A covalently modified hydrogel blend of hyaluronan–methyl cellulose with peptides and growth factors influences neural stem/progenitor cell fate
Roger Y.
Tam
ab,
Michael J.
Cooke
ab and
Molly S.
Shoichet†
*abc
aDepartment of Chemical Engineering and Applied Chemistry, University of Toronto, 200 College Street, Toronto, ON M5S 3E5, Canada
bInstitute of Biomaterials and Biomedical Engineering, 164 College Street, Room 407, Toronto, ON M5S 3G9, Canada
cDepartment of Chemistry, University of Toronto, 80 St. George Street, Toronto, ON M5S 3H6, Canada
First published on 13th July 2012
Abstract
With the goal of controlling stem cell fate, for ultimate application in cell delivery, we synthesized a cell delivery vehicle comprised of an injectable hydrogel of hyaluronan (HA) and methyl cellulose (MC) wherein the MC is modified to allow bio-orthogonal coupling chemistry. Specifically, MC was modified using thiol-maleimide and biotin–streptavidin chemistry to covalently conjugate the cell adhesive peptide, glycine–arginine–glycine–aspartic acid–serine (GRGDS), and the oligodendrocyte-differentiating factor, recombinant platelet-derived growth factor A (rPDGF-A). NSPCs differentiated into significantly more oligodendrocytes when cultured in a HAMC hydrogel modified with both PDGF-A and GRGDS compared to controls of unmodified HAMC hydrogel and HAMC modified with only GRGDS. These results demonstrate both the ability to influence NSPC fate with immobilized growth factor and a platform technology for broad applicability in bioactive factor immobilization.
Introduction
Traumatic spinal cord injury (SCI) is a debilitating injury that has significant consequences as it can dramatically impair sensory and motor function. It is often irreversible, primarily due to the body's limited ability to repair the damaged tissue.1,2 Initial insult to the central nervous system (CNS) causes necrotic cell death,3 and is followed by secondary injury which includes inflammation,4,5 permeation of the blood-spinal cord barrier,6 and lipid peroxidation.7 This causes additional cell death, which subsequently leads to demyelination,8–10 axonal degeneration,11 and formation of a glial scar.12 Demyelination, which impairs the efficient signaling and viability of the surviving neurons, has been shown to be closely associated with the loss of functional capacity in CNS disorders.8,10,13,14A potential therapeutic strategy to replace the damaged oligodendrocytes following SCI is the transplantation of neural stem/progenitor cells (NSPCs).15 These cells have the ability to self-renew and to differentiate into the three major cell types of the CNS: oligodendrocytes, neurons and astrocytes. However, this method is currently limited by the poor survival16,17 and uncontrolled differentiation of transplanted stem cells.15,18 Delivery of stem cells into the target tissue often results in a large amount of cell death due to the harsh inflammatory environment present following injury,17 immune response to foreign cells and differences between microenvironments of the native stem cell niche and target tissue.19 For stem cell therapy to be effective in enabling functional recovery following SCI, cell death must be minimized20,21 and cell differentiation controlled.15,18,22,23
The use of biomaterials, specifically hydrogels, to control stem cell fate has been studied extensively,19,24 with various chemical25–28 and physical28,29 cues incorporated into the material design to increase cell viability and control cell function. For example, the fibronectin-derived peptide sequence RGD has been shown to bind to various cell-surface integrin receptors such as α5β1 and αvβ3,30 and increase cell adhesion and survival.19,31–33 However, while many of these biomaterials show an increase in cell viability and adhesion in vitro, several important design criteria – such as injectability to enable minimally invasive surgery and anti-inflammatory properties – must be considered to enable their use as a cell delivery vehicle in vivo.34 Moreover, the gelation process should exclude common cytotoxic reagents and intermediates used for polymer cross-linking such as acrylates, aldehydes, epoxides and free radicals.35
An injectable cell delivery vehicle composed of hyaluronan (HA) and methyl cellulose (MC) has been shown to improve cell survival and distribution upon transplantation in vivo.36 HA attenuates the inflammatory response and is shear-thinning whereas MC is inverse thermal gelling. Together, this physical blend of HA and MC (HAMC) is injectable through a 30 G needle, allowing for minimally invasive surgery, and quickly gels upon reaching physiological temperatures without the need for additional crosslinking reagents.34 Since HAMC has also demonstrated a pro-survival effect on transplanted cells,36 it is a desirable biomaterial for stem cell transplantation.
To further enhance the properties of the HAMC hydrogel system for controlled differentiation of NSPCs, we investigated its modification with recombinant platelet-derived growth factor-A (rPDGF-A), which has been shown to enhance differentiation of NSPCs to oligodendrocytes, and GRGDS, which has been shown to enhance cell adhesion.37–40 Herein we report a facile strategy to conjugate both GRGDS and recombinant rat PDGF-A (rPDGF-A) to the injectable HAMC hydrogel. Specifically, thiol-maleimide click chemistry was used to covalently immobilize maleimide–GRGDS and maleimide–streptavidin to thiolated-MC polymer. Taking advantage of the strong and selective interaction between streptavidin and biotin, streptavidin-bound MC was subsequently conjugated with biotinylated rPDGF-A. The amount of immobilized rPDGF-A was quantified using a fluorescent analogue. Rat NSPCs cultured in the HAMC–GRGDS/rPDGF-A hydrogel were tested for their differentiation to oligodendrocytes relative to a series of controls (Fig. 1).
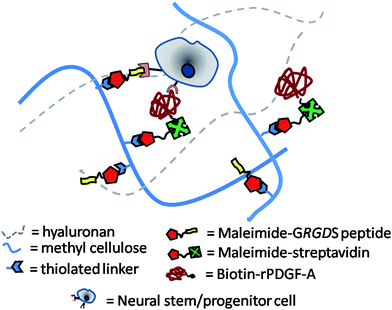 | ||
| Fig. 1 Schematic diagram of a hydrogel composed of hyaluronan (HA) and methyl cellulose (MC) for use as a cell delivery vehicle for neural stem/progenitor cells (NSPCs). MC is chemically modified with a thiolated linker to immobilize recombinant rat platelet-derived growth factor (rPDGF-A) and the cell-adhesive peptide sequence GRGDS to enable the control of NSPC fate. | ||
Materials and methods
Synthesis of sulfhydryl methyl cellulose (4) for peptide and streptavidin conjugation
Methyl cellulose (MC, 1, Mw 310 kg mol−1, degree of substitution (or number of hydroxyl groups that were methylated): 1.8 of 3 hydroxyl groups per glucoside unit, Shin-Etsu Metolose SM-4000, Japan) was chemically converted to its carboxylated derivative (carboxylated-MC, 2) using a previously described protocol.41,42 The product was purified by extensive dialysis (12–14 kDa MWCO membrane) at 4 °C against 0.1 M NaCl (3 times per day, 2 days) and then deionized (DI) water (3 times per day, 2 days). Water was removed from the final polymer by lyophilization to afford 2 as an amorphous white solid. Carboxylated-MC (2, 250 mg) was dissolved in 0.01 M phosphate buffer (150 mL) at pH 7.0 at room temperature, and 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM, 750 mg, 2.7 mmol) was added and vigorously stirred for 20 min. 3,3′-Dithiobis(propionic dihydrazide) (3, 750 mg, 3.2 mmol) was then added and the mixture was stirred overnight at room temperature.43 The reaction mixture was then dialyzed in 0.1 M NaCl (12–14 kDa MWCO membrane, 3 times per day, 1 day) to remove most of the unreacted reagents. The pH of the reaction mixture was then adjusted to pH 8.0 using 0.1 M NaOH and dithiothreitol (DTT, 1.0 g, 6.5 mmol) was added at room temperature for 4 h to afford sulfhydryl-MC (4). Purification was achieved by extensive dialysis with diluted HClaq (pH 5, 3 times per day, 2 days), followed by DI water (3 times per day, 2 days). Lyophilization was used to concentrate polymer 4. Quantification of freely reactive sulfhydryl groups present in 4 was achieved using Ellman's method.44 Cysteine and unmodified methyl cellulose (1) were used for the standard calibration curve. For 1H NMR analyses, samples were dissolved in D2O or DMSO-d6 and spectra were acquired on a Varian Mercury 400 MHz NMR spectrometer at 298 K.Conjugation of maleimide–streptavidin (5) and N-ethylhydroxy maleimide (6a) or synthetic peptide maleimide–GRGDS (6b) to thiolated methyl cellulose (4)
Conjugation of maleimide–streptavidin (5, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol maleimide
