Designs for life: protocell models in the laboratory
Alicja J.
Dzieciol
and
Stephen
Mann
*
Centre for Organized Matter Chemistry, School of Chemistry, University of Bristol, Bristol BS8 1TS, UK. E-mail: s.mann@bris.ac.uk
First published on 27th September 2011
Abstract
Compartmentalization of primitive biochemical reactions within membrane-bound water micro-droplets is considered an essential step in the origin of life. In the absence of complex biochemical machinery, the hypothetical precursors to the first biological cells (protocells) would be dependent on the self-organization of their components and physicochemical conditions of the environment to attain a basic level of autonomy and evolutionary viability. Many researchers consider the self-organization of lipid and fatty acid molecules into bilayer vesicles as a simple form of membrane-based compartmentalization that can be developed for the experimental design and construction of plausible protocell models. In this tutorial review, we highlight some of the recent advances and issues concerning the construction of simple cell-like systems in the laboratory. Overcoming many of the current scientific challenges should lead to new types of chemical bio-reactors and artificial cell-like entities, and bring new insights concerning the possible pathways responsible for the origin of life.
 Alicja J. Dzieciol | Alicja J. Dzieciol was born in 1989 and comes from Lower Silesia, Poland. She is studying for a MSci in Chemistry at the University of Bristol, UK. Currently in her final year, she is pursuing a project focusing on fatty acid vesicles as protocell models under the guidance of Professor Stephen Mann. In her spare time, she enjoys long-distance cycling. |
 Stephen Mann | Stephen Mann is Professor of Chemistry, Director of the Centre for Organized Matter Chemistry, and Principal of the Bristol Centre for Functional Nanomaterials at the University of Bristol, UK. His research interests are focused on the chemical synthesis, characterization and emergence of complex forms of organized matter, including models of protocell assembly. Prof. Mann was elected as a Fellow of the Royal Society, UK, in 2003. In 2011, he was awarded the Royal Society of Chemistry de Gennes Medal, and was a recipient of the Chemical Society of France, French-British Prize. |
1. Introduction
Along with the theory of evolution, the cell theory of life is the most important generalisation in biology.1 The latter theory states that biological cells are the basic components and building blocks of all known living organisms, and that the primary unit of life is represented by the operation of a single cell subjected to Darwinian evolution. However, these two key theories fail to explain the origin of cellularity—how did the first cells emerged in a world devoid of biological evolution? Solving this long-standing mystery is of deep significance as even small advances in our understanding of the origin of life will help to bridge the conspicuous disconnection between non-living and living forms of matter, as well as contribute to the development of a unifying theory for the emergence of biology within a physical universe.The foundation of modern research on the origin of life can be traced back to Oparin who emphasized the concept of molecular evolution as a chemical progenitor of biological evolution.2 Oparin proposed that life developed from simple non-living molecules through a spontaneous and gradual build up of molecular complexity. A commonly accepted hypothesis is that the physicochemical conditions on the early Earth favoured chemical reactions that produced simple organic compounds from inorganic precursors, and that these water-soluble organic molecules underwent subsequent reactions to generate an expanding library of molecular structures with increasing complexity and new properties.3 The hypothesis considers three of these properties—small-molecule catalysis, molecular self-generation, and amphiphilicity—as critical for the emergence of a self-contained chemical system that respectively would provide the basis for the development of coupled reaction cycles (primitive metabolism), self-replication (copying of informational molecules), and compartmentalization (self-assembly of enclosed membranes). How this could be achieved, at least in principle, is the subject of much current research in which plausible chemical and physical pathways are modelled both experimentally and theoretically. This has given rise to various scenarios in which proponents such as ribonucleic acid (RNA world),4 peptides/proteins (metabolic world)5 and lipid/fatty acids (compartmentalization-first hypothesis)6 vie for precedence.
At the same time, it has been increasingly recognised that the integration and collective operation of primitive processes of replication, metabolism and compartmentalization, rather than their individual pre-eminence, could represent a critical step in the emergence of life.7 This view sees life as a systems property that is maintained under non-equilibrium conditions by flows of energy and matter from the surrounding environment. As a consequence, the system functions to increase its own viability through various selection pressures that ultimately lead to a Darwinian evolutionary capacity. Significantly, this notion of a systems-based criticality has focused recent attention on the design and construction of plausible models of autonomous chemical systems—a research field that might be called protobiology. Central to this activity is the concept of the protocell, which has been defined in various ways, ranging from a plausible representation of a hypothetical precursor to the first biological cell, through to the description of a synthetic cell-like entity that contains non-biologically relevant components. For most practical purposes, laboratory investigations aim to construct protocell models involving simple membrane-bound cell-like structures exhibiting at least some of the key characteristics of modern cells. Examples of this approach are highlighted in this tutorial review with particular attention placed on the use of lipid or fatty acid vesicles. Such studies should help to elucidate the types of molecules and physical conditions compatible with the design and construction of protocells, and provide clues about the possible pre-biotic pathways responsible for the formation of cellular progenitors on the early Earth.
