Engineers are from PDMS-land, Biologists are from Polystyrenia
Erwin
Berthier†
a,
Edmond W. K.
Young†
b and
David
Beebe
*b
aDepartment of Medical Microbiology, University of Wisconsin-Madison, Madison, WI, USA
bDepartment of Biomedical Engineering, Wisconsin Institutes for Medical Research, University of Wisconsin-Madison, 1111 Highland Avenue, Room 6009, Madison, WI, USA. E-mail: djbeebe@wisc.edu; Tel: +1-608-262-2260
First published on 8th February 2012
Abstract
As the integration of microfluidics into cell biology research proceeds at an ever-increasing pace, a critical question for those working at the interface of both disciplines is which device material to use for a given application. While PDMS and soft lithography methods offer the engineer rapid prototyping capabilities, PDMS as a material has characteristics that have known adverse effects on cell-based experiments. In contrast, while polystyrene (PS), the most commonly used thermoplastic for laboratory cultureware, has provided decades of grounded and validated research conclusions in cell behavior and function, PS as a material has posed significant challenges in microfabrication. These competing issues have forced microfluidics engineers and biologists to make compromises in how they approach specific research questions, and furthermore, have attenuated the impact of microfluidics on biological research. In this review, we provide a comparison of the attributes of PDMS and PS, and discuss reasons for their popularity in their respective fields. We provide a critical evaluation of the strengths and limitations of PDMS and PS in relation to the advancement and future impact on microfluidic cell-based studies and applications. We believe that engineers have a responsibility to overcome any challenges associated with microfabrication, whether with PS or other materials, and that engineers should provide options and solutions that assist biologists in their experimental design. Our goal is not to advocate for any specific material, but provide guidelines for researchers who desire to choose the most suitable material for their application, and suggest important research directions for engineers working at the interface between microfabrication technology and biological application.
1. Introduction
The development of microscale systems for both fundamental and applied research in cell biology has experienced significant growth and advancement in recent years.1–3 The rapid progress in this area can be partially attributed to increased functionality combined with the small scale of these systems, which offers the advantage of lower volumes, reduced consumption of reagents and potential for increased throughput. For cell-based applications specifically, microscale systems offer a physical scale well suited for studying cells in spatially and temporally controlled microenvironments.4–7 This has importantly led to new advanced methods for probing and understanding cells and their properties, including cell morphology and motility, biomechanical forces, and complex biophysical and biochemical cell-cell interactions (Fig. 1).8–12 In particular, the size of microscale systems are naturally suited for studying cells of limited availability, such as from primary patient samples.13–15 Microscale systems have thus provided new insights into cell behavior and function, and have enabled us to tackle complex biological questions that were once inconceivable with conventional platforms.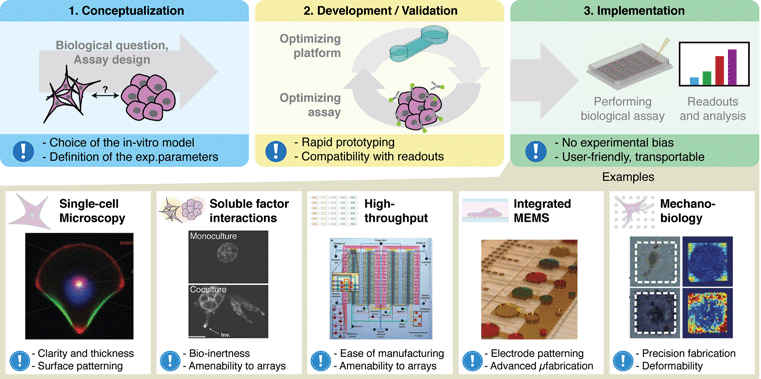 | ||
| Fig. 1 Integration of microscale technologies and cell biology requires collaboration between biologists and engineers from conceptualization to development and validation and finally to implementation. When properly implemented, state-of-the-art microfluidic cell-based systems can drastically enhance our ability to examine the behavior and function of cells. Exclamation mark icon represents key considerations for each section. Examples of these systems have enabled the observation of intracellular organization (reprinted from ref. 8 with permission of the National Academy of Science), the study of multi-cellular soluble factor communication (reproduced in part from ref. 9 with permission of the Royal Society of Chemistry), the creation of high-throughput microfluidic systems (from ref. 10; reprinted with permission from AAAS), the creation of integrated droplet based cell-culture systems (reproduced in part from ref. 11 with permission of the Royal Society of Chemistry) and the measure of mechanical forces exerted by cells (reprinted by permission from Macmillan Publishers Ltd: Nature Methods ref. 12, copyright (2010)). | ||
The current pace of research in the development and application of microscale systems for cell biology has also been facilitated by an increasing number of collaborations between engineers and biologists that are vital to the scientific research enterprise. The interdisciplinary nature of this field necessitates collaborative efforts to advance ideas from concept to assay development and validation, and ultimately to implementation and application (Fig. 1). While many of these collaborations have been successful in advancing science and technology, microscale systems, to a large extent, have not penetrated the cell biology research community, and have yet to become widely accepted platforms for studying cell biology. This is partly attributable to the fact that communication between biologists and engineers continues to be somewhat hindered by subtle yet critical differences between their respective research cultures. The natural tendency of researchers to choose conventional and proven methods rather than novel and potentially more effective methods, which may contain a degree of uncertainty, amplifies this divide.
In the case of microfluidic cell-based systems, this “clash of cultures” has occurred between biologists and engineers over material selection for microdevice fabrication. Microfluidics engineers have become accustomed to using polydimethylsiloxane (PDMS) for rapid prototyping of designs and for fabrication of microdevices because of its many attractive properties over other materials, including its low cost, ease of use, and high compliance. However, a growing number of reports have begun to increase awareness of potential spurious effects associated with culturing and studying cells in PDMS microdevices.16,17 In contrast, biologists have relied heavily in the past fifty years on platforms such as Petri dishes, culture flasks, and microtiter well plates made mostly from polystyrene (PS). The majority of research data collected on biological cells in vitro has been based on cell behavior on PS surfaces. Thus, from the perspective of a biologist, microscale systems would be more attractive if devices were available in more popular and widely used laboratory materials such as PS. Unfortunately, microfabrication in PS and other plastics is considerably more challenging compared to microfabrication in PDMS, limiting the availability of plastic-based microscale systems to those with access to dedicated equipment and expertise.
This contrast in research cultures regarding material selection raises a number of important questions that need to be addressed to further enhance collaborations between engineers and biologists and allow microfluidics research to continue at or above the current pace of discovery. How significant are the adverse effects of PDMS on microscale cell biology studies? What current challenges do engineers face that prevent or limit them from fabricating more in other materials, particularly PS, and how do we overcome these challenges? Importantly, how will increased availability and accessibility of PS microdevices further enhance the integration between microfluidics technology and cell biology?
In this review, we address these questions by directly comparing PDMS and PS in the context of developing and fabricating microscale cell-based systems, specifically for biological studies. The goal is to enhance the collaborative relationships between engineers and biologists, and further accelerate integration between microfluidics and cell biology. First, we provide a historical perspective on the independent developments of cell culture and microfluidics over the past century, and review the recent efforts to integrate these fields for the benefit of interdisciplinary research. Such a historical account will partially help to explain the distinct contrast in research cultures between the respective fields, and potentially help guide future research directions. Second, we discuss the major issues associated with PDMS for microfluidic cell-based systems, their significance, and methods to circumvent these issues. We then review PS fabrication methods and highlight recent advances that may help to alleviate bottlenecks in the process workflow for rapid prototyping in PS. Finally, we address questions raised by both engineers and biologists on the issue of material selection, giving consideration to their respective needs. This review, which discusses topics ranging from microfabrication to cell culture, is intended for integrative bioengineers who aim to create tools that can be translated to biology laboratories. The goal of this review is not to advocate for PDMS, PS, or any other material, but instead to stimulate interest and further dialogue in this important topic of material selection, and allow researchers from the two sides to overcome differences in research culture and come to a mutual understanding. Ultimately, bringing this topic to the forefront will further promote collaborations that will lead to continued progress in research and development of microfluidic cell-based technologies.