1 mol maleimide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol streptavidin, Mw 58 kDa, Sigma-Aldrich) to sulfhydryl-MC (4) was achieved by dissolving 4 (100 mg, 18.5 μmol of reactive thiols) in a solution of 0.1 M phosphate buffer (pH 7.4, 100 mL) that was previously purged for 20 min with inert N2 gas. The solution was cooled to 4 °C and maleimide–streptavidin (5, 1.1 mg, 18.5 nmol) was added and stirred overnight at 4 °C. N-Ethylhydroxy maleimide (6a, 13 mg, 92.5 μmol) or synthetic peptide maleimide–GRGDS45 (6b, 59 mg, 92.5 μmol) was then added to the reaction mixture and mixed overnight at 4 °C. Unbound maleimide-containing reagents were removed by extensive dialysis (100 kDa MWCO membrane) against 0.10 M phosphate buffer (pH 7.0, 3 times per day, 2 days), followed by DI water for another 2 days (3 times per day). Chemically modified-MC (7, 8) was then lyophilized to afford amorphous white solids. Quantification of the amount of amino acids present in 7 and 8 was performed by amino acid analysis. Peptide-containing MC polymers were dissolved in DD H2O and hydrolyzed with 6N HCl at 110 °C for 24 h. Excess HCl was removed by evaporation and the hydrolyzed amino acids were then derivatized at using phenylisothiocyanate (PITC). The phenyl-labeled amino acids were then separated and quantified using HPLC with the following parameters: Acquity UPLC BEH C18 column (2.1 mm × 10 cm); 48 °C column temperature, UV detection at 254 nm. A known amount of streptavidin and GRGDS peptide in a solution of unmodified MC was used as a standard.
mol streptavidin, Mw 58 kDa, Sigma-Aldrich) to sulfhydryl-MC (4) was achieved by dissolving 4 (100 mg, 18.5 μmol of reactive thiols) in a solution of 0.1 M phosphate buffer (pH 7.4, 100 mL) that was previously purged for 20 min with inert N2 gas. The solution was cooled to 4 °C and maleimide–streptavidin (5, 1.1 mg, 18.5 nmol) was added and stirred overnight at 4 °C. N-Ethylhydroxy maleimide (6a, 13 mg, 92.5 μmol) or synthetic peptide maleimide–GRGDS45 (6b, 59 mg, 92.5 μmol) was then added to the reaction mixture and mixed overnight at 4 °C. Unbound maleimide-containing reagents were removed by extensive dialysis (100 kDa MWCO membrane) against 0.10 M phosphate buffer (pH 7.0, 3 times per day, 2 days), followed by DI water for another 2 days (3 times per day). Chemically modified-MC (7, 8) was then lyophilized to afford amorphous white solids. Quantification of the amount of amino acids present in 7 and 8 was performed by amino acid analysis. Peptide-containing MC polymers were dissolved in DD H2O and hydrolyzed with 6N HCl at 110 °C for 24 h. Excess HCl was removed by evaporation and the hydrolyzed amino acids were then derivatized at using phenylisothiocyanate (PITC). The phenyl-labeled amino acids were then separated and quantified using HPLC with the following parameters: Acquity UPLC BEH C18 column (2.1 mm × 10 cm); 48 °C column temperature, UV detection at 254 nm. A known amount of streptavidin and GRGDS peptide in a solution of unmodified MC was used as a standard.
Protein expression of rPDGF-A and biotinylation using biotin ligase
A plasmid was constructed and purchased from Genscript that coded for a hexahistidine tag and a BirA recognition sequence (GLNDIFEAQKIEWHE)46 at the N-terminus, followed by a flexible spacer (EFPKPSTPPGSSGGAP)47 and the rat PDGF-A sequence at the C-terminus in a pET21a(+) plasmid. The recombinant rat PDGF-A plasmid was transformed into BL21 E.coli, which were subsequently cultured in 1.8 L of Terrific Broth media, 4 mL L−1 glycerol and 100 μg mL−1 ampicillin at 37 °C to an optical density of 0.8. Protein expression was induced with 190 μg mL−1 isopropyl-1-thio-β-D-galactopyranoside for 6 h at 37 °C. Cells were harvested and lysed by mixing with denaturing buffer A (6.0 M guanidinium chloride, 100 mM NaH2PO4, 10 mM Tris, 10 mM imidazole, pH 8.0) at room temperature overnight. The mixture was centrifuged and to the supernatant was added Ni-NTA resin (2 mL) to bind to hexahistidine-containing rPDGF-A. The resin was washed several times with denaturing buffer A to remove unbound protein, and rPDGF-A was eluted from the resin using an acidic denaturing buffer (6 M Guanidine HCl, 200 mM glacial acetic acid). Denatured rPDGF-A was then refolded (via dialysis with a 2 kDa MWCO membrane in 0.05 M Tris, 0.154 M NaCl, 0.1 M arginine buffer, pH 8.5, 4 °C, 2 times a day, 4 days) to yield 11 mg rPDGF-A per 1.8 L of terrific broth media. Characterization of the expressed protein was determined using MALDI-TOF. Biotinylation was performed using a biotin-protein ligase kit (EC 6.3.4.15, Avidity) according to the manufacturer's protocol with the following modifications: excess D-biotin (0.47 μmol) and 18 μg of biotin ligase enzyme were added per mg of rPDGF-A, and incubated in a 30 °C water bath overnight. Biotin–rPDGF-A was purified by dialysis using the same conditions as for rPDGF-A.Bioactivity assay of biotinylated-rPDGF-A
A 16-well chamber slide was coated with poly-D-lysine (50 μg mL−1) overnight. Wells were washed with sterile double de-ionized (DD) water. Laminin (5 μg mL−1, from Engelbreth-Holm-Swarm murine sarcoma basement membrane, Sigma) was then added to each well and allowed to adsorb onto the well surface overnight at room temperature. Excess unadsorbed laminin was removed by washing with sterile PBS. Neurobasal media (GIBCO) containing 2% B27 supplement (Invitrogen), 1% L-glutamine (Sigma), 1% penicillin/streptomycin (Sigma), 3 ng mL−1 of epidermal growth factor (EGF), 3 ng mL−1 basic fibroblast growth factor (FGF-2), and 0.3 ng mL−1 heparin were added to each well. Biotinylated rPDGF-A or commercially available recombinant rat PDGF-AA (R&D Systems) was then added to each well at the desired concentration. Neural stem/progenitor cells derived from the subventricular zone of adult Wistar rat brain, as previously described,48 were then seeded at a density of 5000 cells per well, and incubated at 37 °C and 95% CO2 for 5 days.Immobilization of rPDGF-A to streptavidin–methyl cellulose
MC–streptavidin (7 or 8) was dissolved in 0.05 M Tris, 0.1 M NaCl buffer, pH 8.5 and cooled to 4 °C. A solution of biotinylated-rPDGF-A was added and mixed overnight at 4 °C. Upon completion of the reaction, unbound biotinylated-rPDGF-A was removed by extensive dialysis (100 kDa MWCO) in 0.05 M Tris, 0.1 M NaCl, 0.1 M arginine buffer, pH 8.5, followed by several times with distilled water and then with DD water to remove excess salts. The dialyzed mixture was sterile-filtered through a 0.22 μm syringe filter and lyophilized. To quantify the amount of biotinylated-rPDGF-A that was immobilized to MC–streptavidin, biotinylated-rPDGF-A was fluorescently labelled with N-hydroxysuccinimide-Alexa Fluor 633, as previously described.49 Fluorescently tagged biotinylated-rPDGF-A was immobilized to MC–streptavidin as described above. As an adsorption control reaction, fluorescent-rPDGF-A was also incubated with unmodified methyl cellulose. After extensive dialysis, the fluorescence was measured and the amount of protein immobilized quantified.Bioactivity assay for HAMC gels
Hyaluronan and methyl cellulose solutions were sterile-filtered through a 0.22 μm syringe filter and lyophilized. 0.6 wt% per volume of each of HA and MC were dissolved overnight at 4 °C in neurobasal medium with 3 ng mL−1 each of EGF and FGF-2 and 0.3 ng mL−1 of heparin. Cells in neurobasal medium were added to the gels, resulting in a final concentration of 0.5 wt % of each of HA and MC and an even distribution of cells within the gel. 160 μL of this gel was transferred into 8-well chamber slides, and allowed to incubate at 37 °C for 7 days. Cell viability and immunocytochemistry for RIP+ cells were performed and images taken with an Olympus FV1000 confocal microscope. For each sample, three images at 10× (for cell viability assay) and 20× (for immunocytochemistry) magnifications were taken at random locations in each well and counted using ImageJ. The number of cells per well was calculated based upon the surface area covered by the images and extrapolated to the surface area of the well. Three replicates were performed for each sample.Cell viability assay
Cell viability was determined using a LIVE/DEAD cell viability assay kit (Life Technologies). Cells were cultured in HAMC gels as described above. A solution containing calcein AM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution, for live cells), ethidium homodimer (1
500 dilution, for live cells), ethidium homodimer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 dilution, for dead cells) and DAPI (1
250 dilution, for dead cells) and DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 dilution, for cell nuclei) in PBS was prepared, and 30 μL of this solution was added to each well containing cells. Cells were then incubated for 30 min at 37 °C and images were immediately taken.
