Small molecules DNAmethyltransferasesinhibitors†
Nadine
Martinet
*a,
Benoît Y.
Michel
a,
Philippe
Bertrand
b and
Rachid
Benhida
a
aLaboratoire de Chimie des Molécules Bioactives et des Arômes, UMR 6001 CNRS, Institut de Chimie de Nice, Université de Nice-Sophia Antipolis, Parc Valrose, 06108 Nice Cedex 2, France. E-mail: nadine.martinet@inserm.fr; benoit.michel@unice.fr; rachid.benhida@unice.fr; Fax: (+33)0492076125; Tel: (+33)0492076121
bLaboratory of reactivity and synthesis of natural substances, UMR 6514 Poitiers, France. E-mail: philippe.bertrand@univ-poitiers.fr; Fax: (+33)0549453501; Tel: (+33)0549454192
First published on 21st October 2011
Abstract
Methylation catalyzed by the DNAmethyltransferases affects the C5 position of cytosine residues in DNA. This physiological process is active from the embryo conception, throughout all its developmental steps, and also later for the maintenance of the adult organism. Excess methylated cytosine in tumor suppressor genes is a consistent hallmark of human cancers. However, DNA methylation variation is now acknowledged to significantly contribute to genetic and common diseases. DNAmethyltransferases became attractive therapeutic targets as DNA demethylation, in vitro, brought cancer cell differentiation and apoptosis. Inhibitors are already in use, alone or in combination, to treat myeloid malignancies, while clinical assays are ongoing for other diseases. DNA methylation and histone modifications are intimately correlated with epigenetic heritable modifications of gene expression that are independent of changes in the genetic sequence. Common initiatives for epigenetic research have built public databases with useful resources. The recent discovery of 5-hydroxymethyl cytosine has added new questions and challenges for the epigenome community. We review here knowledge about DNA methylation to provide researchers with the information needed to make more active inhibitors for the benefit of patients. Because of space limitations, many important works cannot be cited. We refer the reader to reviews containing these references.
Introduction
In mammals, methylation occurs at the C5 positions of cytosine (C) residues without interfering with usual base pairing. DNA then becomes more stable and methylated C (5mC) protrudes into the major groove of the DNA, impeding the binding of the transcriptional machinery and thus preventing gene expression. Methylated DNA (mDNA) is further bound by methyl binding proteins (MBD), also preventing the transcriptional machinery from accessing the DNA.mDNA is present in restricted genomic regions and domains but also in each DNA strand, giving a complex pattern of DNA epigenetic information that must be passed to daughter cells. 5mC inhibits or facilitates DNA strand separation, depending on methylation level and sequence context.1 In mammals, CpGdinucleotides, the targets of DNAm (DNA methylation) are disseminated throughout the genome. They are present at a lower than expected abundance because during evolution, spontaneous deamination of Cp into T occurred. However, a high concentration is found in “CpG islands” defined as “200 bp regions of DNA where the CpG content is greater than 60%”. CpG islands are localized in the promoter of approximately 50% of human genes and are unmethylated. In a normal adult cell, disseminated CpG present in repetitive and mobile genomic elements are highly methylated. DNAm is only one contributor to the epigenome that controls the transcriptional machinery, but it still depends upon its cofactor availability and also upon the surrounding chromatin architecture. It remains unclear whether DNAm begins or if it rather follows the locking of DNAtranscription. On the one hand, chromatin repressive “closing” could be completed by a secondary DNAm, but on the other hand, DNAm could be “finished up” by a secondary chromatin closure. DNAm controls gene expression during the development of mammal embryos,2 for parental imprinting,3 for X-chromosome inactivation, which silences at random one X chromosome in females,4 and for genome integrity when repressing viral sequence and mobile elements transcriptions.5 Later patterns of DNAm have been found to diverge influenced by the environment, as seen in human monozygotic twins as they age. Thus, DNAm plays several crucial roles in establishing nuclear totipotency for normal human development and for embryonic stem cell (ESC) biotechnology, for animals cloning, and during acquired epimutations in cancer.6
DNAm was first discovered in calf thymus as a result of the activity of DNAmethyltransferases (DNMTs).7 The first prokaryote DNMT was described before the first eukaryote DNMT.8,9 Several mammalian DNMTs have been identified: DNMT1, DNMT2, DNMT3A and DNMT3B. DNMT1a and DNMT1b are thought to be responsible for most of the maintenance of DNAm as well as the de novoDNAm occurring in the somatic cells of vertebrates (hemi-methylation of CpG islands).8DNMT1a is the most abundant form of enzyme found in adult cells. DNMT2, dispensable for de novoDNAm, can function as a tRNA methyltransferase that methylates cytosine 38 in the anticodon loop.9 Recent data suggest an additional role for DNMT2 during the biogenesis of tRNA-derived small RNAs.10DNMT3L is a catalytically inactive DNMT, which associates with both DNMT3A and DNMT3B (interacting through their C-terminal domains) and enhances their de novomethylation activities by docking them onto the DNA.11 All three establish maternal imprinting mainly during gametogenesis.12 The N-terminal domain of DNMT3L binds also the histone H3 amino tail and recruits histone deacetylases (HDAC) to create a more “closed” chromatin and maintain the established imprints.13
DNMTs cellular biology
DNMT1 is ubiquitous; highly expressed in fetal tissues, heart, kidney, placenta, peripheral blood mononuclear cells, and less expressed in spleen, lung, brain, small intestine, colon, liver, and skeletal muscle. Isoform 2 is two thirds less expressed than isoform 1. The abundance of DNMT1 is reduced to non-detectable levels at the G0 phase of the cell cycle, but DNMT1 is induced upon entrance into the S-phase, when it is found at DNA replication forks to maintain the DNAm pattern in the newly synthesized DNA strand. During G2 and M phases, DNMT1 associates with chromatin to maintain the DNAm pattern. During DNA replication, the methylation pattern of the parent strand is transferred to the newly synthesized strand to convert the hemi-methylated to a fully mDNA. The observed high fidelity of the inheritance of DNMT1methylation patterns relies on the interaction with UHRF1 (ubiquitin-like, containing Plant Homeo Domain (PHD) and Really Interesting New Gene (RING), finger domains 1). The maintenance of DNAm is compromised in UHRF1 deficient cells. UHRF1 binds both DNMT1 and DNMT3A/B and hemi-methylated DNAvia a SET and RING-Associated (SRA) domain to help DNMT1 to methylate the daughter DNA strand (crystal structure of unliganded SRA domain of mouse UHRF1,PDB: 2ZKG).14 The SRA domain is thought to recognize the 5mC (and even the 5-hydroxymethyl cytosine or 5hmC), flip it out from the DNA helix and next help it to return to its initial position and finally move the DNMT to the next C. Thus, the adaptor UHFR1, appears as a new therapeutic target for DNAm inhibition.15DNMTs network and post-translational modifications