2. Historical perspective
The use of PS for in vitro biology and PDMS for microfluidics happened through a series of historic events that provided the right environment for their emergence. To understand how PS and PDMS became the materials of choice in the respective areas of cell culture and microfluidics, it is informative to first examine these landmark events that have helped shape the separate fields. Doing so provides us with the necessary background to understand why these materials became popular and which properties of the material contributed to their rise in popularity. Here we provide a brief historical account of the major scientific events over the last century (Fig. 2), and offer insight into how they shaped the research cultures of each field. This insight will hopefully instruct us on how to address necessary compromises in material selection when working at the interface between microfluidics and cell biology.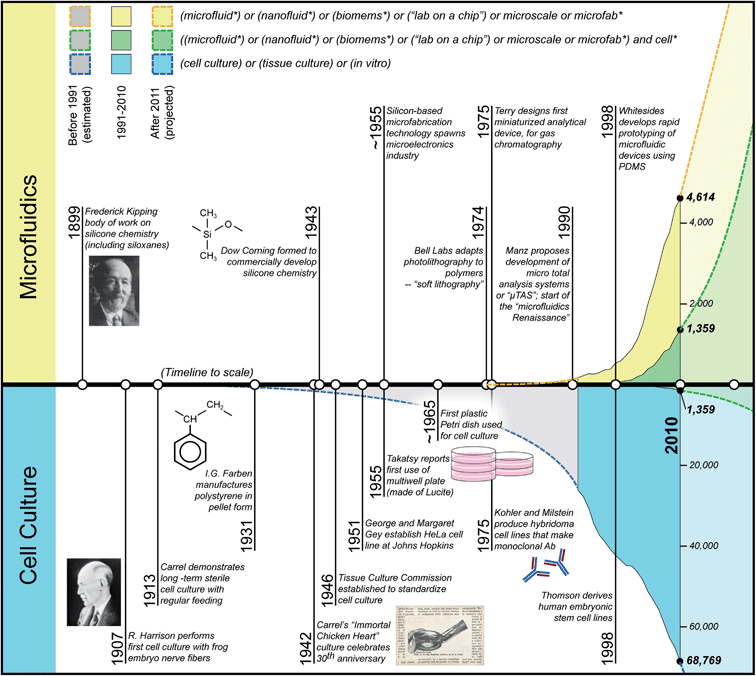 | ||
| Fig. 2 Timelines of major historic events in microfluidics and cell culture. Filled curves represent number of publications found per year, using the Web of Science search terms noted in the figure legend (yellow: microfluidics; blue: cell culture; green: integration of microfluidics and cell culture; search performed September 2011). Note that blue and yellow curves have different scales, and that the green curves are identical, mirror images of each other to show proportion within the independent fields of research. | ||
2.1. Timeline of cell culture
In 1907, Ross Harrison marked the beginning of cell (or tissue) culture and in vitro biology when he first demonstrated outgrowth of nerve fibers from a frog embryo on a glass dish.18 While this type of culture is considered routine today, such a feat – the ability to sustain life at the cellular level outside the whole of an organism – was in fact, at the time, viewed with both amazement and guarded skepticism.19 In the next three decades, Alexis Carrel shaped much of the tissue culture field by introducing aseptic methods, designing the canted-neck culture glassware, and optimizing cell culture techniques. One of the most publicized achievements in this era was perhaps the demonstration of the “immortal chicken heart”, in which Carrel showed that routine replenishment of nutrients and removal of cellular waste products was sufficient and necessary to sustain a culture for thirty years.20The mid-1940s through the 1950s marked an important turning point in the field of cell culture.19 During this period, the potential impact of cell culture, both as a tool for basic research in biology and as a vehicle for mass production of biological substances, began to fully emerge. First, the Tissue Culture Commission (now the Society for In Vitro Biology) was established to standardize cell culture techniques and provide materials and references on cell culture, marking the official scientific recognition of cell culture as a major research field. Second, important immortal cell lines were developed, first by Earle, who established the L929 mouse fibroblast cell line, followed by George and Margaret Gey, who established the well-known HeLa cell line,21 thereby allowing the establishment of cells as standard biological instruments for research and biotechnology. The ability to store, ship, and exchange the cell lines was enabled by the development of cryopreservation techniques, further fueling the expansion of the field. Finally, one of the most impactful developments was the work by Enders and Salk in 1954 that led to the mass production of the polio vaccine using cultured cells as instruments for massive expansion of the vaccine.22
As the field received increasing attention, the exclusive use of glass cultureware became a limiting factor, and alternative materials such as plastics began receiving more consideration as culture surfaces. Replacing glass with plastics helped reduce labor-intensive cleaning processes, lower operational costs, and improve durability of culture equipment, further promoting cell culture as a practical and accessible field of science. Coincidentally, the plastics industry was flourishing, and companies such as Dow Corning and Falcon Plastics developed processes that allowed a significant decrease in the cost of plastic fabrication, particularly PS, thereby enabling its use in myriad applications including household appliances, building materials and electronics equipment.23 Because of these advances, as well as a number of attractive properties (e.g., optical clarity, mechanical strength, durability, and low cost), PS became a reasonable choice as a material for commercial disposable cell culture labware. The final step in the transition to PS as a cell culture substrate was achieved with the development of oxygen plasma surface treatments by Falcon Plastics, which rendered PS more hydrophilic and “glass-like”, and ultimately improved its ability to promote cell adhesion and proliferation.24,25
PS ultimately became the dominant material from the 1960s onward, presumably due to increasing availability provided by the major plastics manufacturing firms.24 While other materials such as poly(methylmethacrylate) (PMMA or “Lucite”) were also used at the time, such as for the first “well plate”,26 their commercial availability eventually declined due to the increased pressure from PS. Over the subsequent fifty years, PS labware became the standard for in vitro experiments, leading to an exponential increase in the number of studies and publications in in vitro biology (Fig. 2). Because of its ubiquitous use, coupled with the accumulation of trusted data over decades of research, it has become difficult for other materials to establish themselves as materials of choice for biological research platforms and become more widespread.
2.2. Timeline of microfluidics
While microfluidics has a much shorter history than cell culture, similarities exist in their paths of development. In the mid-1970s, several key technical advances were reported that laid important groundwork in microfluidics, namely the first miniaturized analytical device designed for gas chromatography,27,28 and the development of soft lithography at Bell Laboratories.29 However, the developments in microfluidics were slow in the subsequent fifteen years, and focused mainly on microfabrication techniques in silicon for various device components.30–35The turning point for microfluidics occurred in the early 1990s with the idea of creating micro total analysis systems (μTAS), or “lab on a chip” devices, that were capable of reducing conventional laboratory equipment onto miniaturized platforms.36 During this “Renaissance” era of microfluidics,37 research focused mostly on applied chemistry and physics at the microscale,38–40with only a small number of pioneering reports demonstrating integration of microfluidics with biology41 and even fewer reports using microfluidics specifically for cell biology research.42–44 The popular material choices for these “first generation” microdevices included silicon and glass, mostly because microfabrication techniques were already established for these materials from microelectronics applications. Thermoplastics such as PMMA and polycarbonate (PC) were also prominent in the early developments of microfluidics because of their low cost and ease of manufacturing. Yet, despite growing interest, research in microfluidics was restricted to laboratories equipped with specialized equipment for performing silicon, glass, and thermoplastic microfabrication. The most significant advancement in microfluidics was the development of soft lithography and the use of PDMS as a material for rapid prototyping of microscale devices.45 In addition to possessing a number of attractive properties (e.g., optical clarity, low cost, reproducibility) that made PDMS competitive with glass and thermoplastics, its most significant advantage was its ease of use, specifically the ability to fabricate devices without the need for expensive equipment, which allowed rapid prototyping and testing of microscale designs, immediate widespread adoption in research laboratories, and an explosion in the number of research teams and publications in the field of microfluidics (Fig. 2).