300 dilution, for cell nuclei) in PBS was prepared, and 30 μL of this solution was added to each well containing cells. Cells were then incubated for 30 min at 37 °C and images were immediately taken.
Immunocytochemistry
For all immunocytochemical procedures, the appropriate controls were obtained by omission of the primary antibody (RIP, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 dilution in 10% FBS, DSHB, University of Iowa, Iowa City).50 Cells were fixed with a solution of PBS containing 4% paraformaldehyde for 20 min (on glass) or 2 h (in the hydrogel) at room temperature and then washed with PBS 3 times. Non-specific binding was then blocked by treating cells with a solution of PBS containing 10% FBS at room temperature for 2 h. A solution of the primary antibody in 10% FBS was added for 1 h at room temperature (on glass) or overnight at 4 °C (in hydrogel), followed by extensive washing with a solution of PBS containing 1% of FBS. Samples were then treated with the secondary antibody (Cy3 Jackson, 1
5 dilution in 10% FBS, DSHB, University of Iowa, Iowa City).50 Cells were fixed with a solution of PBS containing 4% paraformaldehyde for 20 min (on glass) or 2 h (in the hydrogel) at room temperature and then washed with PBS 3 times. Non-specific binding was then blocked by treating cells with a solution of PBS containing 10% FBS at room temperature for 2 h. A solution of the primary antibody in 10% FBS was added for 1 h at room temperature (on glass) or overnight at 4 °C (in hydrogel), followed by extensive washing with a solution of PBS containing 1% of FBS. Samples were then treated with the secondary antibody (Cy3 Jackson, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution in 10% FBS) for 1 h (on glass) or 4 h (in hydrogel) and then washed extensively with 1% FBS. Finally, cell nuclei were counterstained with Vectashield mounting media containing DAPI (Invitrogen).
500 dilution in 10% FBS) for 1 h (on glass) or 4 h (in hydrogel) and then washed extensively with 1% FBS. Finally, cell nuclei were counterstained with Vectashield mounting media containing DAPI (Invitrogen).
Statistical analysis
Data for cell studies were analyzed by one-way ANOVA with Bonferroni-Holm post hoc analysis using Graph Pad. For all studies, p-values of less than 0.05 were considered to be statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).Results and discussion
Chemical modification of methyl cellulose with streptavidin and GRGDS peptide
Methyl cellulose (MC) of HAMC is resorbed more slowly than hyaluronan (HA) in vivo,51 and thus MC was chemically modified with GRGDS and streptavidin (for eventual modification with biotin–rPDGF-A). To synthesize MC–GRGDS and MC–streptavidin, MC was first modified to introduce a reactive thiol42 that would react with maleimide–GRGDS and maleimide–streptavidin by Michael-type addition. Methyl cellulose was first functionalized with reactive carboxylates (2), which were then activated with DMT-MM43 and coupled to 3,3′-dithiobis(propionic dihydrazide) (Scheme 1). Reductive cleavage of the disulfide bond with dithiothreitol (DTT) afforded the reactive thiolated methyl cellulose (4). Solution state 1H NMR confirms the presence of the ethylene groups of the propionyl hydrazide linker (Fig. 2C). Using Ellman's assay,44 we determined that there were 185 μmol of thiol per gram of methyl cellulose, which correlates to approximately one reactive thiol group per 30 glucoside units.![Synthetic scheme for the conjugation of biotin–rPDGF-A and GRGDS peptide to methyl cellulose (MC, 1). The hydroxyls of MC (1) were derivatized to carboxylates (2), which were subsequently coupled with a dithiobis(propionic dihydrazide) linker (3). Free thiols (4, 185 μmolSH per gram) were formed upon reduction with dithiothreitol (DTT) to enable covalent immobilization to maleimide-containing streptavidin (5, 185 nmol per gram) and N-ethylhydroxy maleimide (6a) or bioactive GRGDS peptide (6b, 170 μmol per gram). Conjugation of biotin–rPDGF-A to MC–streptavidin (7,8) results in MC–rPDGF-A (9) and MC–GRGDS/rPDGF-A (10). Reagents and Conditions: (a) Bromo acetic acid (BrCH2CO2H, 10 equiv.), 1.5 M NaOH, 4 °C, overnight; (b) [H2N–NHCO(CH2)2S–]2 (3,21 mM), DMT-MM (18 mM), 0.01 M PBS, pH 7.0, 22 °C, overnight; (c) DTT (43 mM), 0.01 M PBS, pH 8.0, 22 °C, 8 h; (d) maleimide–streptavidin (5, 0.1 mol%), 0.01 M phosphate buffer, pH 7.4, 4 °C, overnight; (e) N-ethylhydroxy maleimide (6a, 5.0 equiv.) or maleimide–GRGDS (6b, 5.0 equiv.), 0.1 M phosphate buffer, pH 7.4, 4 °C, overnight (f) biotin–rPDGF-A (8.0 equiv.), 0.05 M Tris, 0.154 M NaCl, pH 8.5, 4 °C, overnight. DMT-MM = 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride; DTT = dithiothreitol; GRGDS = gly–arg–gly–asp–ser peptide.](/image/article/2012/JM/c2jm33680d/c2jm33680d-s1.gif) | ||
| Scheme 1 Synthetic scheme for the conjugation of biotin–rPDGF-A and GRGDS peptide to methyl cellulose (MC, 1). The hydroxyls of MC (1) were derivatized to carboxylates (2), which were subsequently coupled with a dithiobis(propionic dihydrazide) linker (3). Free thiols (4, 185 μmolSH per gram) were formed upon reduction with dithiothreitol (DTT) to enable covalent immobilization to maleimide-containing streptavidin (5, 185 nmol per gram) and N-ethylhydroxy maleimide (6a) or bioactive GRGDS peptide (6b, 170 μmol per gram). Conjugation of biotin–rPDGF-A to MC–streptavidin (7,8) results in MC–rPDGF-A (9) and MC–GRGDS/rPDGF-A (10). Reagents and Conditions: (a) Bromo acetic acid (BrCH2CO2H, 10 equiv.), 1.5 M NaOH, 4 °C, overnight; (b) [H2N–NHCO(CH2)2S–]2 (3,21 mM), DMT-MM (18 mM), 0.01 M PBS, pH 7.0, 22 °C, overnight; (c) DTT (43 mM), 0.01 M PBS, pH 8.0, 22 °C, 8 h; (d) maleimide–streptavidin (5, 0.1 mol%), 0.01 M phosphate buffer, pH 7.4, 4 °C, overnight; (e) N-ethylhydroxy maleimide (6a, 5.0 equiv.) or maleimide–GRGDS (6b, 5.0 equiv.), 0.1 M phosphate buffer, pH 7.4, 4 °C, overnight (f) biotin–rPDGF-A (8.0 equiv.), 0.05 M Tris, 0.154 M NaCl, pH 8.5, 4 °C, overnight. DMT-MM = 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride; DTT = dithiothreitol; GRGDS = gly–arg–gly–asp–ser peptide. | ||
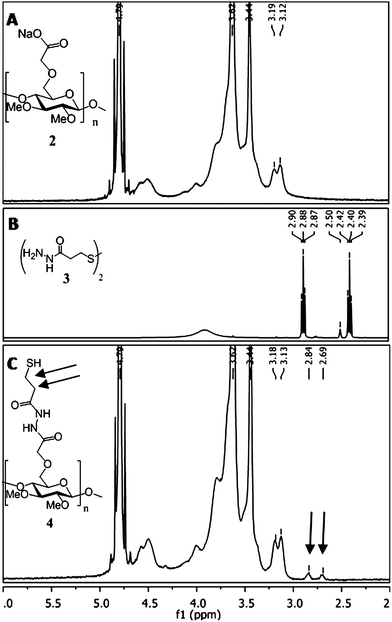 | ||
| Fig. 2 Characterization of methyl cellulose functionalized with a thiolated linker.1H NMR spectra of (A) carboxylated-methyl cellulose (2, in D2O) and (B) 3,3′-dithio-dipropanylhydrazide (3, in (CD3)2SO). (2) and (3) were conjugated together using the coupling reagent DMT-MM to afford sulfhydryl-methyl cellulose (4). (C) 1H NMR spectra of (4) in D2O, showing the inclusion of the ethylene protons (2.84 and 2.69 ppm, black arrows) of the hydrazide linker to the polysaccharide backbone. | ||
