated that nickel levels in E. coli continually increased as a function of increasing extracellular nickel concentration, indicating that a constant intracellular level of nickel ions was not maintained over any range of extracellular nickel concentration.
L-His increases the affinity of Ni(II) for the NikA protein
The nickel transport data (Fig. 2C) support the formation of a Ni-(L-His)2:NikA ternary complex during NikABCDE-dependent Ni-uptake. Different approaches were used to detect the formation of this complex. Size exclusion chromatography of purified NikA with 63Ni in the presence and absence of L-His showed formation of a stable complex only when all three components were present (Fig. S5, ESI†), providing direct qualitative evidence for a role for L-His in the interaction of Ni and the NikA protein. Fluorescence spectroscopy using the native Trp residues of NikA, including two in the binding pocket, showed that L-His, but not D-His, had a significant effect on nickel-dependent quenching of NikA tryptophan fluorescence (Fig. 6). The combination of L-His and Ni resulted in ∼50% fluorescence quenching, compared to the absence of any effect when either ligand was added by itself. This change was substantially greater than previously observed (∼10%) when only nickel was added to NikA.31 This signal change was used to determine the stoichiometry of the Ni-His-NikA complex. The titration of NiCl2 against 6 μM NikA in the presence of 400 μM L-His resulted in maximal fluorescence quenching at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Ni
1 Ni![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NikA (Fig. 6A). The addition of L-His to 6 μM NikA in the presence of 6 μM Ni resulted in maximal fluorescence quenching when the L-His concentration was sufficient to generate a stoichiometric amount of the Ni-His2 species (6 μM; Fig. 6A and B), based upon the formation constant of this species under these buffer conditions,25 which are different than the uptake assay. A linear change in fluorescence as a function of added L-His would require much higher NikA concentrations, and was not studied.
NikA (Fig. 6A). The addition of L-His to 6 μM NikA in the presence of 6 μM Ni resulted in maximal fluorescence quenching when the L-His concentration was sufficient to generate a stoichiometric amount of the Ni-His2 species (6 μM; Fig. 6A and B), based upon the formation constant of this species under these buffer conditions,25 which are different than the uptake assay. A linear change in fluorescence as a function of added L-His would require much higher NikA concentrations, and was not studied.
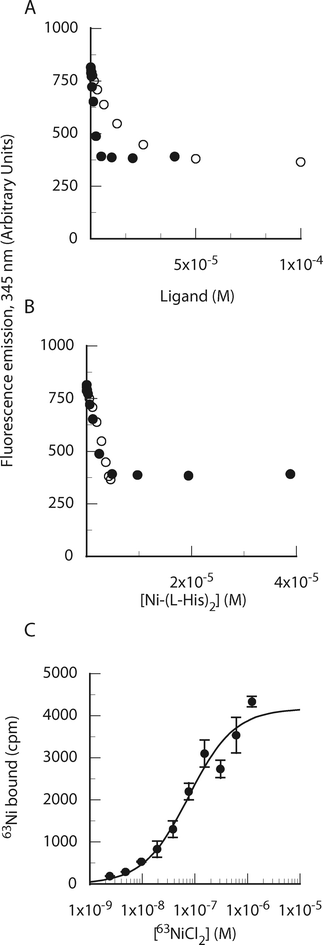 | ||
| Fig. 6 The Ni-(L-His2) complex binds to purified NikA. (A) Determination of binding stoichiometry by fluorescence quenching. Ni(II) or L-His were added to NikA (6 μM) containing a fixed concentration of the other ligand (6 μM Ni(II) or 1 mM L-His). (B) Data plotted as a function of Ni-(L-His2) species using association constants (Ni-His, 106.19; Ni-His2, 105.05) determined in Hepes buffer by Zhang et al.25 (C) Determination of Ni-(L-His2) binding affinity by filter binding. 63NiCl2 was added to NikA (30 nM) and 400 μM L-His. | ||
Filter binding with 63Ni was also used to determine the affinity of the Ni-His2 species for NikA (Fig. 6C). In the presence of 400 μM L-His, the midpoint of the transition as a function of added nickel occurred at 80 nM. At this L-His-concentration, the distribution of nickel-histidine species is almost exclusively shifted to Ni-(L-His)2, indicating a Kd value for the NikA:Ni-(L-His)2 complex of ∼75 nM.
