Photo-catalytic conversion of carbon dioxide to organic acids by a recombinant cyanobacterium incapable of glycogen storage†
Damian
Carrieri‡
,
Troy
Paddock‡
,
Pin-Ching
Maness
,
Michael
Seibert
and
Jianping
Yu
*
National Renewable Energy Laboratory, Biosciences Center, 15013 Denver West Parkway, Golden, CO, USA. E-mail: Jianping.Yu@nrel.gov; Fax: +1 303 384 7839; Tel: +1 303 384 6252
First published on 12th September 2012
Abstract
Deletion of the gene encoding glucose-1-phosphate adenylyltransferase (ΔglgC) in the non-nitrogen-fixing cyanobacterium, Synechocystis sp. PCC 6803, disables glycogen synthesis, arrests cellular biomass accumulation under nitrogen deficiency, and redirects photosynthetically fixed carbon to organic acids (α-ketoglutarate and pyruvate) that appear in the extracellular medium.
Broader contextOxygenic photosynthetic microbes (algae and cyanobacteria) have great potential to produce fuels and valuable organic chemicals from sunlight, water, and carbon dioxide. However, efficient conversion of carbon dioxide to products of interest is in direct competition with cellular biomass accumulation. A better understanding of how cells regulate partitioning of photosynthetically fixed carbon between target products, native storage compounds, and cellular growth is needed. Here we report a mutant strain, ΔglgC, of the cyanobacterium Synechocystis sp. PCC 6803. Under nitrogen depleted conditions, this mutant diverts photosynthetically fixed carbon from glycogen synthesis to production of organic acids (α-ketoglutarate and pyruvate) that appear in the extracellular medium. Also under nitrogen deprivation the strain halts biomass accumulation and does not degrade its phycobilins as is typically observed in wild-type strains of cyanobacteria. This strain provides a novel system to study solar bio-catalysis and regulation of photosynthetic carbon partitioning, and serves as a platform for de novo conversion of carbon dioxide to organic products. |
A fundamental challenge for engineering photosynthetic microbes to produce fuels and chemicals from carbon dioxide and water efficiently lies in the ability to enhance product-to-biomass ratios.1 Cellular growth competes with desirable product synthesis and necessitates disposal or recycling of biomass. Arresting biomass accumulation, while simultaneously redirecting photosynthetically fixed carbon to products of interest, would offer a solution. However, much remains to be learned about how cells regulate the partitioning of photosynthate among competing carbon sinks, such as target product(s), native storage compounds, and cellular growth.
A common method for perturbing metabolism is to limit the macronutrient availability. Upon removal of nitrogen, non-nitrogen fixing cyanobacteria continue photosynthesis at attenuated rates, accumulating biomass, mostly in the form of glycogen.2 After prolonged nitrogen deficiency, photosynthesis decreases to a minimal rate that matches the maintenance demands of stationary cells.3 We and others have hypothesized that blocking glycogen synthesis will divert photosynthetically fixed carbon to alternative sinks in cyanobacteria upon nutrient limitation, thus providing a genetic model for studying regulation of photosynthetic carbon partitioning.
Glucose-1-phosphate adenylyltransferase is a key enzyme in the metabolic pathway leading to glycogen storage. Mutants deficient in its gene, glgC, have been constructed and characterized in Synechocystis sp. PCC 6803 (ref. 4) and Synechococcus sp. PCC 7942;5 however, to our knowledge they were not characterized under nitrogen deprivation. Since the glgC mutant in Synechocystis sp. PCC 6803 previously generated was not available for study, we engineered a new strain of this organism incapable of glycogen synthesis (ΔglgC) and a transcomplemented strain (ΔglgC psbA2::glgC) in which the glgC gene was reinserted into the chromosome behind the psbA2 promoter (ESI Fig. S1†). Unlike the previously published mutant, the ΔglgC features precise open reading frame replacement such that the expression of the inserted antibiotic resistance gene is controlled by the native glgC promoter and ribosome binding site, therefore minimizing possible interference of downstream gene expression (polar effect). Cultures of WT and ΔglgC strains grow at similar rates under moderate continuous light flux (65 μE m−2 s−1, ESI Fig. S2†).