DNMT1 interacts with more than 31 proteins, mainly at its N-terminus, but such binding found in vitro may not be relevant in vivo. The N-terminal is a cargo platform for a multitude of proteins (most are listed in Table 1). The interacting molecules are involved in chromatin organization, DNA repair, cell cycle regulation, and apoptosis including RNA polymerase II, RNA-binding proteins, and DNMT1-inhibitory RNA molecules.16 The C-terminus of DNMT1 interacts with the molecular chaperone p23, which disrupts receptor-mediated transcriptional activation and also interacts with the tumor suppressor p53 in complex with HDAC.14 Several post-translational modifications like phosphorylation can change DNMT1’s ability to bind DNA target sites. Catalytically active CDKL5 phosphorylates the N-terminal region of DNMT1 in the presence of DNA.17,18DNMT1RNA is also the target of several miRNAs (Table 1).| Definitions | Names |
|---|---|
| Reference sequences | NM_ 00113823 and NM_ 001379 |
| Chromosomal location | Chr19: 10305774–10305811 |
| Isoforms | P26358-1, P26358-2, P26358-3 |
| DNMT1a | 1616 amino acids, 183![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 165 Da 165 Da |
| Subunit | homodimer localized in the nucleus |
| Interacting proteins | DNMT3a, DNMT3b, UHFR1, PRC2/EED-EZH2,NR1P1,YWHAZ, PCNA, HDAC1, RB1, AHCY, E2F2, MAT2B, AHCYL2, AMD1, TFDP1, E2F1, UBC9, BAZ2A/TIP5, MBD2 and MBD3 |
| In complexes with | DMAP1/HDAC2 and E2F1, BB1, HDAC1/SUV39H1; |
| Post-translational modifications | Methylation: S143, phosphorylation: K142, protein degradation: K142, protein stabilization: S143 |
| MiRNAs for DNMT1 | has-miR152, has-miR-148b, has-mir1826 and has-miR-148a |
Genetics
The loss of DNMT1 is lethal in mice embryos, but yields viable ESC. The DNMT2 embryos display developmental defects including brain and retina abnormalities. DNMT3A-depleted mice appear normal after birth, but die within 1 month. DNMT3B knockout prevents the formation of embryos. The double mouse knockout DNMT3A/B dies shortly after gastrulation. The phenotype of DNMT3L knockout mice is indistinguishable from that of DNMT3Agerm cell-specific conditional knockout mice, as both have lost de novoDNA imprinting in maternal germ cells, and methylation of mobile elements in paternal germ cells.19,20Imprinting
Imprinted genes are expressed in a parent-of-origin manner because DNAm marks silence either the paternal or maternal allele. In mammals, imprinting is achieved through a timely reshaping of the genomic DNAm marks. Parental imprints are first erased in primordial germ cells to be re-established later during gametogenesis. Fertilization triggers a new genome-wide demethylation while pre-implantation induces de novoDNAm that does not involve imprinted regions. In humans, at least 80 genes are imprinted with a roughly equal maternal or paternal expression. Specific imprinting controls regions that display allele- and parent-specific DNAm regulate this partition. Imprinting controls regions are de novo methylated after birth in oocytes but before meiosis in fetal male germ cells. Imprinting tightly regulates genes affecting the fetal-maternal interface and “eliminates” the mutagenic environment of the paternal germline. As shown by nuclear transfer experiments, lack of maternal imprints compromises early embryonic viability while absence of paternal imprints leads to a retarded lethality. In both cases, the development of the placenta is severely altered, and compromises the embryo development.21,22 During genomeDNA demethylation, the methyl group is actively removed from DNA. In the search for the effector, a novel regulatory paradigm able to modulate the epigenetic landscape emerged. The genome-wide methylation is also a C hydroxymethylation of cytosine and is mediated by the Ten-Eleven Translocation 3 enzyme. Indeed, mammalian genomic DNA contains not only 5mC but also 5hmC, which could be an intermediate in the demethylation of DNA. After fertilization, 5mC is converted into 5hmC in the male but not the female pronucleus. Later 5hmC is associated with genomic enhancers.23 The role of 5hmc is not yet fully understood but the understanding of DNA demethylation is progressing.Mis-imprinting syndromes
Instead of a single disease, such syndromes encompass a complex group of multiple genetic and epigenetic defects with severe mental retardation. Two imprinted genomic domains controlling foetal and postnatal growth lay at chromosome11p15. Each is differentially methylated and further regulated by the additional methylation of their own imprinting control region. A loss of DNAm at 11p15 and/or at the imprinted region of 7p13-p11.2 and 7q31-qter is found in over 50% of patients with Russell–Silver syndrome, whereas a gain or a loss is found in 10% of patients with Beckwith–Wiedemann syndrome. Prader–Willi syndrome is a multiform disorder that is caused by the absence of expression of paternally inherited imprinted genes at 15q11-q13. Angelman syndrome is due to a lack of a normal active maternal copy of ubiquitin protein ligase E3A (subject to genomic imprinting). An increase in mis-imprinting diseases has been found in children born via assisted reproductive technology, stressing the implication of environmental factors in the establishment and the maintenance of allele-specific DNAm.24–28 For details see: http://www.ncbi.nlm.nih.gov/omim.Mis-DNA methylation and diseases
Mis-DNAm is involved in several diseases. The Alpha-thalassemia X-linked mental retardation syndrome changes the methylation of ribosomal DNA, Y-specific and subtelomeric repeats. In Fragile X syndrome, a CGG repeat in the fragile X mental retardation 1 gene 5′ untranslated regionn expands and becomes methylated, causing gene silencing. The immunodeficiency, centromeric region instability and facial anomalies syndrome results in a subset of patients bearing germline mutations in DNMT3B, in DNA hypomethylation of DNA satellites II and III with expansion of juxta centromeric heterochromatin, and formation of complex multiradiate chromosomes. Rett syndrome is due to germline mutations of MeCP2, a MBD protein. Defects in cyclin-dependent kinase like 5 gene, that phosphorylates DNMTs, also cause Rett syndrome. In other genetic diseases also with repeat expansions, aberrant methylation of mobile elements is suspected to play a role. Environmental events and maternal care have marked influences on brain function through epigenetic mechanisms, suggesting that behavorial psychiatric disorders may involve epigenetic modifications, and this is raising important ethical issues such as “nature versus nurture?”29–32DNA methylation and cancer