Maleimide–streptavidin (5) was then conjugated to the thiolated methyl cellulose polysaccharide chain (4) (Scheme 1). To enable the conjugation of multiple peptide/protein components to the same polysaccharide chain, limiting amounts of streptavidin (5, 0.1 mol%) relative to reactive thiols present in methyl cellulose were used. Unreacted free thiols could then react with a second desired maleimide-containing component in a second subsequent step. For conjugation of streptavidin only (7), unreacted free thiols were quenched with an excess of N-ethylhydroxy maleimide (6a) in the second step to prevent polymer precipitation or crosslinking via disulfide bonds. The terminal hydroxyl groups had the added benefit of mimicking the hydroxyl groups present in the unmodified methyl cellulose (1).
For immobilization of the GRGDS peptide to the same polysaccharide chain as maleimide–streptavidin (5), maleimide–GRGDS (6b) was added in excess in the second step to react with the remaining, unreacted thiols. Immobilized streptavidin and GRGDS peptide were detected and quantified using amino acid analysis (Fig. 3). Fig. 3A and B show the presence of the desired amino acids in each respective sample. Using a known amount of streptavidin and GRGDS peptide in a solution of unmodified MC as a standard, we were able to quantify the amount of streptavidin and GRGDS peptide conjugated to the polymer (185 nmol and 170 μmol per gram of MC, respectively). To confirm that our results were due to the successful immobilization reactions, we also performed an adsorption control: unmodified methyl cellulose was incubated with streptavidin and GRGDS, dialyzed in the same manner as above, and analyzed by amino acid analysis. Only trace amounts of each were detected (Fig. 3C).
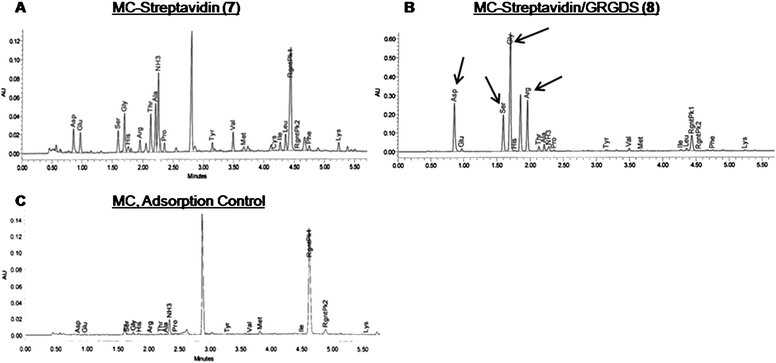 | ||
| Fig. 3 Amino acid HPLC chromatographic traces show (A) streptavidin amino acids of methyl cellulose conjugated with streptavidin and hydroxyethyl groups (7) and (B) GRGDS and streptavidin amino acids of methyl cellulose conjugated with streptavidin and GRGDS (8). To account for the amount of peptide/streptavidin that was adsorbed onto methyl cellulose, (5) and (6b) were dissolved in unmodified methyl cellulose (1), and dialyzed to remove unbound substrates. (C) Spectra shows trace amounts of amino acids that had adsorbed onto (1) following dialysis. | ||
Biotinylation and bioactivity of rPDGF-A
To immobilize recombinant platelet-derived growth factor (rPDGF-A) to the methyl cellulose polymer, we took advantage of the orthogonal binding partners, streptavidin and biotin, which have a kd of 10−15 M.52 With streptavidin covalently conjugated to methyl cellulose (7, 8), we required the biotinylation of rPDGF-A. Although non-specific biotinylation of proteins is routinely performed using conventional chemistry, we were interested in using a site-specific reaction to prevent non-specific modification that could potentially hinder the bioactivity of the expressed rPDGF-A. To this end, we incorporated a 15 amino acid biotin-ligase recognition sequence within the N-terminus of rPDGF-A (Fig. 4A). Upon incubation with the enzyme biotin ligase and excess D-biotin, a biotin moiety was selectively and covalently bound at a single lysine (K) residue within this sequence. MALDI-TOF analysis of rPDGF-A (Fig. 4B) and biotin–rPDGF-A (Fig. 4C) demonstrated the success of the biotinylation reaction. It was observed that for both biotinylated and non-biotinylated protein, the C-terminal arginine residue was cleaved during the MALDI ionization process, which is consistent with previous results of C-terminal arginine residues.53![(A) Site-specific biotinylation of recombinant rat PDGF-A (rPDGF-A) using biotin ligase. Biotin is conjugated specifically to a single lysine residue (K*) within the BirA recognition sequence that is incorporated into the N-terminus of expressed rPDGF-A. MALDI-TOF spectra of (B) expressed rPDGF-A (Calculated for [bn−1 + H2O + NH4]+ 18 390.8 Da, found 18 390.5 Da) and (C) biotinylated rPDGF-A (Calculated for [bn−1 + H2O + NH4]+ 18 617.1 Da, found 18 615.8 Da). An increase in the mass of biotin–rPDGF-A with the loss of a water molecule (226.3 Da) is observed between spectra B and C, indicating the successful biotinylation of a single residue of rPDGF-A.](/image/article/2012/JM/c2jm33680d/c2jm33680d-f4.gif) | ||
Fig. 4 (A) Site-specific biotinylation of recombinant rat PDGF-A (rPDGF-A) using biotin ligase. Biotin is conjugated specifically to a single lysine residue (K*) within the BirA recognition sequence that is incorporated into the N-terminus of expressed rPDGF-A. MALDI-TOF spectra of (B) expressed rPDGF-A (Calculated for [bn−1 + H2O + NH4]+ 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390.8 Da, found 18 390.8 Da, found 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390.5 Da) and (C) biotinylated rPDGF-A (Calculated for [bn−1 + H2O + NH4]+ 18 390.5 Da) and (C) biotinylated rPDGF-A (Calculated for [bn−1 + H2O + NH4]+ 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 617.1 Da, found 18 617.1 Da, found 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 615.8 Da). An increase in the mass of biotin–rPDGF-A with the loss of a water molecule (226.3 Da) is observed between spectra B and C, indicating the successful biotinylation of a single residue of rPDGF-A. 615.8 Da). An increase in the mass of biotin–rPDGF-A with the loss of a water molecule (226.3 Da) is observed between spectra B and C, indicating the successful biotinylation of a single residue of rPDGF-A. | ||
To ensure that the expression and biotinylation of rPDGF-A yielded a bioactive product, its capacity to differentiate rat neural stem/progenitor cells (NSPCs) into oligodendrocytes was compared to that of commercially available recombinant rat PDGF-AA, which has been previously shown to promote differentiation to oligodendrocytes.37 Cells were seeded into wells coated with poly-D-lysine and laminin at an initial density of 5000 cells per well, and allowed to grow for 5 days (Fig. 5). Immunocytochemistry was used to quantify the number of cells that expressed receptor-interaction protein (RIP), which is a specific marker for oligodendrocytes. Fig. 5 shows that in the presence of biotinylated rPDGF-A, the number of oligodendrocytes (7.5 ± 1.5 × 103 per well) is comparable to commercially available PDGF-AA (5.3 ± 1.6 × 103 per well). Statistical analysis shows that these values are not significantly different. Importantly, these values are significantly higher than cells differentiated in the absence of any PDGF-A in the media (1.1 ± 0.7 × 103 per well).