The ΔglgC strain showed unusual responses to nitrogen depletion. Log phase cells of WT, mutant, and transcomplemented strains in nutrient sufficient BG11 medium were collected by centrifugation, washed and resuspended in nitrogen-free BG11 medium. The ΔglgC strain exhibited no growth, while both WT and ΔglgC psbA2::glgC increased in optical density (Fig. 1A). Non-nitrogen fixing cyanobacteria are known to transition from blue-green to yellow-green upon nitrogen depletion because of phycobilin protein degradation and the loss of phycocyanin pigment and chlorophyll.2,3,6,7 While this “bleaching” phenotype is observed for WT and ΔglgC psbA2::glgC cultures, ΔglgC remains blue-green even after four days of nitrogen starvation (Fig. 1B, ESI Fig. S3†). However, chlorophyll absorbance from whole-cell suspensions decreased to ∼70% of initial values in both WT and ΔglgC after four days (data not shown). A few Synechocystis mutants are known to remain blue-green after days of nitrogen starvation. Of these so-called nbl mutants (for non-bleaching), the nblA1 deficient strain has a phenotype that most closely mimics that of ΔglgC, with no growth observed over five days of nitrogen starvation.7 However, the photosynthetic activity of the nblA1 strain diminishes when measured by light-saturated O2-evolution activity to 8 ± 3% of its nitrogen-replete value, compared to 54 ± 8% in WT, after two days of starvation.7 The ΔglgC strain surprisingly maintains O2-evolution rates similar to WT rates during nitrogen starvation treatment, diminishing to 47 ± 5% of initial activity after two days (Fig. 1C).
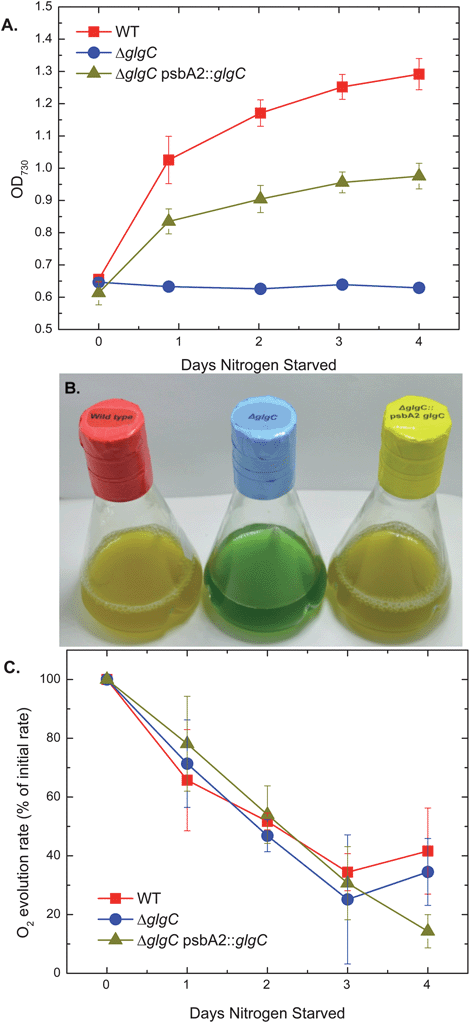 | ||
| Fig. 1 Optical densities (A), photograph (B), and light-saturated, oxygen-evolution rates (C) of Synechocystis sp. PCC 6803 cultures from WT, ΔglgC, and ΔglgC psbA2::glgC over a nitrogen starvation growth regime. Optical densities were measured as light absorbance of whole-cell suspensions at 730 nm. The photograph was of cultures after 4 days of nitrogen starvation. Oxygen-evolution rates were normalized to volume and listed as a percentage of the rates measured from replicate cultures that were not nitrogen starved on day 0. All samples had oxygen evolution activities within 1 standard deviation of each other when normalized to optical density at time zero. | ||
The complete cessation of growth coupled with the retention of WT levels of photosynthetic activity suggested the extracellular medium as a potential repository for photosynthetically derived carbon. Medium in which cells were nitrogen starved was examined by high pressure liquid chromatography (HPLC) and proton nuclear magnetic resonance (NMR) spectroscopy, and α-ketoglutarate (AKG) and pyruvate were identified as metabolites originating from the cells. Additionally, cellular dry weights did not increase in nitrogen-depleted ΔglgC cultures. Dry weights of cells from all three strains, glycogen fractions of this material, and extracellular organic acids from ΔglgC are shown in Fig. 2 for a four-day, nitrogen-starvation time-course. WT and ΔglgC psbA2::glgC produced very low amounts of excreted organic compounds concentrations (<20 μM, not shown) and accumulated biomass mostly in the form of glycogen. In contrast, the ΔglgC strain did not accumulate biomass but produced considerable amounts of excreted AKG and pyruvate (final concentrations in medium >700 μM). A small portion of the cellular biomass that was not glycogen also accumulated in the WT and ΔglgC psbA2::glgC strains.