Alterations in genomemethylation patterns are present in most cancers with a global loss of DNAm, and a localized gain of DNAm in CpG islands. During the pre-invasive stages of lung, prostate and colon cancer, abnormal promoter DNAm of several genes has been reported.33 These epimutations are extremely varied from patient to patient and in each tumor type rendering the emergence of specific biomarkers difficult. However, diagnostic tools to search for such epimutations, could refine the detection of occult lung and colon tumors. Increased levels of DNMT1 are seen in human breast and colon cancer cells.34 In leukemic cells, DNMT1 and DNMT3A are recruited by the oncogenic transcription factor promyelocytic leukemia-retinoic acid receptor α oncoprotein to hypermethylategene promoters.35Chromatin architecture is also changed during cancers. Methyl-H3K9 (histine 3, lysine 9) marks are recognized by M-phasephosphoprotein 8 to induce methylation of tumour suppressor gene and UHRF1.36 Overexpression of histonemethyltransferases and DNMT3B/L coincide in cancer development and progression.37 Identification of DNAm is important in the clinical context. Indeed, hypermethylation of O6-methylguanine-DNA methyltransferase (MGMT) predicts temozolomide resistance of glioma and melanoma.38 Likewise, DNAmismatch repairgenes hypermethylation induces microsatellites instability in colon cancers and predicts tumor agressivity.39 Cancer-testis antigengenes like melanoma antigen family A (MAGE), normally expressed in germ cells and trophoblast tissues, are aberrantly expressed in a variety of human malignancies. Most contain CpG-rich regions that are regulated by DNAm. Being highly selective and immunogenic, combined therapy adding DNMT inhibition and cancer-testis antigen vaccination is currently being actively pursued to treat MAGE containing melanoma.40Deleterious global DNA hypomethylation in tumours reactivates oncogenes normally repressed by methylation, promoting further genomic and chromosomal instability when genomic mobile elements, such as retro transposons able to insert themselves into genes to disrupt their expression, are reactivated. Hypomethylation destabilizes the sites of attachment of the cell mitotic spindle in the centromeric heterochromatin and promotes chromosome instability favouring recombination and causing translocations and deletions. The majority of methylation variability in cancer is associated with hypomethylation of intragenic repetitive elements whose methylation status could be predictive of tumor agressivity.41
Inherited tumor predisposition syndromes and DNAm
Various tumors arise in patients with hereditary retinoblastoma, tuberous sclerosis complex and neurofibromatosis. The current hypothesis is that one allele of the involved gene carries an inherited mutation and that a second hit onto the second allele, like DNA hypermethylation, gives rise to the tumors via dysregulation of other genes. As an illustration, DNMTproteins are not expressed in normal retinas, whereas they are overexpressed in retinoblastomas.42DNAm in ESC
ESCs are able to differentiate into any possible specialized cell. De novomethylation activity provided by DNMT3A and B is sufficient for survival of undifferentiated ESC, but ESC proper differentiation and survival requires maintenance of DNAm patterns by DNMT1. The latter is consistent with the idea that differential DNAm patterns in differentiated cells contribute to their specific gene expression programs. Genome-wide analysis using MethylC-Seq identified a new type of DNAm that is selectively seen in undifferentiated ESCs and occurs in CHG and CHH contexts, where H = A, C or T.43 This non-CG methylation encompasses 25% of the total methylome in ESCs.44Measuring DNAm
Identification of aberrantly mDNA can be achieved on a gene per gene basis or via higher throughput varied technologies. Usually, DNA is treated first by sodium bisulfite to convert Cp to uracil while 5mC remains unaffected. The base change is then identified as a new site for a specific restriction enzyme or/and by PCR with its several variants.45,46 PCR amplifies the modified sequence translating 5mC as C and uracil as T. Reduced-representation bisulfite sequencing reduces the portion of the genome analyzed through an MspI initial DNAdigestion and further fragment size selection. Such sequencing achieves single-base resolution but for only a subset of the genome.47 MethylC-seq is a shotgun sequencing of bisulfitedDNA that provides genome-wide single-nucleotide-polymophism (SNP) resolution data but it is expensive like variant pyrosequencing.48 In MeDIP-seq, an anti-C5mantibody is used to immunoprecipitate methylated single-stranded DNA fragments followed by sequencing with a resolution on the order of hundreds of nucleotides. MBD-seq utilizes the MBD2 methyl-CpG binding domain to enrich for methylated double-stranded DNA fragments before sequencing. Comparable methylation calls are obtained when using the selected DNA for further microarray analysis.49 Dedicated microarray technologies have been developed to identify genome-wide aberrant CpG islandmethylation patterns. SNP microarrays for allelic DNAm analysis confines the detection of variants to those present on the array.50 Sequencing-based methods define the methylation status of mobile and repeat elements for which microarrays are not yet available. Deep sequencing of bisulfitedDNA is also feasible. Ultra-performance liquid chromatography with detection by ion trap tandem mass spectrometry is now the preferred tool to define global DNAm status.51 Several surrogate markers are now used to monitor DNMTinhibitors (DNMTi) activity in patients enrolled in clinical trials. Ideally, demethylation of CmG islands and of repeat elements could be analyzed since in murine cell lines repeat elements are easily demethylated, but CmG islands are demethylated only at effective DNMTi doses.525mC chemistry
Methylation at the C5 position of C is mediated by DNMTs. They catalyze the transfer of methyl groups to DNA from S-adenosine-methionine (AdoMet or SAM); producing S-adenosyl-L-homocysteine (AdoHcy or SAH) plus 5mC (Fig. 1). The methyl group of SAM is bound to a sulfonium atom rendering the methylthiol of the methionine moiety very reactive towards nucleophilic attack. From a mechanistic point of view, DNMT is deemed to form a complex with DNA, flipping out the C to methylate from the DNA base-pairing state, and projecting it out into the DNMTcatalytic pocket.53 The thiol group of the cysteine residue (or its thiolate form) in the active site of DNMTs acts as a nucleophile at the C6-position of the C, therefore generating a covalent Michael-like adduct between the enzyme and the DNA (Fig. 2). It was speculated that the attack on the C6-position is assisted by transient protonation of the C ring at the endocyclic nitrogen atomN3, stabilized by the glutamate residue of the enzyme. It is very likely that this protonation activates the nucleophilic addition and increases the reaction rate. The 5-position of C is ready for a nucleophilic attack on the methyl group of the methyl-donating cofactor SAM. A 5-methyl covalent adduct with SAH is formed. The last step consists of a β-elimination giving the 5mC product and the free DNMT, available for another catalytic cycle. The 5mC then flips back into its original position within the DNA to maintain the Watson–Crick base-pairing.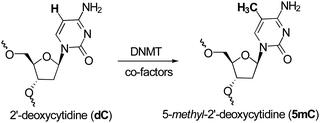 | ||
| Fig. 1 Conversion of dC to 5mC. | ||
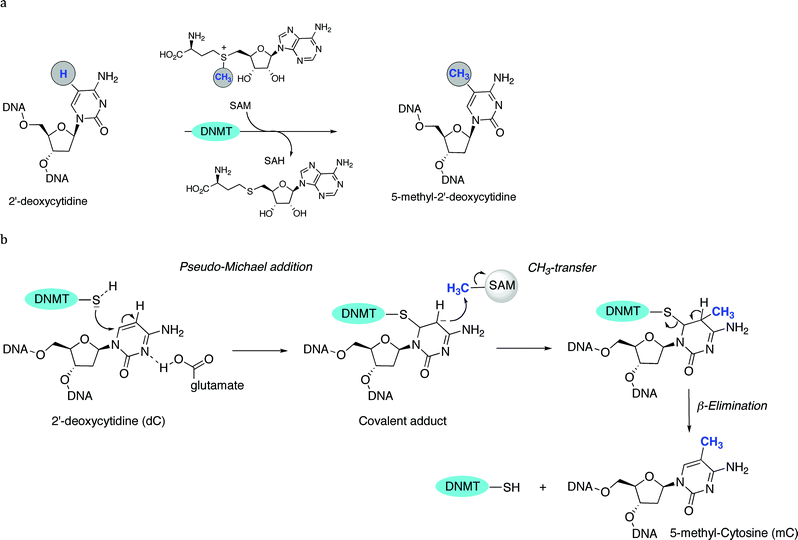 | ||
| Fig. 2 a General mechanism of C5-methylation of 2′-deoxycytidine. b Detailed mechanism (steps 1 to 3): pseudo-Michael addition (1), CH3-transfer (2) and β-elimination (3). | ||
Small molecule DNMTs inhibitors (DNMTi)
Nucleoside analogs
DNMTi are separated into two broad categories: nucleoside and non-nucleoside inhibitors. 5-Azacytidine (5-azaC) and its 2′-deoxy analog (5-azadC) are the first nucleosides to have been identified as DNMTi (Fig. 3). They were synthesized and studied 50 years ago for their classical antiproliferative activity.54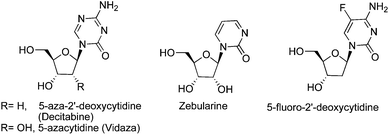 | ||
| Fig. 3 Nucleoside DNMTi. | ||