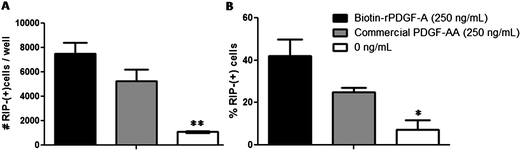 | ||
Fig. 5 Bioactivity of expressed biotinylated rPDGF-A (■) compared to commercially available recombinant rat PDGF-AA (R&D Systems,  ) on rat neural stem/progenitor cells (NSPCs). Expressed biotin–rPDGF-A and commercially available PDGF-AA significantly increase the number and percentage of RIP+ oligodendrocytes compared to samples in the absence of PDGF-A (□). (A) Number and (B) percentage of RIP+ oligodendrocytes are shown, as determined by immunocytochemistry. Cells were plated at 5 × 103 cells per well into a 16-well chamber slide (coated with poly-D-lysine and laminin) for 5 days with or without PDGF-A. Values are shown as mean ± standard deviation (n = 3). One way ANOVA was performed, asterisks represent significant differences compared to 250 ng mL−1 of commercial PDGF-AA and expressed rPDGF-A (*p < 0.05, **p < 0.01). ) on rat neural stem/progenitor cells (NSPCs). Expressed biotin–rPDGF-A and commercially available PDGF-AA significantly increase the number and percentage of RIP+ oligodendrocytes compared to samples in the absence of PDGF-A (□). (A) Number and (B) percentage of RIP+ oligodendrocytes are shown, as determined by immunocytochemistry. Cells were plated at 5 × 103 cells per well into a 16-well chamber slide (coated with poly-D-lysine and laminin) for 5 days with or without PDGF-A. Values are shown as mean ± standard deviation (n = 3). One way ANOVA was performed, asterisks represent significant differences compared to 250 ng mL−1 of commercial PDGF-AA and expressed rPDGF-A (*p < 0.05, **p < 0.01). | ||
The ability of PDGF-AA to induce differentiation of NSPCs into oligodendrocytes has been extensively studied,54–58 and occurs via binding to PDGF receptor-α (PDGFR-α). This causes receptor dimerization and triggers intracellular autophosphorylation, which activates phosphatidylinositol 3-kinase (PI3K)57 and mitogen-activated protein kinase (MAPK)57,58 pathways, and upregulation of Olig2 gene expression.58 As our results show that the number of RIP+ oligodendrocytes are comparable between cells cultured in media containing biotinylated rPDGF-A and commercial PDGF-AA, we can conclude that both the expressed rPDGF-A is bioactive and the incorporation of the biotin-ligase recognition sequence and biotinylation at the N-terminus of rPDGF-A is unlikely to affect receptor-binding and dimerization at 250 ng mL−1 of rPDGF-A. Thus the increased number of RIP+ oligodendrocytes in the presence of rPDGF-A (compared to cells cultured in the absence of any PDGF) is attributed to increased differentiation of NSPCs.
Immobilization of biotinylated-rPDGF-A to streptavidin–methyl cellulose
Recombinant PDGF-A-modified MC (10, 11) was synthesized by incubating an excess of biotin–rPDGF-A with MC–streptavidin (7, 8) (Scheme 1). For quantification purposes, Alexa Fluor 633-modified biotin–rPDGF-A49 was immobilized to MC–streptavidin. To ensure that predominantly covalently bound rPDGF-A was quantified, unbound rPDGF-A was removed from the MC polymer by extensive dialysis. Interestingly, the addition of 0.1 M arginine in the dialysis buffer was important to the effective removal of adsorbed rPDGF-A. Overall, 0.79 μg of rPDGF-A was bound per mg of MC polymer. Previous efforts to immobilize PDGF-AA to hydrogels yielded only 0.175 μg per mg of agarose polymer,37 demonstrating the benefit of the current approach for chemical conjugation.Bioactivity of immobilized rPDGF-A in the HAMC gel
Previous work36 has shown that the physical blend of 0.5/0.5 wt% HAMC biomaterial is not cytotoxic. To ensure chemical modifications of methyl cellulose did not affect cell viability compared to unmodified HAMC, a cell viability assay was performed to compare the effect of chemical modification. There was no significant difference between the percentage of live cells cultured after 7 days between unmodified HAMC and HAMC–rPDGF-A. Therefore, the growth factor modification did not affect cell viability.To assay for the ability of the chemically modified HAMC hydrogels to induce differentiation of NSPCs into oligodendrocytes, an in vitro bioactivity assay was performed. NSPCs were encapsulated within the HAMC–GRGDS/rPDGF-A hydrogel for 7 days of incubation and then characterized for differentiation to RIP+ cells by immunocytochemistry. The RIP monoclonal antibody was used to specifically target both immature and mature oligodendrocytes.50,59,60
Fig. 6A and B show that NSPCs encapsulated within a 0.5/0.5 wt% HAMC–rPDGF-A hydrogel differentiated into 2.4 ± 0.3 × 104 RIP+ cells per well and 40 ± 1% RIP+ cells which is significantly greater differentiation than NSPCs encapsulated in unmodified HAMC (8.7 ± 0.5 × 103 RIP+ cells per well and 23 ± 2% RIP+ cells). Importantly, NSPCs that were cultured in HAMC–rPDGF-A showed comparable differentiation to NSPCs cultured in unmodified HAMC hydrogels with the same concentration of soluble rPDGF-A (2.1 ± 0.2 × 104 RIP+ cells per well and 42 ± 7% RIP+ cells). This is consistent with results from a previous study that reported rat NSPCs cultured with the same concentration of soluble or agarose-immobilized PDGF-AA exhibited the same oligodendrocyte differentiation.37 To ensure that the differentiation observed resulted from chemically immobilized rPDGF-A, we conducted an adsorption control where we observed significantly fewer RIP+ cells than in the presence of either immobilized or soluble rPDGF-A: 7.1 ± 0.9 × 103 RIP+ cells per well and 23 ± 1% RIP+ cells.