The accessibility and ease of use of PDMS are the main reasons for the enormous expansion of cell-based microfluidics research. These properties have enabled many laboratories, including those with modest resources and technical expertise, to quickly set up fabrication processes and rapidly develop novel experimental tools for basic and applied scientific research. This has been especially important in cell biology research because the complexity of cells and cellular systems requires significant optimization of the tools used to probe them, which can only be achieved efficiently with a design process that allows rapid iteration through many designs. While many of the early reports on cell-based microfluidics (as well as some current reports) were focused on proof-of-concept demonstrations showcasing technical capabilities and the wide range of potential applications in biology,46–51 the field has since matured considerably, extending to studies focused on providing novel insights in fundamental cell biology.9,52–56
2.3. Summary
The emergence of PS and PDMS marked major turning points in the histories of cell culture and microfluidics, leading to immense growth for both fields, as can be seen by the sharp increases in publications (Fig. 2). In both cases, these materials emerged from a list of alternatives based on two main practical considerations: accessibility and availability. Indeed, it can be argued that there are currently no other notable materials that can rival the accessibility and availability of PS and PDMS in their respective disciplines over the same breadth of applications. Yet, while both fields have had similar major turning points followed by similar paths, there are also differences in their histories that will likely contribute to their distinct future trends. The most notable difference is the length of their histories: cell culture has more than a century of development while microfluidics spans a mere three decades of sharp progress. Importantly, these histories are linked respectively to two contrasting research cultures that further distinguish the nature of these fields. While technological fields like microfluidics are inherently more dynamic and transient, areas of basic science like biology necessarily build their strength from the gradual process of collecting empirical data and experiences, resulting in a solid foundation of scientific evidence and knowledge that gradually becomes fortified in the literature after years of rigorous testing. While this is a simplified view, given these two disciplines, microfluidics has the fluidity to adapt, evolve, and conform to the more rigid landscape in basic science and biology.In general, while PDMS and PS will likely coexist and continue to play significant roles within their distinct fields, a challenge arises at the interface of these areas: Which material do we use when we do research that integrates microfluidics and cell biology? This important question must be addressed for continued progress in this interdisciplinary research area. Our focus in this review, on discussing the suitability of PDMS and PS for cell-based microfluidics applications, is succinctly summarized in the following quote, which ironically comes from a book on PS:
“Unfortunately the selection of a suitable material, and elimination of unsuitable ones is not so easy. If the designer or fabricator does not consider carefully the properties of the material in terms of what is required of the finished article, he [or she] may make a poor choice.”
----- Teach and Kiessling (1960).23
3. PDMS versus PS
PS and PDMS have many attractive qualities well suited for their separate fields. Yet, these materials also have a number of important limitations that have raised some concerns regarding their usage as the material of choice at the intersection of these fields with respect to cell-based microfluidics applications. In this section, we change our focus from a historical perspective to the technical aspects of these fields, and discuss the main issues with using PS and PDMS at the interface of microfluidics and cell biology, both in terms of in vitro experimentation and microfabrication, and review the current approaches used to overcome these issues.3.1. PDMS for cell-based assays
PDMS has been central to the development of microfluidics in the last fifteen years due to a number of attractive properties. Nevertheless, a number of limitations have recently been cited that raise concerns about PDMS as an appropriate material for cell-based studies. Here we discuss five important properties of PDMS that potentially have adverse effects for microscale cell studies: deformation, evaporation, absorption, leaching of uncrosslinked oligomers, and hydrophobic recovery (Table 1).| Problem | Cause | Applications Affected | Solution | References |
|---|---|---|---|---|

|
- High compliance (low stiffness) | - Endothelial cell response to shear | - Modify curing parameters | - Gervais et al. (2006) |
| - Low aspect ratio | - Avoid low aspect ratios | |||
| - Avoid high pressure flows | ||||

|
- Permeability to water vapor | - Static no-flow experiments | - Coat with Parylene | - Verneuil et al. (2004) |
| - Osmolarity-sensitive experiments | - Ensure environments humidified | - Heo et al. (2007) | ||
| - Cell death from bubble propagation | - Incorporate media baths or sacrificial liquid reservoirs | - Berthier et al. (2008) | ||
| - Lecault et al. (2011) | ||||

|
- High permeability of material | - Soluble factor signaling studies involving small hydrophobic molecules | - Coat with Parylene | - Toepke & Beebe (2006) |
| - Coat with paraffin wax | - Regehr et al. (2009) | |||
| - Ren et al. (2010) | ||||
|
- Uncrosslinked oligomers | - Protein trafficking across membrane | - Coat with Parylene | - Regehr et al. (2009) |
| - Signaling through membrane-bound receptor proteins | - Soxhlet extraction | |||

|
- Surface diffusion of low molecular weight chains | - Unstable surface treatment or functionalization | - Use surface-treated device as soon as possible after treatment | - Eddington et al. (2006) |
| - Use hybrid devices with non-PDMS culture surface |
The compliance of PDMS, however, can become a limitation in applications where the deformation of microchannels and other microfeatures are not desirable. This is particularly true for cell culture experiments that require precise control over shear forces, such as shear stress studies on endothelial cell monolayers.6,51–53,56,66,67 Under certain pressure-driven flow conditions, for instance, microchannels may bulge due to the high pressure acting against compliant PDMS,68 or sag at the ceiling under its own weight when the aspect ratio is too low, causing variations in the expected shear rates. While cross-sectional deformations also occur in thermoplastic devices, typically after thermal bonding (see Section 2.2.3), the issue with PDMS is that deformation occurs dynamically during operation, and changes with varying pressure. Inability to account for these deformations can lead to erroneous estimation of shear stress, and can contribute to bias in data analysis and interpretation. This issue can be partially mitigated by changing PDMS curing parameters (e.g., mixing ratio, curing temperature and time) to achieve higher PDMS stiffnesses.68,69 However, this approach reduces deformation at the cost of introducing additional variability between PDMS devices made with different parameters, further complicating comparisons of biological results, particularly across different laboratories.
Permeability can also become a detriment in various microfluidic applications because of the issue of evaporation. Whereas PS and COP have low water vapor permeability (∼43 μm2 s−1 and 0.86 μm2 s−1, respectively),72 PDMS is extremely permeable to water vapor (∼1,000–6,000 μm2 s−1).75,76 Evaporation is inherently present in regular macroscale cell cultures as well, but the phenomenon becomes more dominant at the microscale where small amounts of evaporation can significantly shift volumes, concentrations, chemical balances, critical gradients and other factors.77–79 In certain cases, evaporation can be exploited at the microscale to reveal new techniques or methods, such as evaporation-induced sample concentration,80 or evaporation-driven continuous flow.81 However, evaporation more commonly leads to important adverse effects in microfluidic cell-based systems, such as osmolarity shifts that affect cell differentiation.75 A particularly challenging mode of evaporation occurs through the bulk PDMS material because of its relatively high water vapor permeability, even though the entire fluidic volume may be contained. While evaporation rates are slow in closed systems, they can cause significant loss of liquid volume, which can lead to bubbles that propagate, block flow, and lyse cells in microchannels, causing dramatic, detrimental effects that can lead to loss of data or result in experiments ending prematurely. In “open” systems where liquid surfaces are exposed to air, such as open-channel devices,82 EWOD-based digital microfluidic platforms,11,83 and passive pumping-based systems,50,84 evaporation may still occur through bulk PDMS layers, but occurs at a higher rate at exposed vapor-liquid interfaces, such as surfaces of open wells and droplets. Since this higher rate of evaporation is due to open microfluidic designs, it can be avoided by simply designing closed microfluidic systems when open design is not necessary for device function.
Methods have been developed to alleviate the adverse effects of PDMS permeability and evaporation. Parylene coating of PDMS surfaces was demonstrated as an effective method of preventing or significantly mitigating evaporation,75 but it requires additional fabrication procedures and equipment, and may hinder subsequent bonding or surface treatment steps. Recently, a report showed that a large isoosmotic media bath helped to maintain desired osmolarity in cell culture chambers that were adjacent to but physically separated from the bath by a PDMS membrane.85 Both these methods, however, are circumventions that are needed because of the use of PDMS.