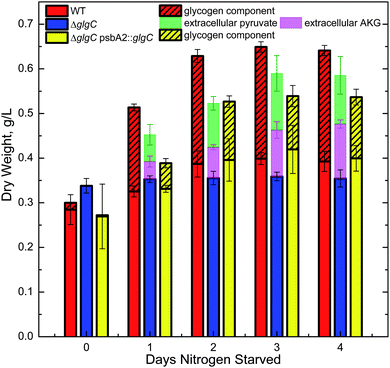 | ||
| Fig. 2 Dry weights of cells and extracellular organic acids from cultures of Synechocystis sp. PCC 6803 WT, ΔglgC, and ΔglgC psbA2::glgC during nitrogen starvation. WT and ΔglgC psbA2::glgC cells accumulated biomass, mostly in the form of glycogen (hashed), while the ΔglgC strain did not accumulate cell biomass or detectable levels of glycogen (limit of detection 0.2% of dry weight), but rather produced extracellular organic acids, α-ketoglutarate (AKG) and pyruvate. Extracellular components produced by cells in quantities lower than 0.08 g L−1 are not plotted as these components are less than the typical standard deviation of the cellular dry weights measured. | ||
We then asked whether the extracellular organic acids were synthesized by ΔglgC cells directly from newly fixed carbon or derived from catabolism of existing biomass (e.g., proteins, lipids). Mid-log phase ΔglgC cells grown in unlabeled (natural abundance 13C, 1.01%) medium were collected by centrifugation and resuspended in nitrogen-free medium with 13.4% 13C-labeled bicarbonate, and labeling of the organic acids accumulated extracellularly was observed over the first 18 hours of nitrogen starvation. A proton NMR spectrum of partially labeled products shows both “parent” and “satellite” peaks from protons attached to 12C and 13C, respectively for pyruvate (Fig. 3A). These peaks were integrated and the 13C labeling ratio8 of the pyruvate was calculated to be 13.5 ± 0.8%. Labeling ratios were also calculated for AKG and matched that of bicarbonate (ESI Table S1†). A carbon-13 NMR spectrum of cell-free medium isolated from the cultures after three days of nitrogen starvation visualized the labeled form of the excreted metabolites and confirmed that the predominant products were indeed AKG and pyruvate (Fig. 3B). Thus three independent methods (HPLC, proton NMR, and carbon NMR) confirm the identity of these metabolites. The carbon NMR spectrum also confirmed that all carbons on AKG and pyruvate were labeled, as each predicted peak is apparent at the expected relative intensity. These results demonstrate that the organic acids measured in the extracellular medium were produced de novo and not from remobilizing the cell material stored previously during growth in nitrogen-replete medium.