The identification of L-His as a requirement for NikABCDE-dependent nickel uptake in E. coli provides a foundation to better understand the structural basis for nickel acquisition by this transporter. The concentration dependence of nickel uptake and the affinity of Ni-His2 for NikA are consistent with a high-affinity interaction similar to those determined for other ABC transporter proteins involved in metal complex uptake in E. coli, such as FepB:ferric enterobactin, Kd 30 nM.32 The range of rates of 63Ni-(L-His2) uptake observed for biologically relevant solute concentrations is consistent with the rapid delivery of sufficient nickel for the assembly of Ni–Fe H2ase isozymes in response to changes in growth conditions. The NikABCDE pathway is unlikely to provide a significant amount of L-His to E. coli, which has a free L-His pool of ∼100 μM22 and a total L-His of 107 molecules per cell.33,34
Previous studies of nickel import in bacteria have used different growth media to examine either accumulation or initial uptake. The only previous study of NikABCDE-dependent 63Ni-uptake7 used cells grown anaerobically in LB then resuspended in phosphate buffer before 63Ni addition (150 nM). An initial uptake rate was not calculated from the data (reported as pmol mg−1 cell dry weight), but a reasonable estimate suggests a low rate of uptake (∼0.5 Ni per cell per s). Our observations suggest that Ni-transport studies must be carried out using a variety of growth conditions. Additionally, quantification of the data is important to identify whether the observed rates of transport are consistent with physiologically relevant rates and to compare the capacities of different transporters (Vmax and Km).
The structural homology between DppA–OppA and NikA11,35 suggests a possible basis for recognition of the Ni-L-His2 complex by NikA. Peptide recognition by DppA and OppA occurs via a main chain H-bond network between protein and ligand that enforces chiral specificity but still allows binding of peptides with different side chains.11,35 We speculate that a similar interaction will be present in the Ni-(L-His)2:NikA complex, where the amino and carboxyl groups that coordinate the Ni ion can also still form H-bond interactions with the main chain atoms of NikA, as is observed in the DppA and OppA structures. The proposed binding mode also explains why D-His is not a suitable substrate for transport or binding, as well as why nickel complexes with polycarboxylate ligands,9,10 which lack a peptide-like spacing of H-bond donor and acceptors, are not functional substrates for transport. An additional consideration is that NikA binds a Ni-(L-His)2 stereoisomer36 other than the trans-imidazole octahedral species that has been observed crystallographically.37
An open question is how NikA enforces metal-specific recognition. The proposed mode of binding would not necessarily require direct contact between the Ni2+ atom and NikA. The other divalent metal complexes tested have different metal–ligand distances (Co2+) and coordination numbers (Cu2+ and Zn2+). The additivity of small differences in the positions of the His atoms not in the primary metal coordination sphere could have a significant effect on affinity. Water molecules may also help in this discrimination between the protein and portions of Ni-(L-His)2 complex, as has been observed in peptide binding to OppA.12 Metal-specificity determinants could also be present at additional steps after binding to NikA, such as in the handoff to the NikBC transmembrane complex and the NikDE cytoplasmic subunits.
The delivery of a nickel–ligand complex to the cytoplasm of E. coli has implications for Ni-enzyme assembly and Ni-dependent gene regulation.38 In particular, the kinetic mechanisms of nickel transfer reactions between intracellular proteins involved in [Ni–Fe] H2ase assembly (HypA/HybF, SlyD, HypB),38 nickel-responsive gene regulation (NikR and RcnR),39 and efflux (RcnA)40 and others41 will likely differ due to the different coordination geometries and binding affinities observed in these different proteins. Kinetic mechanisms of Ni-transfer involving Ni-(L-His)2 have been studied previously.42,43 The results obtained here indicate that the same type of kinetic studies would be relevant to intracellular trafficking in E. coli.
The requirement for exogenous L-His has several implications for the nickel physiology of E. coli. The abundance of extracellular L-His and its effect on nickel uptake will modulate the activity of anaerobically expressed [Ni–Fe] H2ase isozymes.44 Bacteria with only a NikABCDE-type nickel transporter must grow in rich medium to accumulate significant levels of intracellular nickel ions. The abundance of amino acids, in particular L-His, could arise from degradation of proteins in the environmental niche, such as the mammalian gastrointestinal tract,26 or via secretion of L-His in micromolar quantities, which E. coli is not known to do. The gastrointestinal tract is a well-established growth niche for E. coli,45 but growth in nutrient deficient environments is not compatible with nickel accumulation and H2ase expression. Studies of anaerobic gene expression in E. coli using established defined minimal growth medium13 are likely to underestimate the contribution of Ni-dependent enzyme activity to cell physiology.