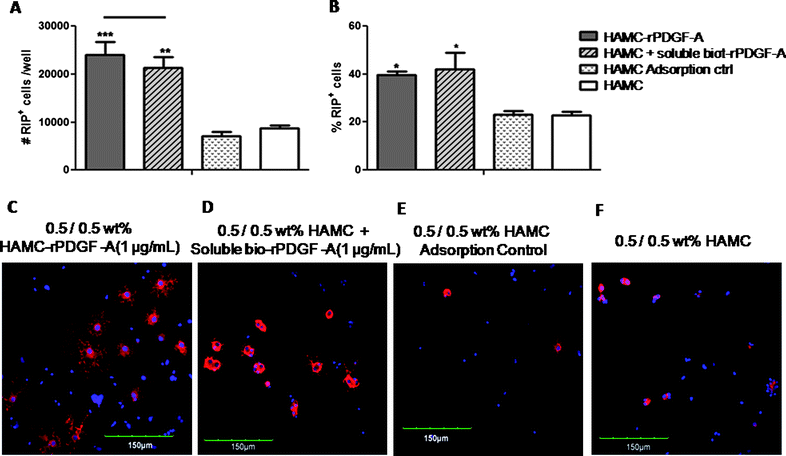 | ||
Fig. 6 Effect of rPDGF-A immobilization to HAMC hydrogels on the differentiation of rat NSPCs. Cells were pre-mixed into the HAMC hydrogel, and then plated at 4 × 104 cells per well into an uncoated 8-well chamber slide for 7 days. Immobilization of rPDGF-A promotes oligodendrocyte differentiation. (A) Number and (B) percentage of RIP+ oligodendrocytes are shown, as determined by immunocytochemistry. Immobilization of rPDGF-A to HAMC (9,  ) show a greater number (***p < 0.001) and percentage (*p < 0.05) of RIP+ cells compared to HAMC in the absence of rPDGF-A (□). Similarly, HAMC with soluble rPDGF-A (▨) had a significantly greater number (**p < 0.01) and percentage (*p < 0.05) of RIP+ cells compared to HAMC in the absence of rPDGF-A (□). However, HAMC immobilized with rPDGF-A ( ) show a greater number (***p < 0.001) and percentage (*p < 0.05) of RIP+ cells compared to HAMC in the absence of rPDGF-A (□). Similarly, HAMC with soluble rPDGF-A (▨) had a significantly greater number (**p < 0.01) and percentage (*p < 0.05) of RIP+ cells compared to HAMC in the absence of rPDGF-A (□). However, HAMC immobilized with rPDGF-A ( ) was not significantly different than a mixture of soluble rPDGF-A in HAMC (▨) in terms of RIP expression. Values are shown as mean ± standard deviation (n = 3). One way ANOVA was performed for all samples. (C-F) Confocal images of rat NSPCs after encapsulation in 0.5/0.5 wt % HAMC gels for 7 days. Cells were stained for anti-RIP (for oligodendrocytes, red) and counterstained with DAPI (for cell nuclei, blue). Immobilization of rPDGF-A to HAMC (C) shows distinct cell morphology compared to HAMC in the absence of immobilized rPDGF-A (E and F). ) was not significantly different than a mixture of soluble rPDGF-A in HAMC (▨) in terms of RIP expression. Values are shown as mean ± standard deviation (n = 3). One way ANOVA was performed for all samples. (C-F) Confocal images of rat NSPCs after encapsulation in 0.5/0.5 wt % HAMC gels for 7 days. Cells were stained for anti-RIP (for oligodendrocytes, red) and counterstained with DAPI (for cell nuclei, blue). Immobilization of rPDGF-A to HAMC (C) shows distinct cell morphology compared to HAMC in the absence of immobilized rPDGF-A (E and F). | ||
In addition, confocal images (Fig. 6C) show that NSPCs cultured in HAMC–rPDGF-A possessed web-like cellular processes characteristic of oligodendrocytes, which are similar to those NSPCs cultured in soluble rPDGF-A (Fig. 6D), yet different from those cultured in adsorption controls (Fig. 6E) and unmodified HAMC (Fig. 6F).50 Importantly, the immobilized rPDGF-A is as bioactive as soluble rPDGF-A, thereby allowing us to pursue HAMC–rPDGF-A as a cell delivery vehicle. Immobilization of rPDGF-A to the HAMC biomaterial will ensure co-localization with cells within the injection site and will prevent rPDGF-A from diffusing away upon transplantation into the injury site.
Several studies have demonstrated the importance of co-immobilizing cell-adhesive and cell-differentiating factors to a biomaterial substrate to guide cell fate;37,61 however, only recently has the importance of the proximity of these factors been elucidated.27,28 To gain further insight, we immobilized both GRGDS and rPDGF-A to the HAMC hydrogel and compared NSPC differentiation in HAMC where both GRGDS and rPDGF-A were immobilized on the same MC (10) (HAMC–GRGDS/rPDGF-A) vs. HAMC where GRGDS and rPDGF-A were immobilized on separate MC chains (8 and 9) that were then simply mixed together with HA (HAMC–GRGDS/MC–rPDGF-A). By conjugating GRGDS and rPDGF-A to separate MC polysaccharide chains, we expected that the average distance between these two components would be increased.27 We questioned whether a change in the proximity between the peptide and growth factor would affect cell differentiation. NSPCs cultured within HAMC–GRGDS/rPDGF-A yielded 3.1 ± 0.4 × 104 RIP+ cells per well and 53 ± 10% RIP+ cells whereas NSPCs cultured in HAMC–GRGDS/MC–rPDGF-A yielded 2.1 ± 0.6 × 104 RIP+ cells per well and 33 ± 6% RIP+ cells (Fig. 7A and B, respectively). While the numbers of oligodendrocytes for these two formulations were not statistically different, the percentage of oligodendrocytes was significantly higher when both GRGDS and rPDGF-A were conjugated to the same polysaccharide chain. Moreover, HAMC–GRGDS/rPDGF-A had significantly greater numbers and percent of RIP+ cells compared to HAMC–GRGDS, where we observed 1.1 ± 0.2 × 104 RIP+ cells per well and 26 ± 0%.