2. Key features of living cells for protocell modelling
We begin by considering some of the key features of living cells. As even the simplest known living cells—such as the parasitic bacterium Mycoplasma genitalium—consist of a highly complex network of hundreds of genes and proteins within the confines of the cell membrane8 only the core criteria of life are of immediate interest for the design and construction of protocell models (Fig. 1). All living cells share the following characteristics:9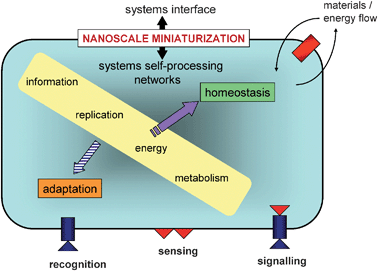 | ||
| Fig. 1 Systems representation of cellular life showing core criteria of immediate interest for the design and construction of protocell models.7 Two primary mechanistic features must emerge for life; (i) a systems interface with the environment based on the cell membrane and embedded protein-based sensors, and (ii) a systems network for internalized self-processing via processes of metabolism, replication etc. Together, these processes constitute a dissipative non-equilibrium system that operates through nanoscale miniaturization and is maintained by continuous active exchange between the intracellular milieu and surrounding environment. The self-referential nature of life is manifest in the steady state of materials and energy fluxes (homeostasis), which necessitates that the hierarchical networks of internal self-processing must be capable of passively or actively assimilating novel environmental inputs into pre-existing processes without undermining viability. In tension with this implicit conservative nature, is the enduring ability of the cell to adapt to novel disturbances by permanent transformations in the systems interface and internal processing networks to maintain and enhance viability. This is not realized at the level of the individual cell but by selection pressures on cell populations (Darwinian evolution). | ||
- a semi-permeable membrane that encloses the cell constituents and acts as a selective barrier between the cell interior and the external environment, regulating the flow of materials in and out of the cell (compartmentalization).
- genetic information carried in the form of double-stranded molecules of DNA which is inherited by daughter cells during cell division.
- template polymerisation to copy hereditary information (replication).
- transcription of the genetic information stored in DNA into RNA, and translation of RNA into proteins (the “central dogma”).
- protein-based catalysts that participate in myriad chemical transformations for self-maintainance and regeneration (metabolism), as well as in transcription, translation and DNA replication. This feedback loop between DNA and proteins is the basis of the self-reproducing capacity of living cells.
- a steady state (homeostasis) based on a non-equilibrium system that requires a continuous influx of energy from the surroundings to sustain life, grow and divide.
Given the scope of the above criteria,10 it is reasonable to pre-suppose that the first cells to appear on the Earth (assuming they were not extra-terrestrial!) were much simpler systems than contemporary cells. Moreover, as primitive cells would have lacked the advanced nanoscale machinery required to process the complex cascades of biochemical reactions observed in modern cells, basic cellular functions would need to be maintained through physicochemical interactions operating between constituent molecules and the surrounding environment. Therefore, it seems reasonable to consider that the first step in the formation of a hypothetical protocell involved the spontaneous self-ordering of a mixture of abiogenic molecules under appropriate conditions. This notion constitutes the basis of bottom-up approaches to the construction of protocell models in the laboratory (Fig. 2). One area of great interest focuses on the self-assembly and self-organization of biological-like structures of increasing complexity from simple molecules that were likely to be present on the pre-biotic Earth. This is a historical approach, and thus the use of biopolymers such as enzymes and polynucleic acids is not countenanced in this approach, as there is no current evidence for the synthesis of such compounds under pre-biotic conditions.3 Alternatively, other researchers use individual biological components, often along with synthetic constituents, to construct protocell models from the bottom-up in order to reconstitute and mimic certain cellular functions under simplified conditions. This strategy exploits modern technologies such as cell-free gene expression to investigate how synthetic cells can be designed and constructed. As a consequence, the focus is more geared towards the notion of artificial cells rather than providing plausible claims concerning the origin of cellularity on the early Earth. In contrast to the bottom-up strategies, there is much current research activity based on exploiting a top-down synthetic biology approach in which contemporary cells are progressively simplified by removing genes that are thought not necessary to sustain the essential properties of cellular life such as self-maintenance and self-reproduction (Fig. 2).11 The goal is to engineer a minimal cell, which represents a stripped-down organism comprising the lowest number of genes necessary to maintain basic cellular functions. Together, the top-down and bottom-up approaches are complementary approaches to protocell modeling, and cover a wide spectrum of complexity and organization ranging from ensembles of simple non-living molecules to retro-engineered modern cells.