The DNMT demethylating property of 5-azaC was described in 1980 by Jones and Taylor, who observed that this compound induced differentiation in various biological systems and that its incorporation into genomic DNA inhibited the methylation process.55 The correlation between DNA demethylation and geneactivation led to intense fundamental and clinical “epigenetic” research, and 5-azaC and 5-azadC received approval in 2004 (Vidaza®) and 2006 (Decitabine/Dacogen®), respectively, for the treatment of patients with myelodysplastic syndromes (MDS).56NucleosideDNMTi are first converted into the active triphosphate form by different kinases and then incorporated into DNA and/or RNA. These analogs are thought to have more than one activity besides demethylation of aberrantly silenced genes, like the induction of DNA damage. AzaC is primarily converted to 5-azaC monophosphate (5-azaC-MP) by uridine-cytidine kinase and is incorporated into DNA and then to diphosphate (5-azaC-DP) and triphosphate (5-azaC-TP) by pyrimidine monophosphate and diphosphate kinases, respectively. 5-azaC-TP is incorporated into RNA and interferes with protein translation. By these sequential enzymatic reactions, about 10% of 5-azaC is incorporated into DNA. 5-azaC-DP is converted by ribonucleotide reductase to its 2′-deoxy analog, 5-azadC-DP, and by nucleoside diphosphate kinases to the triphosphate 5-azadC-TP, the active metabolite in DNA synthesis. 5-azadC is activated by deoxycytidine kinase and is only incorporated into DNA to give more efficient inhibition of DNMTs. In contrast, 5-azaC is mainly incorporated into newly synthesized RNA and induced less cytotoxicity when interfering with protein translation. Therefore, the improved therapeutic efficacy of 5-azadC over 5-azaC may be explained by the selective incorporation of 5-azadC into DNA. Because of the nitrogen substitution in the base ring of 5-azaC and 5-azadC, they form, after being incorporated into DNA during replication, a covalent complex with DNMT that cannot be released, since the β-elimination process is hampered (Fig. 4).57 Consequently, the cellular DNMT is depleted, with a concomitant inhibition of DNAm.58
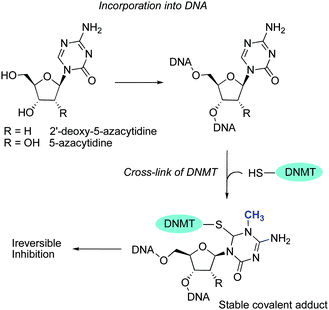 | ||
| Fig. 4 Mechanism of nucleoside DNMTs inhibition. | ||
The cytotoxicity associated with aza-nucleosides creates an increased demand for more effective and selective DNMTi. Zebularine and 5-fluoro-2′-deoxycytidine (5-FdC) are promising nucleoside analogs that mimic reactive intermediates in the mechanism of DNAm (Fig. 3). Zebularine [1-(β-D-ribofuranosyl)-1,2-dihydropyrimidin-2-one] is the first orally active DNMTi, more stable than the parent compound. It has minimal cytotoxicity both in vivo and in vitro. Zebularine was reported to induce selective depletion of DNMT1 over DNMT3A and DMT3B in human cancer cells.59Zebularine was also found to be selectively incorporated into the DNA of tumor cells because of the overexpression of pyrimidine kinases in cancer cells. Developed as a cytidine deaminaseinhibitor, it was shown to form a covalent bond with bacterial methyltransferases at their active sites. Its mechanism of action is similar to the one for aza-analogs and involves the formation of a covalent adduct with DNMTs (Fig. 5) during transcriptionviathiol addition (Cys81)59,60 at the C6 position of the pyrimidinone ring, followed by a concomitant proteasomal pathway-mediated degradation of DNMT1.61,62
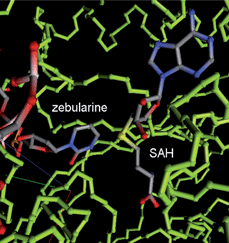 | ||
| Fig. 5 DNMT (PDB code 1MOE) with Zebularine (backbone representation in green sticks). The figure was produced with an in-house PDB file viewer by P. Bertrand. | ||
Zebularine was reported to have poor bioavailability resulting from its metabolism into endogenous inactive compounds. Klecker et al. reported that the metabolism of Zebularine by aldehyde oxidase was the major catabolic pathway in hepatic cytosol, leading to uridine as the primary metabolite, which was then converted to uracil by uridine phosphorylase (Fig. 6).63
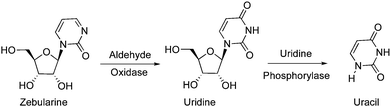 | ||
| Fig. 6 Metabolism of zebularin by aldehyde oxidase. | ||
The metabolic activation of zebularine for DNMT inhibition requires adequate concentration of the active metabolite2′-deoxyzebularine triphosphate (dZTP) following a cascade of complex enzymatic reactions. A quantitative metabolism study of Zebularine in normal and tumor cells was reported by the Kelley and Marquez groups using 14C-labelled zebularine in combination with HPLC, enzymatic and spectroscopic analyses.64 They observed the formation of 5′-mono-, di- and triphosphatemetabolites of zebularine in a dose- and time-dependent manner. In this study, the conversion of zebularine diphosphate (ZDP) to its deoxy analog dZDP by ribonucleotide-diphosphate reductase was shown to be the rate-limiting step that could explain the low potency of zebularine. Accordingly, 2′-deoxyzebularine (dZ) was found to be completely ineffective to overcome this step. One year later, Jones, McGuigan and Marquez described an interesting phosphoramidite approach to bypass the inefficient phosphorylation of dZ and reduction of ZDP to dZDP. Using this strategy, several compounds were identified as more potent DNMTi and stronger inducers of p16 tumor suppresor gene than zebularine (Fig. 7).65
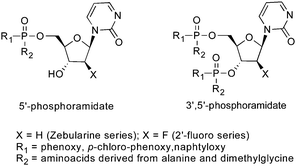 | ||
| Fig. 7 McGuigan phosphoramidite prodrogs of zebularine. | ||
5-Fluoro-2′-deoxycytidine (5-FdC) is another promising DNMTi under clinical investigation in phase I trial for treatment of breast cancer patients (Fig. 3). 5-FdC was reported to inhibit DNAm and induce reexpression of p16 and Ras association domain family member 1 in bladder carcinoma cells and MAGE-1 genes in melanoma cells. Unfortunately, metabolic activation of 5-FdC involves complex enzymatic phosphorylation pathways leading to inactive metabolites. To decrease the in vivo concentration of these unwanted metabolites, 5-FdC could be co-administered with tetrahydrouridine, a cytidine deaminaseinhibitor.
Non-nucleoside analogs
To avoid the toxicity and stability problems associated with nucleosideDNMTi, efforts have focused on the development of non-nucleoside inhibitors. Generally, they inhibit DNAm by binding directly to DNMTs without incorporation into DNA. Some of them are repositioned drugs approved for other indications such as the anti-hypertensive drughydralazine, or the local anesthetic procaine, or even the antiarrhythmic drugprocainamide.RG108 (NSC401077) was first characterized by Brueckner et al. in 2005 and was conceived to interact with the catalytic domain of DNMT1.66 This compound inhibits DNAm and induces the reexpression of different hypermethylation-silenced genes in human cancer cells without affecting the methylation pattern of centromeric satellite sequences.67 Like RG108, the pyridazo-pyridine derivative NSC303530 was discovered by the same group using the NCI chemical compound library and virtual screening based on a 3D model of DNMT1.68Hydralazine is a well-known antihypertensive drug with a DNMTi bioactivity. Recently, Singh et al. reported that hydralazine directly interacts with DNMTvia a complex network of hydrogen bonds with arginine and glutamic acid residues.69 A phase II ongoing clinical study uses hydralazine in combination with standard cytotoxic chemotherapy in order to correlate the reactivation of tumor suppressor genes and DNAm in solid tumors. More than 30 clinical trials are underway using DNMTi alone or in combination with HDACinhibitors, or with chemotherapy (http://clinicaltrials.gov/ct2/results%3Fterm%20%3D%20DNA%2Bmethylation%2Binhibitors).