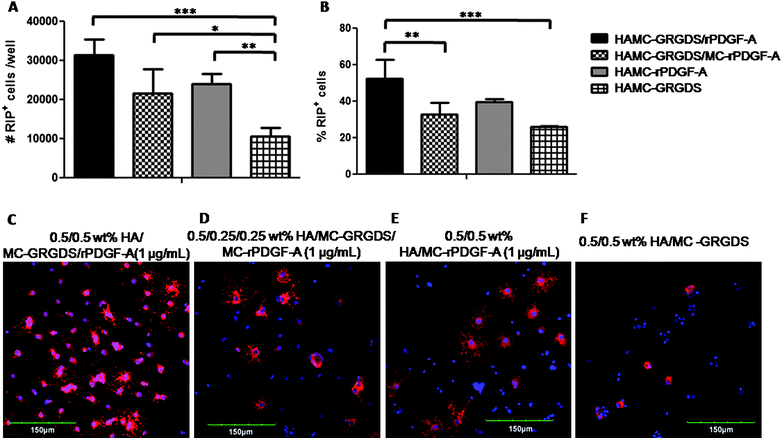 | ||
Fig. 7 Effect of immobilization of GRGDS peptide to HAMC hydrogels on the differentiation of rat NSPCs. Cells were pre-mixed into the HAMC gel, and then plated at 4 × 104 cells per well into an uncoated 8-well chamber slide for 7 days prior to characterization. Co-immobilization of rPDGF-A and GRGDS to the same MC polysaccharide chain (10, ■) further enhances the effects of rPDGF-A and promotes oligodendrocyte differentiation. (A) Number and (B) percentage of RIP+ oligodendrocytes are shown, as determined by immunocytochemistry. A physical mixture of rPDGF-A and GRGDS immobilized on separate polysaccharide chains (![[square tinted (heavy)]](https://www.rsc.org/images/entities/char_e069.gif) ) showed a lower percentage of RIP+ cells compared to HAMC co-immobilized with rPDGF-A and GRGDS on the same chain (10, ■), while the number of RIP+ cells were not significantly different. Values are shown as mean ± standard deviation (n = 3). One way ANOVA was performed for all samples; *p < 0.05, **p < 0.01, ***p < 0.001. (C-F) Confocal images of rat NSPCs after encapsulation in 0.5/0.5 wt % HAMC gels for 7 days. Cells were stained for anti-RIP (for oligodendrocytes, red) and counterstained with DAPI (for cell nuclei, blue). ) showed a lower percentage of RIP+ cells compared to HAMC co-immobilized with rPDGF-A and GRGDS on the same chain (10, ■), while the number of RIP+ cells were not significantly different. Values are shown as mean ± standard deviation (n = 3). One way ANOVA was performed for all samples; *p < 0.05, **p < 0.01, ***p < 0.001. (C-F) Confocal images of rat NSPCs after encapsulation in 0.5/0.5 wt % HAMC gels for 7 days. Cells were stained for anti-RIP (for oligodendrocytes, red) and counterstained with DAPI (for cell nuclei, blue). | ||
These data suggest that decreasing the distance between GRGDS and rPDGF-A enhances the differentiation of NSPCs into oligodendrocytes. The close association between the cell surface receptors for these ligands (integrin62 and PDGFR-α,56 respectively) may cause a synergistic signal activation63–65 to increase cell differentiation.54,57,66–69 Ffrench-Constant and co-workers have previously reported57,66,67 that the proliferation of oligodendrocyte precursor cells is promoted by an adhesion-dependent phosphatidylinositol 3-kinase (PI3K) pathway, which is stimulated by integrin activation in an integrin-PDGF-AA receptor complex. Our results are also consistent with recent work27 published by Hubbell and co-workers, who reported that the use of fibrin hydrogels containing an integrin-binding and a growth factor-binding domain on the same polypeptide chain significantly increased sprouting and bone regeneration of smooth muscle cells and mesechymal stem cells, respectively, compared to hydrogels that incorporated these two components together, but on separate polypeptide chains. Synergistic signal activation between peptide and differentiating factor may account for the increased differentiation of rat NSPCs to oligodendrocytes observed for HAMC–GRGDS/r-PDGF-A where GRGDS and rPDGF-A are immobilized on the same MC chain vs. HAMC–GRGDS/MC–rPDGF-A where they are immobilized on separate MC chains.
Herein, we demonstrated that immobilization of both cell adhesive peptide, GRGDS, and oligodendrocyte-differentiating factor, rPDGF-A, to an injectable hydrogel composed of HA and MC significantly increased the number and percent of NSPCs that preferentially differentiated to RIP+ oligodendrocytes compared to unmodified HAMC and HAMC–GRGDS. Previous reports have demonstrated increased cell viability of transplanted stem cells into the subretinal space when delivered in HAMC vs. in saline.36 Together, these properties suggest that HAMC is a promising cell delivery vehicle for stem cell therapy. As demyelination is a major pathological consequence following spinal cord injury, we hypothesize that HAMC–GRGDS/rPDGF-A will enhance both survival and control the differentiation of transplanted NSPCs into oligodendrocytes which will promote functional repair.
Conclusions
By taking advantage of thiol maleimide and biotin–streptavidin bioorthogonal chemistry, we designed a methodology to immobilize peptides and proteins to an injectable hydrogel for use as a cell delivery vehicle. We demonstrated that the immobilization of both the cell-adhesive peptide, GRGDS, and the cytokine platelet-derived growth factor, rPDGF-A, to the HAMC hydrogel increases the differentiation of rat NSPCs into oligodendrocytes. This lays the framework for future studies of these biomaterials for cell delivery.Acknowledgements
The authors thank Dr Ying Fang Chen for the isolation and propagation of neural stem/progenitor cells, Drs Ryan Wylie and Nic Liepzig for help with protein design and expression, Katarina Vulic, Nuno Silva, Drs Shawn Owen and Tasneem Zahir for helpful discussions, Rey Interior and Li Zhang from the Advanced Protein Technology Centre at Toronto's Hospital for Sick Children for amino acid and MALDI-TOF analyses. We are grateful to funding from: the Ontario Ministry of Research and Innovation (post-doctoral fellowship to RYT), the Ontario Neurotrauma Foundation (post-doctoral fellowship to MJC) and Stem Cell Network (MJC); the Canadian Institute of Health Research and the Natural Sciences and Engineering Research Council (MSS).References
- M. E. Schwab and D. Bartholdi, Physiol. Rev., 1996, 76, 319–368 CAS.
- L. H. Sekhon and M. G. Fehlings, Spine, 2001, 26, S2–S12 CrossRef CAS.
- C. H. Tator, Brain Pathol., 1995, 4, 407–413 CrossRef.
- T. B. Jones, E. E. McDaniel and P. G. Popovich, Curr. Pharm. Des., 2005, 11, 1223–1236 CrossRef CAS.
- P. G. Popovich and T. B. Jones, Trends Pharmacol. Sci., 2003, 24, 13–17 CrossRef CAS.
- P. G. Popovich, P. J. Horner, B. B. Mullin and T. S. Bradford, Exp. Neurol., 1996, 142, 258–275 CrossRef CAS.
- E. D. Hall, P. A. Yonkers, P. K. Andrus, J. W. Cox and D. K. Anderson, J. Neurotrauma, 1992, 9(Suppl), S425–S442 Search PubMed.
- B. Wu and X. Ren, J. Neurotrauma, 2009, 26, 1847–1856 CrossRef.
- M. Mekhail, G. Almazan and M. Tabrizian, Prog. Neurobiol., 2012, 96, 322–339 CrossRef CAS.
- J. W. McDonald and V. Belegu, J. Neurotrauma, 2006, 23, 345–359 CrossRef.
- M. C. Raff, A. V. Whitmore and J. T. Finn, Science, 2002, 296, 868–871 CrossRef CAS.
- J. W. Fawcett and R. A. Asher, Brain Res. Bull., 1999, 49, 371–391 CrossRef.
- O. Raineteau and M. E. Schwab, Nat. Rev. Neurosci., 2001, 2, 263–273 CrossRef CAS.
- M. E. Schwab, Science, 2002, 295, 1029–1031 CrossRef CAS.
- D. J. Mooney and H. Vandenburgh, Cell Stem Cell, 2008, 2, 205–213 CrossRef CAS.
- A. Bakshi, C. A. Keck, V. S. Koshkin, D. G. LeBold, R. Siman, E. Y. Snyder and T. K. McIntosh, Brain Res., 2005, 1065, 8–19 CrossRef CAS.
- S. Karimi-Abdolrazaee, E. Eftekharpour, J. Wang, C. M. Morshead and M. G. Fehlings, J. Neurosci., 2006, 26, 3377–3389 CrossRef.
- Q.-L. Cao, R. M. Howard, J. B. Dennison and S. R. Whittemore, Exp. Neurol., 2002, 177, 349–359 CrossRef CAS.
- M. J. Cooke, K. Vulic and M. S. Shoichet, Soft Matter, 2010, 6, 4988–4998 RSC.
- A. M. Parr, Neuroscience, 2008, 155, 760–770 CrossRef CAS.