The use of Ni–ligand complexes in nickel transport is likely not restricted to NikABCDE-dependent transport. H. pylori uses the exbBD–TonB transducing system to import nickel via the FrpB4 outer membrane receptor46 coupled to an ABC-transporter with homology to known iron-complex transporters,47 providing strong evidence for a stable nickel–ligand complex. This complex has not been identified, but the tonB-independent import of Ni-(L-His)2 uptake in E. coli suggests that H. pylori recognizes a different Ni–ligand species. Whether H. pylori synthesizes a nickelophore or captures an already present Ni–ligand complex is also unknown. H. pylori and E. coli have distinct growth environments, which undoubtedly influences the distribution of Ni-complexes present. H. pylori requires nickel for survival in the acidic environment of the stomach and may employ different transporters to tune nickel acquisition with the intracellular demand and environmental pH.
The results reported here should allow more focused studies of the molecular recognition of nickel ions in the environment by organisms that require them for growth. Functional and bioinformatics studies have identified several distinct structural classes of microbial nickel transporters.48 Microbes, such as some marine cyanobacteria,49 that require on nickel in relatively spartan growth environments may transport Ni(II) in the absence of complexing ligands. Yet, several bacteria encode more than one structural type of nickel transporter. The functional benefits of these distinct transporters remain to be elucidated, but may reflect different structural mechanisms for nickel acquisition under changing growth conditions, such as pH and organic molecule concentration and composition.
Acknowledgements
This work was supported by NSF grant MCB0520877. We thank Kathleen Postle for the gift of the ΔtonB E. coli strain.Notes and References
- C. E. Outten and T. V. O'Halloran, Science, 2001, 292, 2488–2492 CrossRef CAS.
- Z. Ma, F. E. Jacobsen and D. P. Giedroc, Chem. Rev., 2009, 109, 4644–4681 CrossRef CAS.
- D. C. Rees, E. Johnson and O. Lewinson, Nat. Rev. Mol. Cell Biol., 2009, 10, 218–227 CrossRef CAS.
- R. Tam and M. H. Saier, Jr., Microbiol. Rev., 1993, 57, 320–346 CAS.
- W. Koster and V. Braun, J. Biol. Chem., 1990, 265, 21407–21410 CAS.
- H. Li and G. Jogl, J. Mol. Biol., 2007, 368, 1358–1366 CrossRef CAS.
- C. Navarro, L.-F. Wu and M.-A. Mandrand-Berthelot, Mol. Microbiol., 1993, 9, 1181–1191 CrossRef CAS.
- J. Heddle, D. J. Scott, S. Unzai, S. Y. Park and J. R. Tame, J. Biol. Chem., 2003, 278, 50322–50329 CrossRef CAS.
- M. V. Cherrier, L. Martin, C. Cavazza, L. Jacquamet, D. Lemaire, J. Gaillard and J. C. Fontecilla-Camps, J. Am. Chem. Soc., 2005, 127, 10075–10082 CrossRef CAS.
- M. V. Cherrier, C. Cavazza, C. Bochot, D. Lemaire and J. C. Fontecilla-Camps, Biochemistry, 2008, 47, 9937–9943 CrossRef CAS.
- P. Dunten and S. L. Mowbray, Protein Sci., 1995, 4, 2327–2334 CrossRef CAS.
- J. R. Tame, G. N. Murshudov, E. J. Dodson, T. K. Neil, G. G. Dodson, C. F. Higgins and A. J. Wilkinson, Science, 1994, 264, 1578–1581 CAS.
- F. C. Neidhardt, P. L. Bloch and D. F. Smith, J. Bacteriol., 1974, 119, 736–747 CAS.
- J. L. Rowe, G. L. Starnes and P. T. Chivers, J. Bacteriol., 2005, 187, 6317–6323 CrossRef CAS.
- M. Choe and W. S. Reznikoff, J. Bacteriol., 1991, 173, 6139–6146 CAS.
- K. Datsenko and B. Wanner, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6640–6645 CrossRef CAS.
- Current Protocols in Molecular Biology, ed. F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Siedman, J. A. Smith and K. Struhl, Wiley, New York, 1989 Search PubMed.
- R. A. Larsen, M. G. Thomas and K. Postle, Mol. Microbiol., 1999, 31, 1809–1824 CrossRef CAS.
- D. M. LeMaster and F. M. Richards, Biochemistry, 1985, 24, 7263–7268 CrossRef CAS.
- J. S. Iwig, J. L. Rowe and P. T. Chivers, Mol. Microbiol., 2006, 62, 252–262 CrossRef CAS.
- F. W. Studier, Protein Expression Purif., 2005, 41, 207–234 CrossRef CAS.
- G. F. Ames, Arch. Biochem. Biophys., 1964, 104, 1–18 CrossRef CAS.