The 4-amino-benzoic derivatives procaine and procainamide are known as local anesthetic and antiarrhythmic drugs, respectively. Their activity as DNMTi was attributed to their direct binding to CpG-rich sequences. They may act as competitive DNMT1i for its substrates: DNA and SAM.70 Recently, other constrained analogs with higher affinity and selectivity were reported (Fig. 8).71
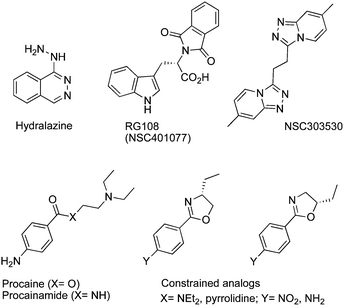 | ||
| Fig. 8 Chemical structure of hydralazine, RG108 and their analogs NSC303530, procaine, procainamide and their constrained analogs. | ||
Several natural products are associated with DNAm inhibition.72 The main polyphenol compound from green tea, epigallocatechin-3-gallate (EGCG), could block the catalytic pocket of DNMT1. However, its catabolism generates hydrogen peroxide that could oxidize DNMTs and other proteins to induce the observed DNA damage and cell death.73 Other tea polyphenols, such as catechin and epicatechin, and the bioflavonoids quercetin, fisetin, and myricetin, have also been implicated in DNAm inhibition. The cathechol containing dietary epicatechin has been suggested to inhibit DNAm indirectly owing to the increased formation of SAH, as a consequence of cathechol-O-methyltransferase-mediated O-methylation of epicatechin.74 Other dietary cathecols with similar mechanism of action are the polyphenols from coffee: caffeic acid and chlorogenic acid.75 Apple polyphenols76 and the major isoflavone from soybean, genistein, and two other isoflavones (biochanin A and daidzein77) have also been reported to have DNA demethylating activity. Their mechanisms of action remain unknown.
Mahanine, a plant-derived carbazolealkaloid, has been reported to inhibit DNMT.78 Psammaplin A and several other disulfide bromotyrosine derivatives isolated from the marine sponge Pseudoceratina purpurea79 among other marine natural products, the peyssonenynes A and B isolated from the red algae Peyssonnelia caulifera,80 and parthenolide, the principal sesquiterpenelactone of feverfew, have been described as DNMT1i (Fig. 9).81Mitoxantrone, an antibiotic, is an inhibitor of topoisomerase II that inhibits DNAm in cell-based assays.82 The antibioticmithramycin A has also been described to inhibit DNAmin vitro.83 More recently, nanaomycin A, a quinineantibiotic, has been described as the first non-SAH analog acting as a DNMT3B-selective inhibitor.84
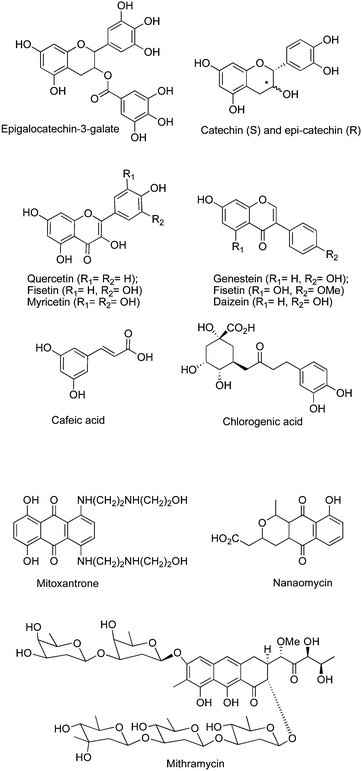 | ||
| Fig. 9 Chemical structure of natural productsDNMTi. | ||
A synthetic quinoline-based compound, SGI-1027, has been described as a DNMT1, DNMT3A and DNMT3Binhibitor, able to degrade the enzymes and compete with SAM.85 SAH analogs presumably binding in the cofactor binding pocket have also been reported as selective DNMTi and putative antimalaria drugs.86 Others have been identified by docking-based virtual screening.87Disulfuram has been shown to be a DNMTi, possibly active on the catalyticcystein.88 Molecular modelling led to several putative DNMTi shown in Fig.10.89,90
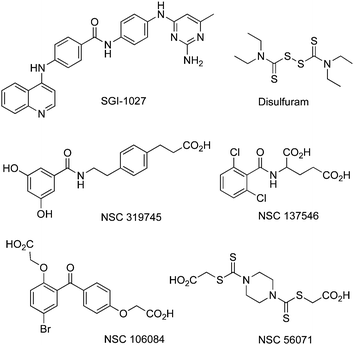 | ||
| Fig. 10 Chemical structures of other DNMTi as all NSC compounds were obtained from the NCI/DTP OpenChemical Repository (http://dtp.cancer.gov). All are for DNMT1 and DNMT3B inhibition, except for NSC319745 which is solely for DNMT1. | ||
DNMTs biochemistry
DNAm depends upon the availability of folate, vitamin B6, vitamin B12 and SAM, which serve as essential cofactors. Maternal deficiency in folate during pregnancy can reduce genomic DNAm. The catabolic clearance of homocysteine to cysteine requires vitamin B6, the deficiency of which results in SAH accumulation, which inhibits DNAm.91All DNMTs have the same conserved catalytic domain (which is highly similar to prokaryotic DNA-(cytosine-5)-methyltransferases with similar subdomains: the binding pocket for the targeted C, the AdoMetcofactor binding domain and the recognition site for the DNA target sequence. DNMT1 has ten aligned conserved sequence motifs: motifs I–III form the cofactorAdoMet binding pocket, motif IV has the catalyticcysteine, motifs VI, VIII and X are for the substrate binding domain and motifs V and VII for the target recognition site.92 In motif VIII, there is a cysteine-rich zinc binding domain (CXXC-type zinc finger) with two zincclusters and a conserved cystein residue involved in DNA binding. Accumulated evidence suggests that the Zn-finger domain could be the allosteric site that leads to inhibition by an excess of unmethylated DNA. The N-terminal part is required for homodimerization (amino acids 310–629) and acts as an allosteric domain. It contains a poly homo bromo domain with a tandem of bromo adjacent homology domains mediating chromatin interactions. This region is also involved in the delivery of the enzyme into the cell nucleus. Connecting the N- and C-termini is a glycine-lysine repeat hinge region. The isolated catalytic domain does not methylateDNA, neither alone nor in combination with other domains.92 The transmission of the methylation pattern of the DNA requires a DNMT with a high preference for methylation of hemi-methylated DNA. Externally added fully mDNA can activate DNMT1 in methylation reactions on unmethylated substrates.93 With fully mDNA, DNMT1 is almost as effective as a maintenance as a de novomethyltransferase. The allosteric activation of DNMT by mDNA could explain the spreading of DNAm seen with aging and cancer.94
Assays for DNMT and models
In vivo, DNA substrates are longer than the short single-site duplexes used for DNMT screening using DNAlabeling with [methyl-3H] AdoMet. They also contain several methylation sites and work with partner proteins. Thus, in vitromethylation of short single-site duplexes does not take into account the processivity of the enzyme able to act several times on the same substrate.95 An ultrasensitive luciferase-linked continuous assay that converts the SAH product of DNAm to a quantifiable luminescent signal has been developed.96 The best system to study such enzymes should be living cells and organisms, where the native substrate and all potential interaction partners are both available. Studies of DNAm rely on several model organisms: arabidopsis,97 yeast.98 fly,99 the popular zebra fish100 and the mouse, especially the agouti phenotype whose color coat depends upon activation of a retrosposon by demethylation.Designing better DNMTi