3.2. PS in microfabrication
While PDMS and soft lithography have enabled the rapid expansion of microscale technologies and helped accelerate the advancement of microfluidic platforms, the inherent limitations of PDMS described above have raised concerns over its suitability in certain cell-based applications. Because most of these limitations are specific to PDMS and generally do not apply to thermoplastics (except for leaching of plasticizers, which has also been reported in PS),91 there has been renewed interest within the research community to develop methods and platforms in plastic materials. In this context, PS is particularly attractive not only because it is unaffected by PDMS-specific drawbacks, but also because of its importance in biology and reputation as a ubiquitous material for tissue culture plasticware. Furthermore, plastics continue to garner special attention in applications that require microscale devices that are manufacturable in large quantities and disposable.92 Therefore, PS has potential to help bridge the current gap between the state-of-the-art in microfluidics technology and the needs of biologists.Although microfabrication in thermoplastics has been well studied,93–106 thermoplastics continue to be less popular than PDMS as device materials for microscale cell-based systems. This trend can be attributed to challenges associated with achieving reliable and repeatable fabrication processes, and to the higher costs associated with thermoplastic microfabrication compared to soft lithography, especially for low production volume applications that are typically found in academic laboratories. PS microfabrication suffers from multiple bottlenecks along the typical process workflow, including the need to (1) make molds capable of resisting high temperatures and pressures that are commonly found in hot embossing processes, (2) create inlet and outlet access ports for world-to-chip interfacing, and (3) overcome challenges associated with bonding thermoplastic materials. Recently renewed interest in thermoplastic materials for microfluidics has triggered the development of improved fabrication methods to alleviate these bottlenecks, as well as other novel methods that may contribute to increasing their popularity. In this section, we review these recent developments, and provide evidence that the microfluidics community has recognized a growing need to develop platforms more immediately suitable to the needs of the biologist.
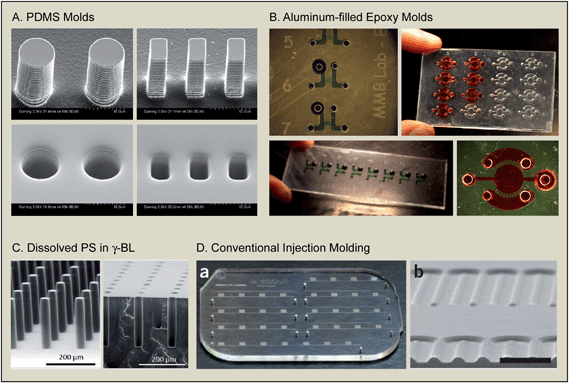 | ||
| Fig. 3 New wave of PS-based microfabrication. (A) The use of PDMS molds to directly emboss into PS. Reproduced in part from ref. 111 with permission from the Journal of Micromechanics and Microengineering (IOP Publishing, Ltd.). (B) The use of aluminum-filled epoxy molds and through-hole embossing to produce arrayed microfluidic cell-based systems in PS. Reprinted with permission from ref. 110. Copyright (2011) American Chemical Society. (C) Fabrication of high-aspect ratio PS features by dissolving solvent into PS before casting into compliant PDMS molds. Reproduced in part from ref. 112 with permission of The Royal Society of Chemistry. (D) PS device fabricated by conventional injection molding. The device shown has been used for clinical diagnostics in the third world, demonstrating the progress made in the application of plastic microfluidic devices. Reprinted by permission from Macmillan Publishers Ltd: Nature Medicine, ref. 109, copyright (2011). | ||
Recent work has begun to demonstrate more practical, low-cost methods for making molds, and have done so by relying on soft lithography methods as a framework, and adapting the process to meet the needs of thermoplastic fabrication. To produce low-cost micromolds, high strength epoxies have been used in place of metal molds to eliminate the need for micromachining and polishing. Positive relief epoxy micromolds for hot embossing can be easily created by casting the epoxy into negative-relief PDMS mold that were in turn originally cast from SU8-silicon molds.61 Use of composite aluminum filled epoxies has since improved mold durability at high temperatures and pressures.110 PS has also been directly embossed from a positive-relief PDMS mold (Fig. 3A),111 which takes advantage of the compliance of PDMS to facilitate demolding. The use of compliant PDMS molds was also demonstrated in a method where PS was dissolved in solvent, poured into a PDMS mold, heated on a hot plate to evaporate the solvent, and then finally removed from the PDMS mold, producing high aspect ratio features (Fig. 3C).112 Additionally, polyurethane plastic molds have also been cast directly from existing PDMS devices to re-create rigid masters.113 These methods only add one extra step to the mold fabrication process, and do not require the use of additional equipment beyond a heated press or hot plate.
Epoxy molds have several important advantages including significantly lower cost, faster turnaround times, and smoother surfaces compared to micromachined metal molds, and can be easily integrated with existing soft lithography methods and workflow, facilitating rapid prototyping in PS. In terms of disadvantages, epoxy molds have lower mechanical strength and durability compared to tool steel, and as a result, fewer devices per mold are produced before mechanical failure of the mold. This is more acceptable in an academic setting than in an industrial setting because designs are frequently modified during development, and additional epoxy molds can be quickly created from the original negative-relief PDMS mold. Epoxy molds are, however, limited in the aspect ratio and density of features that can be achieved because of the demolding process. Therefore, rigid molds typically require draft angles on the vertical faces of the mold features that are challenging to fabricate when relying only on soft lithography methods. Nevertheless, process workflow for thermoplastic fabrication has become sufficiently streamlined that rapid prototyping is no longer prohibitive, making PS fabrication more accessible to academic labs, especially in the design and assay development phases (Fig. 1).
A second popular method is thermal diffusion bonding, which relies on heat and pressure to promote reorganization, diffusion, and physical entanglement of polymeric chains at the interface of bonded surfaces.106,110 Although it is simple to implement and circumvents solvent-related issues, thermal bonding becomes increasingly difficult with larger device dimensions because of the challenge of maintaining uniform interfacial contact over large areas using high enough pressure to permit thermal diffusion, but low enough pressure so that microfeatures are not deformed. Low temperature bonding has recently been demonstrated, though it requires activation of the surfaces with ultraviolet or ozone treatment.101
Other methods, such as laser welding and ultrasonic welding, have also been developed, but are not as frequently found in an academic setting due to the need for expensive specialized equipment (for more details, the reader is referred to the aforementioned review).106 Notably, some interesting methods have been attempted to alleviate some of the problems mentioned. These include the use of permeant-assisted diffusion,123 or the use of two similar thermoplastics with slightly different Tg, in which a thin layer (<10 μm) of the low Tg plastic is applied on a layer with a higher Tg, creating an “adhesive” bonding layer.124
In summary, there are many methods for achieving bonding and sealing of plastic microdevices. The overarching issue is that the bonding process for thermoplastics is significantly more challenging than for PDMS, which can easily be bonded (reversibly or irreversibly) to many other surfaces. While progress has been made, bonding is still regarded as one of the main bottlenecks in the thermoplastic fabrication workflow. Therefore, more pragmatic, reliable, and easy to implement solutions, which are also compatible with cell culture, are necessary to develop a workflow for thermoplastic microfabrication that can be competitive with that of PDMS.
Surface hydrophilization also facilitates preparation of microscale cell-based systems, allowing capillary flow-mediated filling while minimizing bubble generation. This aspect is important because it ultimately reflects on the ease-of-use of the device, particularly by users without engineering or microfluidic training. Despite limited options for surface functionalization, PS remains advantageous compared to PDMS because hydrophobic recovery is minimal in comparison,130 thereby enabling the fabrication of devices well in advance of experiments. The ability to prepare devices in advance, ship, and store them is an attractive characteristic that is likely to further enhance collaboration that typically involves coordinating multiple researchers from different laboratories.