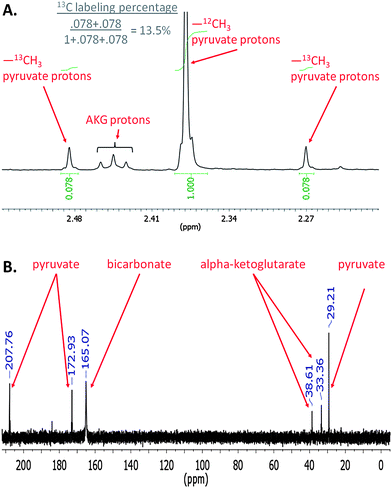 | ||
| Fig. 3 Proton (A) and carbon (B) NMR spectra of 13C-labeled, nitrogen-free medium after incubation of ΔglgC cells. Proton spectra were taken from medium with 13.4% 13C labeling in which cells were incubated for 18 hours under illumination. The area ratios of the 12C and 13C methyl group protons of pyruvate give the labeling ratio of pyruvate. A carbon NMR of nitrogen-free medium in which ΔglgC cells were incubated for 3 days under illumination shows all the predicted peaks for pyruvate and α-ketoglutarate, indicating all carbon atoms were labeled on these organic acids. | ||
Compared to WT, photosynthetic carbon flux in ΔglgC under nitrogen deprivation is redirected from synthesis of glycogen and other cellular biomass to AKG and pyruvate. This may indicate that ΔglgC cells are overwhelmed with the influx of carbon from photosynthesis and excrete these products in response, as is the case with some metabolic responses in other bacteria. For example, in cultures of Streptomyces, a genera of Gram positive bacteria, excretion of AKG and pyruvate is often observed when cells are grown in medium with a rapidly utilized carbon source such as glucose.9 In Cupriavidus necator, a (Gram-negative) proteobacterium, pyruvate excretion results from genetic impairment of the accumulation of poly β-hydroxybutyric acid (PHB), a major storage compound, in nutrient-limited medium when excess carbon is available;10 even more analogously, in this same mutant, pyruvate, malate, and citrate are excreted when these cells are supplied with H2, CO2, and O2 (sufficient substrates for carbon fixation in this species) under nitrogen-limited conditions.11 We propose that AKG and pyruvate excretion into the medium of ΔglgC cultures under nitrogen deficiency is a necessary metabolic adaptation, because the cultures continue photosynthesis even when the native pathway for excess carbon storage is lost. In the above literature examples, cells were supplied with either a reduced carbon substrate or H2 as a reductant, which they readily consume. However our results with ΔglgC cells are observed in the absence of such reduced chemical species. Instead, reductant must be generated via oxygenic photosynthesis. This photo-catalytic mode of ΔglgC-cell metabolism is therefore particularly interesting for biotechnological applications for reducing CO2 from water and sunlight.
A scheme illustrating our findings is presented in Scheme 1. Under nutrient replete conditions, a majority of the photosynthetically fixed carbon is directed toward biomass accumulation. In the absence of nitrogen, WT redirects a majority of carbon flux to glycogen, while mobilization of nitrogen stored in phycobilins during bleaching presumably allows for de novo protein synthesis and therefore additional cellular biomass accumulation.12,13 In contrast, ΔglgC cells lack the ability to accumulate glycogen and apparently the ability to mobilize nitrogen reserves. Still, photosynthesis proceeds at a rate similar to WT with the flux of recently fixed carbon diverted to AKG and pyruvate synthesis.