The major difficulty for designing DNMTi is the lack of available crystallographic data for the human DNMTcatalytic site with inhibitors bound. Recently obtained structures of human DNMT2 and DNMT1 in complex with DNA confirm that unmethylated CpG are occluded from DNMT active site to ensure that only hemi-methylated CG dinucleotides undergo methylation. A summary of available protein structures is shown in Table 2.| DNMT | Species | PDB code | Remarks |
|---|---|---|---|
| a Resources for epigenetic research are online and are listed non-exhaustively in Table 3. | |||
| DNMT | Haemophilus Haemolyticus | 1M0E 101 | With DNA and zebularine |
| DNMT1 | Mouse | 3AV6 102 | With Adomet |
| 3AV5 102 | With Adohcy | ||
| 3AV4,102 | (731–1602) free state | ||
| 3PT9 103 | (650–1602) with DNA | ||
| 3PT6 103 | |||
| DNMT1 | Human | 3EPZ 104 | replication foci-targeting sequence |
| 3PTA 103 | (646–1600) with DNA | ||
| 3OS5 101 | with Set79 and K142me1 | ||
| DNMT2 | Human | 1G55 105 | |
| DNMT3 | B, mouse | 1KHC 106 | PWWP domain |
| 3QKJ 107 | PWWP domain | ||
| DNMT3 | A–B, human | 3LLR (SGC) | PWWP domain |
| 3A1A 108 | Add domain | ||
| 3A1B 108 | Complex with H3 | ||
| DNMT3 | A–L, human | 2QRV 108 | C-terminal domain |
| DNMT3 | L, human | 2PV0 109 | DNMT3L like protein with |
| 2PVC 109 | unmethylated H3K4 |
Perspectives
Although more than 20 years have elapsed since mDNA discovery, we are just beginning to envision the process of DNA demethylation. We feel that human genetic diseases cell lines which are available via the orphanet network could be useful models to understand the bioactivity of DNMTi and maybe to offer the donor patients a hope for treatment.| Definitions | Addresses |
|---|---|
| Epigenome studies | http://www.genome.gov/gwastudies |
| The Biomarkers Consortium | http://www.biomarkersconsortium.org/ |
| NIH′s Roadmap Epigenomics Program | http://www.roadmapepigenomics.org |
| Epigenome Network of Excellence | http://www.epigenome-noe.net/WWW/index.php |
| The SMARTER initiative (drugs for epigenetic) | http://www.smarter-chromatin.eu/ |
| The EPITRON initiative (cancerology) | http://www.epitron.eu/ |
| The HEROIC program (mouse epigenome) | http://www.heroic-ip.eu/ |
| The Australian Alliance for Epigenetics | http://www.epialliance.org.au/ |
| National Center for Biotechnology Information | http://www.ncbi.nlm.nih.gov/epigenomics |
| International Cancer Genome Consortium | http://www.icgc.org/ |
| Epigenome (human diseases and environmental effects) | http://nihroadmap.%20nih.gov/epigenomics/fundedresearch.asp |
| The International Human Epigenome Consortium | http://ihec-epigenomes.org/ |
| Orphanet network | http://www.orpha.net/consor/cgi-bin/index.php%3Flng%20%3D%20FR |
| Atlas of genetic diseases | http://www.ncbi.nlm.nih.gov/omim |
| NCI clinical trials | http://clinicaltrials.gov/ct2/results%3Fterm%20%3D%20DNA%2Bmethylation%2Binhibitors |
| NCI/DTP Open Chemical Repository | http://dtp.cancer.gov/ |
Acknowledgements
COST TD 905 “Epigenetic from bench to bedside” and Agence Nationale pour la Recherche.Reference
- P. M. Severin, X. Zou, H. E. Gaub and K. Schulten, Nucleic Acids Res., 2011, 39, 13–18.
- M. Jeanblanc, J. Salvaing, K. Mason, P. Debey and N. Beaujean, Gynecol., Obstet. Fertil., 2008, 36, 1126–1132 Search PubMed.
- A. C. Ferguson-Smith, Nat. Rev. Genet., 2011, 12, 565–575 CrossRef CAS.
- C. Morey and P. Avner, Ann. N. Y. Acad. Sci., 2010, 1214, 18–33 CrossRef.
- T. A. Rauch, X. Zhong, X. Wu, M. Wang, K. H. Kernstine, Z. Wang, A. D. Riggs and G. P. Pfeifer, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 252–257 CrossRef CAS.
- W. Reik, W. Dean and J. Walter, Science, 2001, 293, 1089–1093 CrossRef CAS.
- P. H. Roy and A. Weissbach, Nucleic Acids Res., 1975, 2, 1669–1684 CrossRef CAS.
- T. H. Bestor, Genetics, 1988, 74, 9–12 CAS.
- M. G. Goll, F. Kirpekar, K. A. Maggert, J. A. Yoder, C. L. Hsieh, X. Zhang, K. G. Golic, S. E. Jacobsen and T. H. Bestor, Science, 2006, 311, 395–398 CrossRef CAS.
- M. Schaefer, T. Pollex, K. Hanna, F. Tuorto, M. Meusburger, M. Helm and F. Lyko, Genes Dev., 2010, 24, 1590–1595 CrossRef CAS.
- A. Jeltsch, ChemBioChem, 2011, 12, 206–222 CrossRef CAS.
- J. R. Weaver, M. Susiarjo and M. S. Bartolomei, Mamm. Genome, 2009, 20, 532–543 CrossRef.
- F. Chédin, Prog. Mol. Biol. Transl. Sci., 2011, 101, 255–285 Search PubMed.
- C. Bronner, Sci. Signaling, 2011, 4, e3.
- C. Frauer, T. Hoffmann, S. Bultmann, V. Casa, M. C. Cardoso, I. Antes and H. Leonhardt, PLoS One, 2011, 6, e21306 CrossRef CAS.
- Ž. M. Svedružić, Prog. Mol. Biol. Transl. Sci., 2011, 101, 221–254 Search PubMed.
- X. Cheng and R. M. Blumenthal, Structure, 2008, 16, 341–350 CrossRef CAS.
- K. T. Rigbolt, T. A. Prokhorova, V. Akimov, J. Henningsen, P. T. Johansen, I. Kratchmarova, M. Kassem, M. Mann, J. V. Olsen and B. Blagoev, Sci. Signaling, 2011, 4, e3.
- A. R. Karpf and S. Matsui, Cancer Res., 2005, 65, 8635–8639 CrossRef CAS.
- H. W. Lim, M. Iwatani, N. Hattori, S. Tanaka, S. Yagi and K. Shiota, J. Reprod. Dev., 2010, 56, 86–93 CrossRef CAS.
- J. R. Weaver, M. Susiarjo and M. S. Bartolomei, Mamm. Genome, 2009, 20, 532–543 CrossRef.
- B. Novakovic, N. C. Wong, M. Sibson, H. K. Ng, R. Morley, U. Manuelpillai, T. Down, V. K. Rakyan, S. Beck, S. Hiendleder, C. T. Roberts, J. M. Craig and R. Saffery, J. Biol. Chem., 2010, 285, 9583–9593 CrossRef CAS.
- M. Münzel, D. Globisch and T. Carell, Angew. Chem., Int. Ed., 2011, 50, 6460–6468 CrossRef.
- P. Arnaud and R. Feil, Birth Defects Res., Part C, 2005, 75, 81–97 Search PubMed.
- R. Hirasawa and R. Feil, Essays Biochem., 2010, 48, 187–200 CrossRef CAS.
- J. Demars, M. E. Shmela, S. Rossignol, J. Okabe, I. Netchine, S. Azzi, S. Cabrol, C. Le Caignec, A. David, Y. Le Bouc, A. El-Osta and C. Gicquel, Hum. Mol. Genet., 2010, 19, 803–814 CrossRef CAS.
- J. Walter and M. Paulsen, Semin. Cell Dev. Biol., 2003, 14, 101–110 CrossRef CAS.
- K. M. Cornish, K. M. Gray and N. J. Rinehart, Adv. Child Dev. Behav., 2010, 39, 211–235 Search PubMed.
- J. L. Calles, Pediatr. Clin. North Am., 2011, 58, 189–203 CrossRef.
- M. P. Hitchins, Adv. Genet., 2010, 70, 201–243 Search PubMed.
- H. van Bokhoven and J. M. Kramer, Neurobiol. Dis., 2010, 39, 3–12 CrossRef CAS.
- J. I. Martín-Subero and M. Esteller, Adv. Exp. Med. Biol., 2011, 711, 162–177 CrossRef.
- S. Khandige, V. V. Shanbhogue, S. Chakrabarty and S. Kapettu, Oncol. Res., 2011, 19, 105–110 CrossRef.
- W. S. el-Deiry, B. D. Nelkin, P. Celano, R. W. Yen, J. P. Falco, S. R. Hamilton and S. B. Baylin, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 3470–3474 CrossRef CAS.