- S. Letoffe, P. Delepelaire and C. Wandersman, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12891–12896 CrossRef CAS.
- H. Nikaido, J. Biol. Chem., 1994, 269, 3905–3908 CAS.
- Y. Zhang, S. Akilesh and D. E. Wilcox, Inorg. Chem., 2000, 39, 3057–3064 CrossRef CAS.
- S. A. Adibi and D. W. Mercer, J. Clin. Invest., 1973, 52, 1586–1594 CrossRef CAS.
- P. Trumbo, A. A. Yates, S. Schlicker and M. Poos, J. Am. Diet. Assoc., 2001, 101, 294–301 CrossRef CAS.
- M. M. Harding and H. A. Long, J. Chem. Soc. A, 1968, 2554–2559 RSC.
- R. H. Kretsinger, F. A. Cotton and R. F. Bryan, Acta Crystallogr., 1963, 16, 651–657 CrossRef CAS.
- P. Deschamps, P. P. Kulkarni and B. Sarkar, Inorg. Chem., 2004, 43, 3338–3340 CrossRef CAS.
- K. de Pina, C. Navarro, L. McWalter, D. H. Boxer, N. C. Price, S. M. Kelly, M. A. Mandrand-Berthelot and L. F. Wu, Eur. J. Biochem., 1995, 227, 857–865 CrossRef CAS.
- C. Sprencel, Z. Cao, Z. Qi, D. C. Scott, M. A. Montague, N. Ivanoff, J. Xu, K. M. Raymond, S. M. Newton and P. E. Klebba, J. Bacteriol., 2000, 182, 5359–5364 CrossRef CAS.
- B. D. Bennett, E. H. Kimball, M. Gao, R. Osterhout, S. J. Van Dien and J. D. Rabinowitz, Nat. Chem. Biol., 2009, 5, 593–599 CrossRef CAS.
- F. C. Neidhardt, J. L. Ingraham and M. Schaechter, Physiology of the Bacterial Cell – A Molecular Approach, Sinauer Associates, Inc., Sunderland, MA, 1990 Search PubMed.
- S. H. Sleigh, P. R. Seavers, A. J. Wilkinson, J. E. Ladbury and J. R. Tame, J. Mol. Biol., 1999, 291, 393–415 CrossRef CAS.
- R. J. Sundberg and R. B. Martin, Chem. Rev., 1974, 74, 471–517 CrossRef CAS.
- K. A. Fraser and M. M. Harding, J. Chem. Soc. A, 1967, 415–420 RSC.
- Y. Li and D. B. Zamble, Chem. Rev., 2009, 109, 4617–4643 CrossRef CAS.
- J. S. Iwig and P. T. Chivers, Nat. Prod. Rep., 2010, 27, 658–667 RSC.
- A. Rodrigue, G. Effantin and M. A. Mandrand-Berthelot, J. Bacteriol., 2005, 187, 2912–2916 CrossRef CAS.
- S. Wang, Y. Wu and F. W. Outten, J. Bacteriol., 2011, 193, 563–574 CrossRef CAS.
- M. Tabata and B. Sarkar, J. Inorg. Biochem., 1992, 45, 93–104 CrossRef CAS.
- J. D. Glennon and B. Sarkar, Biochem. J., 1982, 203, 15–23 CAS.
- R. G. Sawers, S. P. Ballantine and D. H. Boxer, J. Bacteriol., 1985, 164, 1324–1331 CAS.
- M. Schaechter, Microbiol. Mol. Biol. Rev., 2001, 65, 119–130 CrossRef CAS.
- K. Schauer, B. Gouget, M. Carriere, A. Labigne and H. de Reuse, Mol. Microbiol., 2007, 63, 1054–1068 CrossRef CAS.
- J. Stoof, E. J. Kuipers, G. Klaver and A. H. van Vliet, Infect. Immun., 2010, 78, 4261–4267 CrossRef CAS.
- D. A. Rodionov, P. Hebbeln, M. S. Gelfand and T. Eitinger, J. Bacteriol., 2006, 188, 317–327 CrossRef CAS.
- C. L. Dupont, K. Barbeau and B. Palenik, Appl. Environ. Microbiol., 2008, 74, 23–31 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2mt20139a |
| ‡ Current address: Department of Chemistry and Biochemistry, Oberlin College, Oberlin, OH 44074, USA. Fax: +1 400-775-6682; Tel: +1 440-775-8149. |
| § Current address: Department of Molecular and Cellular Biology, University of California, Berkeley, Berkeley, CA 94720, USA. |
| This journal is © The Royal Society of Chemistry 2012 |