- V. Patel, Neuropathology, 2010, 27, 789–801 Search PubMed.
- K. Watanabe, M. Nakamura, A. Iwanami, Y. Fujita, Y. Kanemura, Y. Toyama and H. Okano, Dev. Neurosci., 2004, 26, 275–287 CrossRef CAS.
- W. Zhao and J. M. Karp, ChemBioChem, 2009, 10, 2308–2310 CrossRef CAS.
- M. S. Shoichet, Macromolecules, 2010, 43, 581–591 CrossRef CAS.
- H. Kim, T. Zahir, C. H. Tator and M. S. Shoichet, PLoS One, 2011, 6, e21744 CAS.
- D. S. W. Benoit, M. P. Schwartz, A. R. Durney and K. S. Anseth, Nat. Mater., 2008, 10, 816–823 CrossRef.
- M. M. Martino, F. Tortelli, M. Mochizuki, S. Traub, D. Ben-David, G. A. Kuhn, R. Müller, E. Livne, S. A. Eming and J. A. Hubbell, Sci. Transl. Med., 2011, 3, 100ra189 CrossRef.
- S. X. Hsiong, P. Carampin, H.-J. Kong, K.-Y. Lee and D. J. Mooney, J. Biomed. Mater. Res., Part A, 2007, 85A, 145–156 CrossRef.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS.
- E. Ruoslahti, Annu. Rev. Cell Dev. Biol., 1996, 12, 697–715 CrossRef CAS.
- M. D. Pierschbacher and E. Ruoslahti, Nature, 1984, 309, 30–33 CrossRef CAS.
- E. Ruoslahti and M. D. Pierschbacher, Science, 1987, 238, 491–497 CAS.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS.
- D. Gupta, C. H. Tator and M. S. Shoichet, Biomaterials, 2006, 27, 2370–2379 CrossRef CAS.
- F. Bischoff, Clin. Chem., 1972, 18, 869–894 CAS.
- B. G. Ballios, M. J. Cooke, D. V. D. Kooy and M. S. Shoichet, Biomaterials, 2010, 31, 2555–2564 CrossRef CAS.
- Y. Aizawa, N. Leipzig, T. Zahir and M. S. Shoichet, Biomaterials, 2008, 29, 4676–4683 CrossRef CAS.
- J. G. Hu, S. I. Fu, Y. X. Want, X. Y. Jiang and X. F. Wang, Neuroscience, 2008, 151, 138–147 CrossRef CAS.
- Y. Miyamoto, J. Yamauchi and A. Tanoue, J. Neurosci., 2008, 28, 8326–8337 CrossRef CAS.
- W. D. Richardson, N. Pringle, M. J. Mosley, B. Westermark and M. Dubois-Dalcq, Cell, 1988, 53, 309–319 CrossRef CAS.
- G. Hermanson, in Bioconjugate Techniques, Academic Press, San Diego, CA, 2nd edn, 1996, pp. 1–168 Search PubMed.
- K. Vulic and M. S. Shoichet, J. Am. Chem. Soc., 2012, 134, 882–885 CrossRef CAS.
- M. Kunishima, C. Kawachi, J. Morita, K. Terao, F. Iwasaki and S. Tani, Tetrahedron, 1999, 55, 13159–13170 CrossRef CAS.
- G. D. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–72 CrossRef CAS.
- N. S. Silva, M. J. Cooke, R. Y. Tam, N. Sousa, A. J. Salgado, R. L. Reis and M. S. Shoichet, Biomaterials, 2012, 33, 6345–6354 CrossRef CAS.
- D. Beckett, E. Kovaleva and P. J. Schatz, Protein Sci., 1999, 8, 921–929 CrossRef CAS.
- S. M. Deyev, R. Waibel, E. N. Lebedenko, A. P. Schubiger and A. Pluckthun, Nat. Biotechnol., 2003, 21, 1486–1492 CrossRef CAS.
- C. M. Morshead, B. A. Reynolds, C. G. Craig, M. W. McBurney, W. A. Staines and D. Morassutti, Neuron, 1994, 13, 1071–1082 CrossRef CAS.
- R. G. Wylie and M. S. Shoichet, Nat. Mater., 2011, 10, 799–806 CrossRef CAS.
- B. Friedman, S. Hockfield, J. A. Black, K. A. Woodruff and S. G. Waxman, Glia, 1989, 2, 380–390 CrossRef CAS.
- C. E. Kang, P. C. Poon, C. H. Tator and M. S. Shoichet, Tissue Eng., 2009, 15, 595–604 CAS.
- N. M. Green, Adv. Protein Chem., 1975, 29, 85–133 CrossRef CAS.
- M. Dupré, S. Cantel, J. Martinez and C. Enjalbal, J. Am. Soc. Mass Spectrom., 2012, 23, 330–346 CrossRef.
- N. P. Pringle, H. S. Mudhar, E. J. Collarini and W. D. Richardson, Development, 1992, 115, 535–551 CAS.
- J. Andrae, R. Gallini and C. Betsholtz, Genes Dev., 2008, 22, 1276–1312 CrossRef CAS.
- R. A. Seifert, C. E. Hart, P. E. Phillips, J. W. Forstrom, R. Ross, M. J. Murray and D. F. Bowen-Pope, J. Biol. Chem., 1989, 264, 8771–8778 CAS.
- H. Colognato, W. Baron, V. Avellana-Adalid, J. B. Relvas, A. B.-V. Evercooren, E. Georges-Labouesse and C. ffrench-Constant, Nat. Cell Biol., 2002, 4, 833–841 CrossRef CAS.
- J.-G. Hu, S.-L. Fu, Y.-X. Wang, Y. Li, X.-Y. Jiang, X.-F. Wang, M.-S. Qui, P.-H. Lu and X.-M. Xu, Neuroscience, 2008, 151, 138–147 CrossRef CAS.
- J. Hsieh, J. B. Aimone, B. K. Kaspar, T. Kuwabara, K. Nakashima and F. H. Gage, J. Cell Biol., 2004, 164, 111–122 CrossRef CAS.
- S. Jhaveri, R. S. Erzurumlu, B. Friedman and G. E. Schneider, Glia, 1992, 6, 138–148 CrossRef CAS.
- S. Y. Yoo, A. Merzlyak and S.-W. Lee, Soft Matter, 2011, 7, 1660–1666 RSC.
- P. Schaffner and M. M. Dard, Cell. Mol. Life Sci., 2003, 60, 119–132 CrossRef CAS.
- P. M. Comoglio, C. Boccaccio and L. Trusolino, Curr. Opin. Cell Biol., 2003, 15, 565–571 CrossRef CAS.
- F. G. Giancotti and G. Tarone, Annu. Rev. Cell Dev. Biol., 2003, 19, 173–206 CrossRef CAS.
- C. K. Miranti and J. S. Brugge, Nat. Cell Biol., 2002, 4, 83–90 CrossRef.
- W. Baron, L. Decker, H. Colognato and C. ffrench-Constant, Curr. Biol., 2002, 13, 151–155 CrossRef.
- W. Baron, S. J. Shattil and C. ffrench-Constant, EMBO J., 2002, 21, 1957–1966 CrossRef CAS.
- J. A. Ellison and J. deVellis, J. Neurosci. Res., 1994, 37, 116–128 CrossRef CAS.
- A. Hall, N. A. Giese and W. D. Richardson, Development, 1996, 122, 4085–4094 CAS.
Footnote |
| † Corresponding Author: University of Toronto, Donnelly Centre for Cellular & Biomolecular Research, 160 College Street, Room 514 Toronto, Ontario M5S 3E1, Canada. Phone: +1 416 978 1460; Fax: +1 416 978 4317; E-mail: E-mail: molly.shoichet@utoronto.ca |
| This journal is © The Royal Society of Chemistry 2012 |