- L. A. Godley, J. Cunningham, M. E. Dolan, R. S. Huang, S. Gurbuxani, M. E. McNerney, R. A. Larson, H. Leong, Y. Lussier, K. Onel, O. Odenike, W. Stock, K. P. White and M. M. Le Beau, Semin. Oncol., 2011, 38, 215–224 CrossRef CAS.
- K. Kokura, L. Sun, M. T. Bedford and J. Fang, EMBO J., 2010, 29, 3673–3687 CrossRef CAS.
- M. Y. Kang, B. B. Lee, Y. H. Kim, D. K. Chang, S. Kyu Park, H. K. Chun, S. Y. Song, J. Park and D. H. Kim, Int. J. Cancer, 2007, 121, 2192–2197 CrossRef CAS.
- E. M. Goellner, B. Grimme, A. R. Brown, Y. C. Lin, X. H. Wang, K. F. Sugrue, L. Mitchell, R. N. Trivedi, J. B. Tang and R. W. Sobol, Cancer Res., 2011, 71, 2308–2317 CrossRef CAS.
- M. Agostini, M. V. Enzo, L. Morandi, C. Bedin, S. Pizzini, S. Mason, R. Bertorelle, E. Urso, C. Mescoli, M. Lise, S. Pucciarelli and D. Nitti, Cancer Biomark., 2010, 6, 49–61 Search PubMed.
- M. F. Gjerstorff, J. Burns and H. J. Ditzel, Expert Opin. Biol. Ther., 2010, 10, 1061–1075 Search PubMed.
- L. Sigalotti, E. Fratta, E. Bidoli, A. Covre, G. Parisi, F. Colizzi, S. Coral, S. Massarut, J. M. Kirkwood and M. Maio, J. Transl. Med., 2011, 9, 78–88 CrossRef CAS.
- Y. Qu, G. Mu, Y. Wu, X. Dai, F. Zhou, Y. Wang and F. Wei, Am. J. Clin. Pathol., 2010, 134, 826–834 CrossRef CAS.
- A. Meissner, Nat. Biotechnol., 2010, 28, 1079–1088 CrossRef CAS.
- Y. Yamada and A. Watanabe, Adv. Genet., 2010, 70, 177–199 Search PubMed.
- V. Beier, C. Mund and J. D. Hoheisel, Adv. Biochem. Eng. Biotechnol., 2007, 104, 1–11 CAS.
- Y. Y. Degenhardt, R. Wooster, R. W. McCombie, R. Lucito and S. Powers, Curr. Opin. Genet. Dev., 2008, 18, 1868–1872.
- L. Wang, J. Sun, H. Wu, S. Liu, J. Wang, B. Wu, S. Huang, N. Li, J. Wang and X. Zhang DOI:10.1016/j.jbiotec.2011.06.034.
- J. R. Ecker, W. Li, P. J. Farnham, R. A. Waterland, A. Meissner, M. A. Marra, M. Hirst, A. Milosavljevic and J. F. Costello, Nat. Biotechnol., 2010, 28, 1097–1105 CrossRef.
- N. Pälmke, D. Santacruz and J. Walter, Methods, 2011, 53, 175–184 CrossRef.
- Y. Wang, J. Hayakawa, F. Long, Q. Yu, A. H. Cho, G. Rondeau, J. Welsh, S. Mittal, I. De Belle, E. Adamson, M. McClelland and D. Mercola, Ann. N. Y. Acad. Sci., 2005, 1058, 162–185 CrossRef CAS.
- J. J. Zhang, L. Zhang, K. Zhou, X. Ye, C. Liu, L. Zhang, J. Kang and C. Cai, Anal. Biochem., 2011, 413, 164–170 CrossRef CAS.
- H. W. Lim, M. Iwatani, N. Hattori, S. Tanaka, S. Yagi and K. Shiota, J. Reprod. Dev., 2010, 56, 86–93 CrossRef CAS.
- G. C. Burdge and K. A. Lillycrop, Annu. Rev. Nutr., 2010, 30, 315–339 CrossRef CAS.
- (a) F. Sorm, A. Piskala, A. Cihak and J. Vesely, Experientia, 1964, 20, 85–93 CrossRef; (b) A. Cihák, Oncology, 1974, 30, 505–522.
- P. A. Jones and S. M. Taylor, Cell, 1980, 20, 85–93 CrossRef CAS.
- (a) E. Kaminskas, A. Farrell, S. Abraham, A. Baird, L. S. Hsieh, S. L. Lee, J. K. Leighton, H. Patel, A. Rahman, R. Sridhara, Y. C. Wang and R. Pazdur, Clin. Cancer Res., 2005, 11, 3604–3608 CrossRef CAS; (b) H. Kantarjian, J. P. Issa, C. S. Rosenfeld, J. M. Bennett, M. Albitar, J. DiPersio, V. Klimkev, J. Slack, C. De Castro, F. Ravandi, R. Helmer, L. Shen, S. D. Nimer, R. Leavitt, A. Raza and H. Saba, Cancer, 2006, 106, 1794–803 CrossRef CAS.
- H. W. Lim, M. Iwatani, N. Hattori, S. Tanaka, S. Yagi and K. Shiota, J. Reprod. Dev., 2010, 56, 86–93 CrossRef CAS.
- T. Robak, A. Szmigielska-Kapłon, A. Pluta, O. Grzybowska-Izydorczyk, A. Wolska, M. Czemerska and A. Wierzbowska, Curr. Med. Chem., 2011, 18, 638–66 CrossRef CAS.
- C. Champion, D. Guianvarc'h, S. Sénamaud-Beaufort, R. Z. Jurkowska, A. Jeltsch, L. Ponger, P. B. Arimondo and A. L. Guieysse-Peugeot, PLoS One, 2010, 5, e12388 CrossRef.
- J. C. Cheng, C. B. Matsen, F. A. Gonzales, W. Ye and S. Greer, J. Natl. Cancer Inst., 2003, 95, 399–409 CrossRef CAS.
- M. Suzuki, F. Shinohara, M. Endo, M. Sugazaki and S. Echigo, Cancer Chemother. Pharmacol., 2009, 64, 223–232 CrossRef CAS.
- D. M. van Bemmel, A. S. Brank, R. Eritja, V. E. Marquez and J. K. Christman, Biochem. Pharmacol., 2009, 78, 633–641 CrossRef CAS.
- R. W. Klecker, R. L. Cysyk and J. M. Collins, Bioorg. Med. Chem., 2006, 14, 62–66 CrossRef CAS.
- T. Ben-Kasus, Z. Ben-Zvi, V. E. Marquez, J. A. Kelley and R. Agbaria, Biochem. Pharmacol., 2005, 70, 121–133 CrossRef CAS.
- C. B. Yoo, R. Valente, C. Congiatu, F. Gavazza, A. Angel, M. A. Siddiqui, P. A. Jones, C. McGuigan and V. E. Marquez, J. Med. Chem., 2008, 51, 7593–7601 CrossRef CAS.
- B. Brueckner, B. R. Garcia, P. Siedlecki, T. Musch, H. C. Kliem, P. Zielenkiewicz, S. Suhai, M. Wiessler and F. Lyko, Cancer Res., 2005, 65, 6305–6311 CrossRef CAS.
- A. F. Holloway and P. C. Oakford, Curr. Med. Chem., 2007, 14, 2540–7 CrossRef CAS.
- P. Siedlecki, R. G. Boy, T. Musch, B. Brueckner, S. Suhai, F. Lyko and P. Zielenkiewicz, J. Med. Chem., 2006, 49, 678–683 CrossRef CAS.
- N. Singh, A. Dueñas-González, F. Lyko and J. L. Medina-Franco, ChemMedChem, 2009, 4, 792–9 CrossRef CAS.