4. Discussion and outlook
It has become increasingly clear based on trends in scientific research that microscale cell-based systems have had – and will likely continue to have – a significant role in cell biology research. However, there also exists a significant gap in cultures between engineering and biology research that has somewhat hindered the widespread integration and adoption of microscale cell-based systems by the biology community. One particularly important example of the existence of this gap, as argued in the current review, is in device material selection where PDMS and PS have independently become the popular and standard choices in microengineering and cell biology, respectively. To enhance integration of microfluidic systems in biology, further facilitate collaborations between bioengineers and cell biology researchers, and ultimately enable accelerated advancement of in vitro biology, it is imperative that we examine the critical issues related to how we choose between PDMS and PS to fabricate our systems. Here, we reviewed both the historical trajectories of microfluidics and cell culture, and the practical and technical issues associated with material selection, and argued that the choice of material can have significant impact on the long-term adoption of microfluidic systems by the biological research community.First, a historical perspective revealed differences in how engineers and biologists typically approach the adoption of new technologies. On one hand, cell biology has been built mostly from incremental advances that stem from hypothesis-driven experimentation, and have only adopted technologies gradually when it was either necessary (e.g., the transition from glassware to plasticware in cell culture), or exceptionally enabling (e.g., the development of Transwell inserts, or the development of the Zigmond chamber). In contrast, cell-based microscale engineering is a relatively nascent field marked inherently by frequent disruptive developments in technological advancement (e.g., emergence of PDMS-based soft lithography, dynamic substrates131) caused mainly by the natural and constant need for innovation. Because of this difference in research cultures, the integration of new engineered technologies into the biology community is inherently challenging. In this context, the use of PS for microdevice fabrication can potentially help to ease the transition of microfluidics into biology by presenting the technology within the more familiar setting of PS-based formats. While a brief look at our history has proven useful, we also recognize that history and culture alone do not sufficiently explain the limited adoption of microscale cell-based systems in biology. Indeed, the biology community in the past has shown a willingness to embrace other enabling platforms when they have proven to be impactful. We believe that an open discussion on the differences in research culture, within the context of material selection for microfluidic systems, will lead to a recognition that PS-based formats are critical to facilitate and enhance integration, and will likely help reduce existing barriers to widespread adoption.
Second, and perhaps more importantly, we discussed practical and technical issues related to the use of PDMS and PS, and argued that the choice of material can have significant biological implications for many microscale cell-based applications. While the properties of PDMS, such as its compliance and permeability, have been leveraged to add a number of important functionalities to microscale systems,132 issues of microchannel bulging, sagging, and evaporation have also been linked to negative effects on cell-based studies.68,77,78 Mounting evidence suggests more subtle yet fundamental interactions between PDMS and cells in microscale culture due to absorption of small hydrophobic molecules and incorporation of leached PDMS monomers into cellular membranes.16 While the extent and significance of these interactions have yet to be fully explored, these concerns add uncertainty and experimental bias that contribute to hindering widespread adoption of PDMS-based microdevices. Our current understanding suggests these biases are particularly present in studies involving cell signaling, cell-cell communication, and cytokine and drug induction where concentration and function of proteins, small molecules, and growth factors can be affected. While PS and other plasticware may also suffer from some material biases based on recent reports that are bringing this issue to light,91 these biases appear to be much less dramatic in their impact on experiments compared to PDMS-derived issues. We acknowledge that many biological applications, such as PCR, cell sorting, cell capture, and short-term (i.e., <1 h) cell culture studies are not as significantly affected by PDMS-derived issues as cell signaling studies, and as such can be performed equally well in PDMS-based systems. However, we contend that in order to pursue increasingly complex cell biology questions from concept through development to implementation and analysis (Fig. 1), we must become increasingly aware of all such material biases as they pertain to our application of interest. This awareness will likely dictate how we design and validate systems, analyze data, interpret results, and make conclusions, and ultimately determine how successful we will be at integrating technology with basic science.
While the underlying focus of this review has been on device fabrication within an academic setting, an additional confounding factor yet to be discussed is that of manufacturability and its importance on commercialization and integration on a broader scale. Academic laboratories typically serve as the initial sources of newly developed technological platforms, benefiting from the freedom of a flexible research environment to explore innovative ideas and iterate through designs via rapid prototyping. In such a setting, PDMS-based microfabrication remains a useful and powerful methodology because of its accessibility, while in contrast PS-based fabrication is still more challenging despite recent reports aimed at achieving rapid prototyping of PS-based devices (Fig. 3).109–112 From the perspective of commercialization, however, PDMS becomes a major limiting factor because manufacturing costs and production volumes do not scale up favorably with soft lithography, and shipping, packaging, and storage of surface-treated PDMS microdevices remains challenging given that hydrophobic recovery tends to revert surface treatments to their original state. Thus, when manufacturability is factored into design, PS and other plastics have a distinctive edge over PDMS because manufacturing costs for plastics decrease significantly for high production volumes, and plastics do not degrade during long-term storage.
Based on all these perspectives, it is evident that the issue of material selection is far from trivial. Material selection has implications on experimental research, and ultimately on the ease of adoption by collaborating scientists. While our focus has been on comparing PDMS to PS, it should be recognized that the issue of material selection in general is far broader given that a growing number of reports are demonstrating advantages of various other materials, including cyclo-olefin copolymer (COC),17,61,101,106,133 and Teflon.134 Additionally, interesting developments have also been made with the use of paper in microfluidics.135–140 Essentially, choosing which material is appropriate for your cell-based application depends on several key aspects: (1) manufacturability or ease of fabrication, (2) effects on the biology of cultured cells, and (3) suitability of the material for the desired analyses and readouts (Fig. 4). Even with a growing list of alternatives, PDMS remains a leading option because of its convenience, reliability, and its ability to enable unmatched functionalities including flexible valves, deflectable microposts, and cell-stretching membranes. However, for more advanced biological investigations such as long-term cultures involving cell signaling, the PDMS biases must be either circumvented or completely avoided by choosing another material. In these more biologically oriented applications, PS stands clearly above other alternatives due to the wealth of knowledge accumulated on PS over the years and the fact that PS does not suffer from the same key limitations as PDMS. Moreover, as mentioned its manufacturability is superior to that of PDMS, even though its ease of fabrication in low production volume, rapid prototyping settings is less attractive. This is where microfabrication engineers have refocused their research, and continued progress in this area will further close the gap between PDMS and PS on the convenience of fabrication. In the case of other specific applications, such as high-resolution microscopy, alternative materials such as glass and COC are highly recommended. For this reason, COC has also been gaining popularity because most fabrication methods developed for PS are easily translatable to COC. Glass, having the advantage of being the first material used in both microfluidics and in vitro biology, offers well-grounded knowledge, chemical inertness, high quality optics, and the ability to integrate patterned electronics. However, the significantly higher price for microsystems made out of glass, as well as the necessity to re-utilize them, make it a very specialized material. Finally, the most recent addition to our list of material alternatives is Teflon,134 which comprises a unique set of properties that give it potential in niche applications, but has not yet been used widely enough to be competitive.
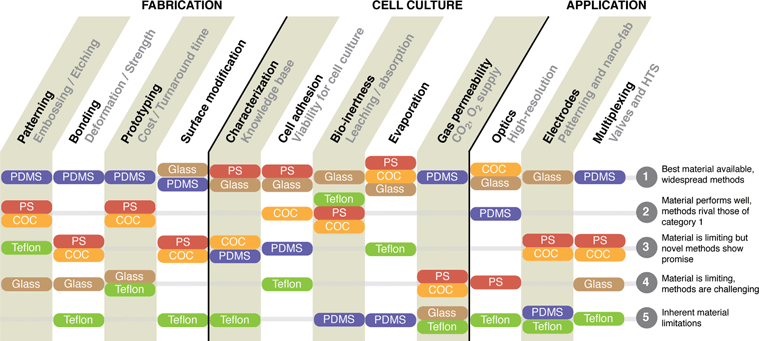 | ||
| Fig. 4 Comparative strengths and weaknesses of materials used for microfluidic cell-based device fabrication. The materials are ranked for their abilities for various properties important in cell-based experiments, which are grouped under three general categories: The ability to fabricate the microsystems, the ability to perform controllable cell-based experiments, and the potential for integrated micro-engineering applications. | ||
Ultimately, adoption of microscale technologies by biologists hinges on two main aspects: (1) successful collaborations between engineers and biologists in academic settings where the technology is first conceived, and (2) the establishment of standard platforms that, because of manufacturability, are widely accessible and available to the biology community at large. The responsibility to develop an acceptable platform relies partially on the engineer to adapt and integrate materials and technologies in a manner that suits the needs of the biologist. This task can and will be facilitated by an expanded repertoire of fabrication methods for different materials, particularly those that can compete with PDMS. Specifically, for cell-based applications, increasingly accessible PS fabrication methods are beginning to emerge. More work in this research area is still required to ease the transition for biologists toward microscale cell-based systems. It is our hope that the increased exposure of the issue of material selection in cell-based studies can further lead to new and improved fabrication methods suited for enabling applications.