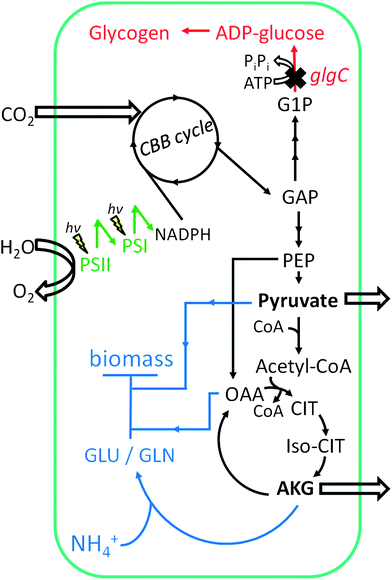 | ||
| Scheme 1 Metabolic scheme for Synechocystis sp. PCC 6803 ΔglgC cells under nitrogen-deprived conditions. Carbon (as CO2) enters the Calvin–Benson–Bassham (CBB) cycle to produce metabolic intermediate carbon compounds (black arrows). In the absence of a reduced nitrogen source, glutamate and glutamine (GLU/GLN) synthesis halted, as is biomass accumulation (blue arrows). Because carbon cannot be stored as glycogen (red arrows), pyruvate and AKG accumulate and are excreted from the cell. Abbreviations: G1P, glucose-1-phosphate; GAP, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; OAA, oxaloacetic acid; CIT, citric acid; Iso-CIT, isocitric acid; AKG, α-ketoglutarate; GLU/GLN, glutamate/glutamine. Light reactions of photosynthesis are indicated by “hν” symbols at reaction centers. | ||
The link between the block in glycogen synthesis and bleaching is yet elusive, though the phenotype observed correlates with the production of AKG in ΔglgC. Cyanobacteria utilize complex mechanisms for carbon/nitrogen (C/N) balance. The central signaling molecule for C/N status in cyanobacteria is AKG, which modulates expression of the global nitrogen response regulator, NtcA.14 High AKG levels are predicted to down-regulate photosynthesis via NtcA.15 A cluster of genes involved in photosynthesis are also shown to be down-regulated during nitrogen-limited growth in Synechocystis.16 Because intracellular levels of AKG are high in ΔglgC (between 0.5 and 2% cell dry weight by GC/MS after 1 day of nitrogen starvation), it is conceivable that the sensory mechanism reliant on this metabolite is disrupted. In addition, sugar metabolism intermediates could interfere with bleaching in Synechocystis.12 Future transcriptomic and proteomic analysis of Synechocystis WT and ΔglgC strains may help discern proteins related to bleaching and a possible link to levels of AKG and sugar metabolites.
For biotechnological applications, it will be preferable to maintain high photosynthetic efficiencies in a photocatalytic mode. Whether the high AKG levels are responsible for the decline in maximal photosynthetic capacity of cells during nitrogen starvation is yet to be determined. A simple solution to decreased photosynthetic efficiencies that are coupled to nitrogen deprivation may be to supply small quantities of nitrogen during the nitrogen-limited regime by re-addition of nitrate or ammonium. This type of approach has been successfully demonstrated for cultures of the green alga Chlamydomonas reinhardtii producing hydrogen under sulfur-limited conditions by re-addition of sulfate to increase photosynthetic efficiencies.17 This approach could lead to higher yields of AKG and pyruvate, which are likely limited only by photosynthesis/carbon fixation and not, for example, by the initial dry weight of the cells upon nitrogen limitation.
While our emphasis has been to highlight the importance of a model system that photo-catalytically converts CO2 to carbon products, we note the biotechnological applications of a system that produces AKG and pyruvate. AKG is a platform chemical for organic synthesis and these chemicals have value in sports nutrition and weight-loss markets. We imagine that carbon flux could be redirected to pathways that produce more valuable chemicals and fuels in the near future. For example, AKG is the immediate metabolic precursor to ethylene in a reaction involving the Ethylene Forming Enzyme (EFE); pyruvate is the carbon precursor to most fermentative products such as acetate, lactate, and ethanol. Furthermore, pyruvate is the immediate precursor to acetyl-CoA which is the substrate for lipids and bio-plastics.