- M. Szyf, Drug Resist. Updates, 2003, 6, 6341–6353.
- S. Castellano, D. Kuck, M. Sala, E. Novellino, F. Lyko and G. Sbardella, J. Med. Chem., 2008, 51, 2321–2325 CrossRef CAS.
- F.-F. Cai, C. Kohler, B. Zhang, M.-H. Wang, W.-J. Chen and X.-Y. Zhong, Int. J. Mol. Sci., 2011, 12, 4465–4476 Search PubMed.
- M. Z. Fang, Y. Wang, N. Ai, Z. Hou, Y. Sun, H. Lu, W. Welsh and C. S. Yang, Cancer Res., 2003, 63, 7563–7570 CAS.
- W. J. Lee and B. T. Zhu, Carcinogenesis, 2006, 27, 269–277 CrossRef CAS.
- L. Fini, M. Selgrad, V. Fogliano, G. Graziani, M. Romano, E. Hotchkiss, Y. A. Daoud, E. B. De Vol, C. R. Boland and L. Ricciardiello, J. Nutr., 2007, 137, 2622–2628 CAS.
- A. Hsu, T. M. Bray, W. G. Helferich, D. R. Doerge and E. Ho, Exp. Biol. Med., 2010, 235, 90–97 Search PubMed.
- M. Z. Fang, D. Chen, Y. Sun, Z. Jin, J. K. Christman and C. S. Yang, Clin. Cancer Res., 2005, 11, 7033–7041 CrossRef CAS.
- A. Link, F. Balaguer and A. Goel, Biochem. Pharmacol., 2010, 80, 1771–1792 CrossRef CAS.
- A. M. Godert, N. Angelino, A. Woloszynska-Read, S. R. Morey, S. R. James, A. R. Karpf and J. R. Sufrin, Bioorg. Med. Chem. Lett., 2006, 16, 3330–3333 CrossRef CAS.
- K. L. McPhail, D. France, S. Cornell-Kennon and W. H. Gerwick, J. Nat. Prod., 2004, 67, 1010–1013 CrossRef CAS.
- Z. Liu, S. Liu, Z. Xie, R. E. Pavlovicz, J. Wu, P. Chen, J. Aimiuwu, J. Pang, D. Bhasin, P. Neviani, J. R. Fuchs, C. Plass, P. K. Li, C. Li, T. H. Huang, L. C. Wu, L. Rush, H. Wang, D. Perrotti, G. Marcucci and K. K. Chan, J. Pharmacol. Exp. Ther., 2009, 329, 505–514 CrossRef CAS.
- B. J. Evison, F. Chiu, G. Pezzoni, D. R. Phillips and S. M. Cutts, Mol. Pharmacol., 2008, 74, 184–194 CrossRef CAS . Epub 2008 Apr 15.
- R. K. Lin, C. H. Hsu and Y. Wang, Anti-Cancer Drugs, 2007, 18, 1157–1164 CrossRef CAS.
- D. Kuck, T. Caulfield, F. Lyko and J. L. Medina-Franco, Mol. Cancer Ther., 2010, 9, 3015–3023 CrossRef CAS.
- J. Datta, K. Ghoshal, W. A. Denny, S. A. Gamage, D. G. Brooke, P. Phiasivongsa, S. Redkar and S. Jacob, Cancer Res., 2009, 69, 4277–4285 CrossRef CAS.
- D. Wyczechowska and K. Fabianowska-Majewska, Biochem. Pharmacol., 2003, 65, 219–225 CrossRef CAS.
- J. M. Bujnicki, S. T. Prigge, D. Caridha and P. K. Chiang, Proteins: Struct., Funct., Genet., 2003, 52, 624–632 CrossRef CAS.
- J. Lin, M. C. Haffner, Y. Zhang, B. H. Lee, W. N. Brennen, J. Britton, S. K. Kachhap, J. S. Shim, J. O. Liu, W. G. Nelson, S. Yegnasubramanian and M. A. Carducci, Prostate, 2011, 71, 333–343 CrossRef CAS.
- D. Kuck, N. Singh, F. Lyko and J. L. Medina-Franco, Bioorg. Med. Chem., 2010, 8, 22–29.
- K. C. Kim, S. Friso and S. W. Choi, J. Nutr. Biochem., 2009, 20, 917–926 CrossRef CAS.
- A. Hermann, H. Gowher and A. Jeltsch, Cell. Mol. Life Sci., 2004, 61, 2571–2587 CrossRef CAS.
- R. Z. Jurkowska, T. P. Jurkowski and A. Jeltsch, ChemBioChem, 2011, 12, 206–222 CrossRef CAS.
- X. Cheng and R. M. Blumenthal, Structure, 2008, 16, 341–350 CrossRef CAS.
- Ž. M. Svedružić, Prog. Mol. Biol. Transl. Sci., 2011, 101, 221–254 Search PubMed.
- I. Hemeon, J. A. Gutierrez, M. C. Ho and V. L. Schramm, Anal. Chem., 2011, 83, 4996–5004 CrossRef CAS.
- C. Mund, T. Musch, M. Strödicke, B. Assmann, E. Li and F. Lyko, Biochem. J., 2004, 378, 763–768 CrossRef CAS.
- M. Rea, M. Chen, S. Luan, D. Bhangu, M. Braud and W. Xiao, J. Vis. Exp., 2011, 47, 2327–2337 Search PubMed.
- I. Olsson and P. Bjerling, Curr. Genet., 2011, 57, 1–12 CrossRef CAS.
- R. Lyko, C. Beisel, J. Marhold and R. Paro, Curr. Top. Microbiol. Immunol., 2006, 310, 23–44 Search PubMed.
- A. Dong, J. A. Yoder, X. Zhang, L. Zhou, T. H. Bestor and X. Cheng, Nucleic Acids Res., 2001, 29, 439–448 CrossRef CAS.
- P. O. Esteve, Y. Chang, M. Samaranayake, A. K. Upadhyay, J. R. Horton, G. R. Feehery, X. Cheng and S. Pradhan, Nat. Struct. Mol. Biol., 2011, 18, 42–48 CrossRef CAS.
- K. Takeshita, I. Suetake, E. Yamashita, M. Suga, H. Narita, A. Nakagawa and S. Tajima, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9055–9059 CrossRef CAS.
- J. Song, O. Rechkoblit, T. H. Bestor and D. J. Patel, Science, 2011, 331, 1036–1040 CrossRef CAS.
- C. Qiu, K. Sawada, X. Zhang and X. Cheng, Nat. Struct. Biol., 2002, 9, 217–224 CAS.
- Y. Wang, K. J. Addess, J. Chen, L. Y. Geer, J. He, S. He, S. Lu, T. Madej, A. Marchler-Bauer, P. A. Thiessen, N. Zhang and S. H. Bryant, Nucleic Acids Res., 2007, 35, 298–300.
- D. Jia, R. Z. Jurkowska, X. Zhang, A. Jeltsch and X. Cheng, Nature, 2007, 449, 248–251 CrossRef CAS.
- H. Wu, H. Zeng, R. Lam, W. Tempel, M. F. Amaya, C. Xu, L. Dombrovski, W. Qiu, Y. Wang and J. Min, PlosOne., 2011, 6, E1891.
- J. Otani, T. Nankumo, K. Arita, S. Inamoto, M. Ariyoshi and M. Shirakawa, EMBO Rep., 2009, 10, 1235–1241 CrossRef CAS.
- S. K. Ooi, C. Qiu, E. Bernstein, K. Li, D. Jia, Z. Yang, H. Erdjument-Bromage, P. Tempst, S. P. Lin, C. D. Allis, X. Cheng and T. H. Bestor, Nature, 2007, 448, 714–717 CrossRef CAS.
Footnote |
| † This article is part of a MedChemComm web theme issue on epigenetics. |
| This journal is © The Royal Society of Chemistry 2012 |