In conclusion, material selection, especially between PDMS and PS, needs to be considered more frequently and more seriously by engineers and biologists alike. The choice of material is particularly important when new technology is being introduced into a well-grounded and complex discipline such as cell biology that involves many unknown interactions and biases. While PDMS is currently the overwhelming choice for microfluidic engineers, developments in PS fabrication methods have begun to improve its accessibility to the point where PS warrants consideration as the new material of choice for certain cell-based applications. The ability for microfluidics engineers to offer platforms in both PS and PDMS may play an important role in achieving further integration between microfluidics and cell biology, and further promote fruitful collaborations between engineers and biologists. Indeed, material considerations constitute only one aspect contributing to limited adoption of microsystems for cell-based research. Other aspects, such as the challenge of integrating with existing infrastructure,50 and the lack of a killer application,141 also deserve some attention from the microfluidics community, especially those involved in cell biology applications. An increased focus on thermoplastic fabrication will hopefully bring new solutions for making PS fabrication more accessible and reliable, expand our choices for microfabrication, and, ultimately stimulate collaborations between biologists and engineers.
References
- A. Folch and M. Toner, Annu. Rev. Biomed. Eng., 2000, 2, 227–256 CrossRef CAS.
- J. El-Ali, P. Sorger and K. Jensen, Nature, 2006, 442, 403–411 CrossRef CAS.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS.
- A. Paguirigan and D. J. Beebe, BioEssays, 2008, 30, 811–821 CrossRef CAS.
- T. Keenan and A. Folch, Lab Chip, 2008, 8, 34–57 RSC.
- E. W. K. Young and C. A. Simmons, Lab Chip, 2010, 10, 143–160 RSC.
- E. W. K. Young and D. J. Beebe, Chem. Soc. Rev., 2010, 39, 1036–1048 RSC.
- M. Thery, V. Racine, M. Piel, A. Pepin, A. Dimitrov, Y. Chen, J.-B. Sibarita and M. Bornens, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(52), 19771–19776 CrossRef CAS.
- K. E. Sung, N. Yang, C. Pehlke, P. J. Keely, K. W. Eliceiri, A. Friedl and D. J. Beebe, Integr. Biol., 2011, 3, 439–450 RSC.
- T. Thorsen, S. J. Maerkl and S. R. Quake, Science, 2002, 298, 580–584 CrossRef CAS.
- I. Barbulovic-Nad, S. H. Au and A. R. Wheeler, Lab Chip, 2010, 10, 1536–1542 RSC.
- J. Fu, Y.-K. Wang, M. T. Yang, R. A. Desai, X. Yu, Z. Liu and C. S. Chen, Nat. Methods, 2010, 7, 733–U95 CrossRef CAS.
- E. Berthier, J. Surfus, J. Verbsky, A. Huttenlocher and D. Beebe, Integr. Biol., 2010, 2, 630–638 RSC.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U. J. Balis, R. G. Tompkins, D. A. Haber and M. Toner, Nature, 2007, 450, 1235–U10 CrossRef CAS.
- K. T. Kotz, W. Xiao, C. Miller-Graziano, W.-J. Qian, A. Russom, E. A. Warner, L. L. Moldawer, A. De, P. E. Bankey, B. O. Petritis, I. Camp, G. David, A. E. Rosenbach, J. Goverman, S. P. Fagan, B. H. Brownstein, D. Irimia, W. Xu, J. Wilhelmy, M. N. Mindrinos, R. D. Smith, R. W. Davis, R. G. Tompkins and M. Toner, Nat. Med., 2010, 16, 1042–U142 CrossRef CAS.
- K. J. Regehr, M. Domenech, J. T. Koepsel, K. C. Carver, S. J. Ellison-Zelski, W. L. Murphy, L. A. Schuler, E. T. Alarid and D. Beebe, Lab Chip, 2009, 9, 2132–2139 RSC.
- X. Su, E. W. K. Young, H. A. S. Underkofler, T. J. Kamp, C. T. January and D. J. Beebe, J. Biomol. Screening, 2011, 16, 101–111 CrossRef CAS.
- R. G. Harrison, Science, 1911, 34, 279–281 CAS.
- H. Landecker, Culturing life: how cells became technologies, Harvard University Press, Cambridge, MA, 2007 Search PubMed.
- A. H. Ebeling, Sci. Am., 1942, 166, 22–24 CrossRef.
- R. Skloot, The Immortal Life of Henrietta Lacks, Crown Publishers, Random House, Inc., New York, NY, 2010 Search PubMed.
- D. M. Oshinsky, Polio: An American Story, Oxford University Press, New York, NY, 2005 Search PubMed.
- W. C. Teach, G. C. Kiessling, Polystyrene, Reinhold Publishing Corp, New York, NY, 1960 Search PubMed.
- A. Curtis, J. Forrester, C. Mcinnes and F. Lawrie, J. Cell Biol., 1983, 97, 1500–1506 CrossRef CAS.
- S. Barker and P. LaRocca, Methods in Cell Science, 1994, 16, 151–153 Search PubMed.
- G. Takatsy, Acta Microbiologica et Immunologica Hungarica, 1955, 3, 191–202 CAS.
- S. C. Terry, A Gas Chromatography System Fabricated On A Silicon Wafer Using Integrated Circuit Technology, Ph.D. Thesis, Stanford University, 1975.
- S. Terry, J. Jerman and J. Angell, IEEE Trans. Electron Devices, 1979, 26, 1880–1886 CrossRef.
- G. Aumiller, Ea. Chandros, Wj. Tomlinso and H. Weber, J. Appl. Phys., 1974, 45, 4557–4562 CrossRef CAS.
- F. Vandepol, D. Wonnink, M. Elwenspoek and J. Fluitman, Sens. Actuators, 1989, 17, 139–143 CrossRef.
- F. Vandepol, H. Vanlintel, M. Elwenspoek and J. Fluitman, Sens. Actuators, A, 1990, 21, 198–202 CrossRef.
- H. Vanlintel, F. Vandepol and S. Bouwstra, Sens. Actuators, 1988, 15, 153–167 CrossRef.
- M. Esashi, S. Shoji and A. Nakano, Sens. Actuators, 1989, 20, 163–169 CrossRef.
- M. Esashi, Sens. Actuators, A, 1990, 21, 161–167 CrossRef.
- S. Shoji, S. Nakagawa and M. Esashi, Sens. Actuators, A, 1990, 21, 189–192 CrossRef.
- A. Manz, N. Graber and H. Widmer, Sens. Actuators, B, 1990, 1, 244–248 CrossRef.
- D. Reyes, D. Iossifidis, P. Auroux and A. Manz, Anal. Chem., 2002, 74, 2623–2636 CrossRef CAS.
- D. Harrison, A. Manz, Z. Fan, H. Ludi and H. Widmer, Anal. Chem., 1992, 64, 1926–1932 CrossRef CAS.
- J. Pfahler, J. Harley, H. Bau and J. Zemel, Sens. Actuators, A, 1990, 22, 431–434 CrossRef.
- I. Papautsky, J. Brazzle, T. Ameel and A. Frazier, Sens. Actuators, A, 1999, 73, 101–108 CrossRef.
- C. Effenhauser, A. Paulus, A. Manz and H. Widmer, Anal. Chem., 1994, 66, 2949–2953 CrossRef CAS.
- S. Masuda, M. Washizu and T. Nanba, IEEE Trans. Ind. Appl., 1989, 25, 732–737 CrossRef.
- M. Washizu, T. Nanba and S. Masuda, IEEE Trans. Ind. Appl., 1990, 26, 352–358 CrossRef.
- P. Li and D. Harrison, Anal. Chem., 1997, 69, 1564–1568 CrossRef CAS.