Conclusions
The observation that ΔglgC cells do not accumulate cellular glycogen yet convert CO2 to substantial amounts of organic acids (in our case up to 65% of the cell dry weight after 4 days of nitrogen starvation; see Fig. 2) provides a novel system to study both solar bio-catalysis and regulation of photosynthetic carbon partitioning. With the revelation of a dramatic redirection of carbon allocation from glycogen to organic acid production, we have demonstrated a system engaged in active photosynthesis, while blocked in cellular biomass accumulation. This was achieved by nitrogen depletion to limit growth and the ΔglgC mutation to redirect metabolism. This new tool will facilitate future research on photosynthetic production of biofuels and chemicals as well as gaining new insights into global regulatory mechanisms in cyanobacterial carbon allocation.Acknowledgements
This work was supported by the Laboratory Directed Research and Development Program at the National Renewable Energy Laboratory (to J.Y. and M.S.). T.P. and J.Y. also acknowledge financial support from Department of Energy, Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division under contract no. DE-AC36-08-GO28308 with the National Renewable Energy Laboratory, and M.S. acknowledges partial support from the NREL pension program. Some data referenced in this paper are presented in ESI figures and tables†. The authors thank Dr Teruo Ogawa for sharing the wild-type strain, Dr Wim Vermaas for sharing the psbA2 plasmid, and Dr Carrie Eckert for its modification. Drs Karen Wawrousek, Carrie Eckert, Justin Ungerer, Phil Pienkos, Al Darzins, Christopher Chang, Ambarish Nag, Paul King, and Maria Ghirardi contributed technical expertise and engaged in helpful discussions. Drs Ghada Ajlani and Christopher Johnson also engaged in helpful discussions. The authors are also grateful to Dr Mark Davis, Dr Erica Gjersing, and Ms Renee Happs for NMR facility access and assistance. Dr Erica Gjersing assisted with NMR auto-sampling and provided helpful technical advice with NMR spectroscopy.Notes and references
- A. Melis, Energy Environ. Sci., 2012, 5, 5531–5539 CAS.
- R. Schwarz and K. Forchhammer, Microbiology, 2005, 151, 2503–2514 CrossRef CAS.
- J. Sauer, U. Schreiber, R. Schmid, U. Volker and K. Forchhammer, Plant Physiol., 2001, 126, 233–243 CrossRef CAS.
- X. L. Miao, Q. Y. Wu, G. F. Wui and N. M. Zhaol, Biotechnol. Lett., 2003, 25, 391–396 CrossRef CAS.
- E. Suzuki, H. Ohkawa, K. Moriya, T. Matsubara, Y. Nagaike, I. Iwasaki, S. Fujiwara, M. Tsuzuki and Y. Nakamura, Appl. Environ. Microbiol., 2010, 76, 3153–3159 CrossRef CAS.
- V. Krasikov, E. Aguirre von Wobeser, H. L. Dekker, J. Huisman and H. C. Matthijs, Physiol. Plant., 2012, 145, 426–439 CrossRef CAS.
- H. Li and L. A. Sherman, Arch. Microbiol., 2002, 178, 256–266 CrossRef CAS.
- D. Carrieri, K. McNeely, A. C. De Roo, N. Bennette, I. Pelczer and G. C. Dismukes, Magn. Reson. Chem., 2009, 47(suppl. 1), S138–S146 CrossRef CAS.
- T. Madden, J. M. Ward and A. P. Ison, Microbiology, 1996, 142, 3181–3185 CrossRef CAS.
- A. Steinbüchel and H. G. Schlegel, Appl. Microbiol. Biotechnol., 1989, 31, 168–175 CrossRef.
- A. M. Cook and H. G. Schlegel, Arch. Microbiol., 1978, 119, 231–235 CrossRef CAS.
- K. Elmorjani and M. Herdman, J. Gen. Microbiol., 1987, 133, 1685–1694 CAS.
- C. Richaud, G. Zabulon, A. Joder and J. C. Thomas, J. Bacteriol., 2001, 183, 2989–2994 CrossRef CAS.
- M. I. Muro-Pastor, J. C. Reyes and F. J. Florencio, J. Biol. Chem., 2001, 276, 38320–38328 CAS.
- Z. Su, F. Mao, P. Dam, H. Wu, V. Olman, I. T. Paulsen, B. Palenik and Y. Xu, Nucleic Acids Res., 2006, 34, 1050–1065 CrossRef CAS.
- E. Aguirre von Wobeser, B. W. Ibelings, J. Bok, V. Krasikov, J. Huisman and H. C. Matthijs, Plant Physiol., 2011, 155, 1445–1457 CrossRef.
- S. Kosourov, A. Tsygankov, M. Seibert and M. L. Ghirardi, Biotechnol. Bioeng., 2002, 78, 731–740 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and methods, Fig. S1–S3, Tables S1 and S2, and ref. 18–21. See DOI: 10.1039/c2ee23181f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2012 |