- D. Duffy, J. McDonald, O. Schueller and G. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS.
- A. Fu, C. Spence, A. Scherer, F. Arnold and S. Quake, Nat. Biotechnol., 1999, 17, 1109–1111 CrossRef CAS.
- S. Takayama, E. Ostuni, P. LeDuc, K. Naruse, D. Ingber and G. Whitesides, Nature, 2001, 411, 1016–1016 CrossRef CAS.
- P. Hung, P. Lee, P. Sabounchi, N. Aghdam, R. Lin and L. Lee, Lab Chip, 2005, 5, 44–48 RSC.
- H. Yu, C. Alexander and D. Beebe, Lab Chip, 2007, 7, 388–391 RSC.
- I. Meyvantsson, J. Warrick, S. Hayes, A. Skoien and D. Beebe, Lab Chip, 2008, 8, 717–724 RSC.
- E. W. K. Young, M. W. L. Watson, S. Srigunapalan, A. R. Wheeler and C. A. Simmons, Anal. Chem., 2010, 82, 808–816 CrossRef CAS.
- E. W. K. Young, A. R. Wheeler and C. A. Simmons, Lab Chip, 2007, 7, 1759–1766 RSC.
- J. Song, S. Cavnar, A. Walker, K. Luker, M. Gupta, Y.-C. Tung, G. Luker and S. Takayama, PLoS One, 2009, 4, e5756 Search PubMed.
- R. Sudo, S. Chung, I. K. Zervantonakis, V. Vickerman, Y. Toshimitsu, L. G. Griffith and R. D. Kamm, FASEB J., 2009, 23, 2155–2164 CrossRef CAS.
- P. J. Cavnar, E. Berthier, D. J. Beebe and A. Huttenlocher, J. Cell Biol., 2011, 193, 465–473 CrossRef CAS.
- J. W. Song and L. L. Munn, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15342–15347 CrossRef CAS.
- K. Chung, Y. Kim, J. S. Kanodia, E. Gong, S. Y. Shvartsman and H. Lu, Nat. Methods, 2011, 8, 171–U103 CrossRef CAS.
- J. Tan, J. Tien, D. Pirone, D. Gray, K. Bhadriraju and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1484–1489 CrossRef CAS.
- M. Murrell, R. Kamm and P. Matsudaira, Biophys. J., 2011, 101, 297–306 CrossRef CAS.
- C. Moraes, J.-H. Chen, Y. Sun and C. A. Simmons, Lab Chip, 2010, 10, 227–234 RSC.
- G. Mehta, J. Lee, W. Cha, Y. Tung, J. Linderman and S. Takayama, Anal. Chem., 2009, 81, 3714–3722 CrossRef CAS.
- W. Gu, X. Zhu, N. Futai, B. Cho and S. Takayama, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15861–15866 CrossRef CAS.
- J. Song, W. Gu, N. Futai, K. Warner, J. Nor and S. Takayama, Anal. Chem., 2005, 77, 3993–3999 CrossRef CAS.
- S. Tay, J. J. Hughey, T. K. Lee, T. Lipniacki, S. R. Quake and M. W. Covert, Nature, 2010, 466, 267–U149 CrossRef CAS.
- M. Unger, H. Chou, T. Thorsen, A. Scherer and S. Quake, Science, 2000, 288, 113–116 CrossRef CAS.
- C. Ku, T. Oblak and D. Spence, Anal. Chem., 2008, 80, 7543–7548 CrossRef CAS.
- J. Shao, L. Wu, J. Wu, Y. Zheng, H. Zhao, Q. Jin and J. Zhao, Lab Chip, 2009, 9, 3118–3125 RSC.
- T. Gervais, J. El-Ali, A. Gunther and K. Jensen, Lab Chip, 2006, 6, 500–507 RSC.
- J. Y. Park, S. J. Yoo, E.-J. Lee, D. H. Lee, J. Y. Kim and S.-H. Lee, BioChip J., 2010, 4, 230–236 CrossRef CAS.
- H. Shiku, T. Saito, C. Wu, T. Yasukawa, M. Yokoo, H. Abe, T. Matsue and H. Yamada, Chem. Lett., 2006, 35, 234–235 CrossRef CAS.
- S. Charati and S. Stern, Macromolecules, 1998, 31, 5529–5535 CrossRef CAS.
- W. D. Niles and P. J. Coassin, Assay Drug Dev. Technol., 2008, 6, 577–590 CrossRef CAS.
- J. S. Gewandter, R. J. Staversky and M. A. O'Reilly, Free Radical Biol. Med., 2009, 47, 1742–1752 CrossRef CAS.
- Y. Tang, E. A. Scheef, Z. Gurel, C. M. Sorenson, C. R. Jefcoate and N. Sheibani, Am. J. Physiol.: Cell Physiol., 2010, 298, C665–C678 CrossRef CAS.
- Y. Heo, L. Cabrera, J. Song, N. Futai, Y. Tung, G. Smith and S. Takayama, Anal. Chem., 2007, 79, 1126–1134 CrossRef CAS.
- Y. Zhang, M. Ishida, Y. Kazoe, Y. Sato and N. Miki, IEEJ Trans. Electr. Electron. Eng., 2009, 4, 442–449 CrossRef CAS.
- E. Berthier, J. Warrick, H. Yu and D. Beebe, Lab Chip, 2008, 8, 852–859 RSC.
- E. Berthier, J. Warrick, H. Yu and D. Beebe, Lab Chip, 2008, 8, 860–864 RSC.
- N. Lynn, C. Henry and D. Dandy, Lab Chip, 2009, 9, 1780–1788 RSC.
- G. Walker and D. Beebe, Lab Chip, 2002, 2, 57–61 RSC.
- M. Zimmermann, S. Bentley, H. Schmid, P. Hunziker and E. Delamarche, Lab Chip, 2005, 5, 1355–1359 RSC.
- L. Millet, M. Stewart, J. Sweedler, R. Nuzzo and M. Gillette, Lab Chip, 2007, 7, 987–994 RSC.
- I. Barbulovic-Nad, H. Yang, P. Park and A. Wheeler, Lab Chip, 2008, 8, 519–526 RSC.
- M. Domenech, H. Yu, J. Warrick, N. Badders, I. Meyvantsson, C. Alexander and D. Beebe, Integr. Biol., 2009, 1, 267–274 RSC.
- V. Lecault, M. VanInsberghe, S. Sekulovic, D. J. H. F. Knapp, S. Wohrer, W. Bowden, F. Viel, T. McLaughlin, A. Jarandehei, M. Miller, D. Falconnet, A. K. White, D. G. Kent, M. R. Copley, F. Taghipour, C. J. Eaves, R. K. Humphries, J. M. Piret and C. L. Hansen, Nat. Methods, 2011, 8, 581–586 CrossRef CAS.
- M. Toepke and D. Beebe, Lab Chip, 2006, 6, 1484–1486 RSC.
- H. Sasaki, H. Onoe, T. Osaki, R. Kawano and S. Takeuchi, Sens. Actuators, B, 2010, 150, 478–482 CrossRef.
- K. Ren, Y. Zhao, J. Su, D. Ryan and H. Wu, Anal. Chem., 2010, 82, 5965–5971 CrossRef CAS.
- J. Lee, C. Park and G. Whitesides, Anal. Chem., 2003, 75, 6544–6554 CrossRef CAS.
- D. Eddington, J. Puccinelli and D. Beebe, Sens. Actuators, B, 2006, 114, 170–172 CrossRef.
- G. R. McDonald, A. L. Hudson, S. M. J. Dunn, H. You, G. B. Baker, R. M. Whittal, J. W. Martin, A. Jha, D. E. Edmondson and A. Holt, Science, 2008, 322, 917–917 CrossRef CAS.
- J. S. Kuo and D. T. Chiu, Lab Chip, 2011, 11, 2656–2665 RSC.
- S. Soper, S. Ford, S. Qi, R. McCarley, K. Kelly and M. Murphy, Anal. Chem., 2000, 72, 642A–651A CrossRef.
- H. Klank, J. Kutter and O. Geschke, Lab Chip, 2002, 2, 242–246 RSC.
- C. Ahn, J. Choi, G. Beaucage, J. Nevin, J. Lee, A. Puntambekar and J. Lee, Proc. IEEE, 2004, 92, 154–173 CrossRef CAS.
- Y. Yang, C. Li, J. Kameoka, K. Lee and H. Craighead, Lab Chip, 2005, 5, 869–876 RSC.
- L. Brown, T. Koerner, J. Horton and R. Oleschuk, Lab Chip, 2006, 6, 66–73 RSC.
- B. Flachsbart, K. Wong, J. Iannacone, E. Abante, R. Vlach, P. Rauchfuss, P. Bohn, J. Sweedler and M. Shannon, Lab Chip, 2006, 6, 667–674 RSC.
- Y. Sun, Y. Kwok and N. Nguyen, J. Micromech. Microeng., 2006, 16, 1681–1688 CrossRef CAS.
- D. S. Kim, S. H. Lee, C. H. Ahn, J. Y. Lee and T. H. Kwon, Lab Chip, 2006, 6, 794–802 RSC.
- C. W. Tsao, L. Hromada, J. Liu, P. Kumar and D. L. DeVoe, Lab Chip, 2007, 7, 499–505 RSC.
- J. Do, S. Lee, J. Han, J. Kai, C.-C. Hong, C. Gao, J. H. Nevin, G. Beaucage and C. H. Ahn, Lab Chip, 2008, 8, 2113–2120 RSC.
- C. Chen, D. Breslauer, J. Luna, A. Grimes, W. Chin, L. Leeb and M. Khine, Lab Chip, 2008, 8, 622–624 RSC.
- Y. Chen, L. Zhang and G. Chen, Electrophoresis, 2008, 29, 1801–1814 CrossRef CAS.
- H. Becker and C. Gaertner, Anal. Bioanal. Chem., 2008, 390, 89–111 CrossRef CAS.
- C. Tsao and D. DeVoe, Microfluid. Nanofluid., 2009, 6, 1–16 CrossRef CAS.
- D. A. Mair, E. Geiger, A. P. Pisano, J. M. J. Frechet and F. Svec, Lab Chip, 2006, 6, 1346–1354 RSC.
- Y.-C. Tung, A. Y. Hsiao, S. G. Allen, Y.-s. Torisawa, M. Ho and S. Takayama, Analyst, 2011, 136, 473–478 RSC.
- C. D. Chin, T. Laksanasopin, Y. K. Cheung, D. Steinmiller, V. Linder, H. Parsa, J. Wang, H. Moore, R. Rouse, G. Umviligihozo, E. Karita, L. Mwambarangwe, S. L. Braunstein, J. van de Wijgert, R. Sahabo, J. E. Justman, W. El-Sadr and S. K. Sia, Nat. Med., 2011, 17, 1015–U138 CrossRef CAS.
- E. W. K. Young, E. Berthier, D. J. Guckenberger, E. Sackmann, C. Lamers, I. Meyvantsson, A. Huttenocher and D. J. Beebe, Anal. Chem., 2011, 83, 1408–1417 CrossRef CAS.
- V. N. Goral, Y.-C. Hsieh, O. N. Petzold, R. A. Faris and P. K. Yuen, J. Micromech. Microeng., 2011, 21 DOI:10.1088/0960-1317/21/1/017002.
- Y. Wang, J. Balowski, C. Phillips, R. Phillips, C. E. Sims and N. L. Allbritton, Lab Chip, 2011, 11, 3089–3097 RSC.
- S. P. Desai, D. M. Freeman and J. Voldman, Lab Chip, 2009, 9, 1631–1637 RSC.
- C. Fredrickson and Z. Fan, Lab Chip, 2004, 4, 526–533 RSC.
- M. L. Hupert, W. J. Guy, S. D. Llopis, H. Shadpour, S. Rani, D. E. Nikitopoulos and S. A. Soper, Microfluid. Nanofluid., 2007, 3, 1–11 CrossRef CAS.
- S. K. Njoroge, M. A. Witek, M. L. Hupert and S. A. Soper, Electrophoresis, 2010, 31, 981–990 CrossRef CAS.
- S. Selimovic, F. Piraino, H. Bae, M. Rasponi, A. Redaelli and A. Khademhosseini, Lab Chip, 2011, 11, 2325–2332 RSC.
- B. Jo, L. V. Lerberghe, K. Motsegood and D. Beebe, J. Microelectromech. Syst., 2000, 9, 76–81 CrossRef CAS.
- M. Worgull, Hot Embossing: Theory and Technology of Microreplication, Elsevier Sci Ltd., Burlington, MA, 2009 Search PubMed.
- P. Zhou, L. Young and Z. Chen, Biomed. Microdevices, 2010, 12, 821–832 CrossRef.
- D. Ogonczyk, J. Wegrzyn, P. Jankowski, B. Dabrowski and P. Garstecki, Lab Chip, 2010, 10, 1324–1327 RSC.
- M. Ghisari and E. C. Bonefeld-Jorgensen, Toxicol. Lett., 2009, 189, 67–77 CrossRef CAS.
- T. I. Wallow, A. M. Morales, B. A. Simmons, M. C. Hunter, K. L. Krafcik, L. A. Domeier, S. M. Sickafoose, K. D. Patel and A. Gardea, Lab Chip, 2007, 7, 1825–1831 RSC.
- J. Steigert, S. Haeberle, T. Brenner, C. Mueller, C. P. Steinert, P. Koltay, N. Gottschlich, H. Reinecke, J. Ruehe, R. Zengerle and J. Ducree, J. Micromech. Microeng., 2007, 17, 333–341 CrossRef CAS.
- T. Horbett, J. Waldburger, B. Ratner and A. Hoffman, J. Biomed. Mater. Res., 1988, 22, 383–404 CrossRef CAS.
- M. Shen and T. Horbett, J. Biomed. Mater. Res., 2001, 57, 336–345 CrossRef CAS.
- J. Grace and L. Gerenser, J. Dispersion Sci. Technol., 2003, 24, 305–341 CrossRef CAS.
- I. Beaulieu, M. Geissler and J. Mauzeroll, Langmuir, 2009, 25, 7169–7176 CrossRef CAS.
- D. Irvine, A. Mayes and L. Griffith, Biomacromolecules, 2001, 2, 85–94 CrossRef CAS.
- E. Occhiello, M. Morra, P. Cinquina and F. Garbassi, Polymer, 1992, 33, 3007–3015 CrossRef CAS.
- M. Mrksich, MRS Bull., 2005, 30, 180–184 CrossRef CAS.
- T. Thorsen, S. Maerkl and S. Quake, Science, 2002, 298, 580–584 CrossRef CAS.
- J. Greener, W. Li, J. Ren, D. Voicu, V. Pakharenko, T. Tang and E. Kumacheva, Lab Chip, 2010, 10, 522–524 RSC.
- K. Ren, W. Dai, J. Zhou, J. Su and H. Wu, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 8162–8166 CrossRef CAS.
- E. Carrilho, A. W. Martinez and G. M. Whitesides, Anal. Chem., 2009, 81, 7091–7095 CrossRef CAS.
- A. W. Martinez, S. T. Phillips and G. M. Whitesides, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 19606–19611 CrossRef CAS.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS.
- J. L. Osborn, B. Lutz, E. Fu, P. Kauffman, D. Y. Stevens and P. Yager, Lab Chip, 2010, 10, 2659–2665 RSC.
- R. Derda, A. Laromaine, A. Mammoto, S. K. Y. Tang, T. Mammoto, D. E. Ingber and G. M. Whitesides, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 18457–18462 CrossRef CAS.
- R. Derda, S. K. Y. Tang, A. Laromaine, B. Mosadegh, E. Hong, M. Mwangi, A. Mammoto, D. E. Ingber and G. M. Whitesides, PLoS One, 2011, 6, e18940, DOI:10.1371/journal.pone.0018940.
- H. Becker, Lab Chip, 2009, 9, 2119–2122 RSC.
Footnote |
| † Contributed equally |
| This journal is © The Royal Society of Chemistry 2012 |