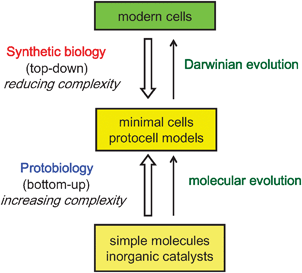 | ||
| Fig. 2 Approaches to the design and construction of primitive cells. See text for details. | ||
3. Membranes as cellular boundaries
A key element of the bottom-up approach is the development of appropriate models of protocellular membrane formation and function. The emergence of discrete, perhaps free-floating, membrane-bound water droplets suspended in an aqueous medium is considered crucial for the appearance of life on the early Earth for several reasons.12 Firstly, an enclosed, semi-permeable barrier restricts the transport and accumulation of nutrient molecules from the environment into the protocell interior. As a consequence, only certain molecules of small size and low polarity have sufficient membrane permeability to become enriched within the interior of the droplets, and this opens up the potential for novel chemistries within the protocell that are separated from but connected to the ambient conditions. Secondly, partitioning of molecules between the inside and outside of the protocell membrane can result in concentration and electrochemical gradients across the protocell membrane, which can be used to drive the internalized reactions against free energy to produce energy-rich molecules and anabolic products such as polymer chains that have low membrane permeability. Thirdly, the presence of the membrane enables retention of the polymerized products, providing an enclosed environment for the self-assembly and entrapment of supramolecular and nanoscale structures that extend the chemistry beyond that possible with small molecules—for example, compartmentalization is considered necessary to facilitate the evolution of polymerases.13 The key step, which is not understood, is how this novel chemistry led to a protocellular system with self-referential properties—that is, the internalized products are generated by a system whose integrity is critically dependent on the making of the products. This form of organizational logic—what has been described using various terms, such as autopoiesis (self-production), autocatalytic self-maintaining metabolic networks, or metabolic closure, is perhaps the ultimate criterion that defines life. It is the basis for autonomy at the level of a single protocell, and when applied to a population of cells is a fundamental selection pressure that drives Darwinian evolution. From the chemical perspective, the key issue is to go beyond the current focus on self-assembly and self-organization, which lead to the spontaneous onset of structure and function, to a phenomenon that explicitly maintains the viability of the system itself—what could perhaps be called “self-determining chemistry”.How should we model the hypothetical protocell membrane? Modern cell membranes are nanometer-thin bilayer assemblies of complex mixtures of amphiphilic long chain molecules such as phospholipids and glycolipids, which consist of a hydrophilic head group and two hydrophobic acyl chains (Fig. 3a). The hydrophobic interior of the phospholipid membrane is a strong barrier to ions and many polar molecules, preventing their free diffusion between the inside and outside of the compartment and therefore maintaining non-equilibrium concentrations of molecules in the cell interior. In modern cells, integral membrane proteins form pores and energy-dependent pumps that are essential for selective transfer of nutrients and waste molecules across the membrane. In addition, growth and division of the membrane in contemporary cells are dependent on a complex series of biochemical reactions that presumably arose during the process of cellular evolution. It seems self-evident therefore that the membranes delineating the first cells were much simpler, and more likely to have been derived from single chain amphiphiles such as fatty acids (Fig. 3b).14 As a consequence, one of the major goals of laboratory investigations on the origins of cellular life is the demonstration of how membrane semi-permeability, growth and division could have been achieved without advanced biochemical machinery.
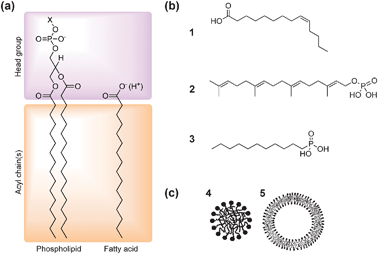 | ||
| Fig. 3 (a) Molecular structures of phospholipids and fatty acids. Phospholipids are double-chained amphiphiles possessing a negatively charged phosphate ester headgroup coupled to another polar functional group (X) such as choline, glycerol, ethanolamine and serine. (b) Examples of single-chain amphiphiles of plausible prebiotic relevance: (1) fatty acid (myristoleic acid), (2) polyprenyl phosphate (geranylgeranyl phosphoric acid), (3) mono-n-alkyl phosphonate (n-decylphosphonic acid). (c) Supramolecular structures formed by spontaneous self-assembly of amphiphiles in water. Single-chain amphiphiles such as fatty acids form spherical micelles (4) with an inner hydrophobic core and outer shell of polar headgroups. In contrast, double-chain phospholipids assembly into spherical vesicles comprising a closed bilayer membrane and entrapped aqueous micro-compartment (5). (Figures (a) and (b) adapted from ref. 12 and 16; Figure (c) adapted from ref. 17). | ||
Significantly, it has been long recognized that phospholipid molecules can spontaneously self-assemble in water into closed spherical vesicles that are delineated by a bilayer membrane approximately 4–5 nm in thickness. Formation of these supramolecular aggregates results from a balance of attractive and repulsive forces, with the entropic hydrophobic effect being the main driving force for self-assembly. Aggregation of the nonpolar tails, rather than solvation of the individual lipid molecules, minimizes disruption to the hydrogen bonding network of water, and therefore lowers the free energy. Moreover, steric and coulombic repulsions between the lipid headgroups prohibit complete phase separation, such that dispersed colloids of membrane-bound droplets rather than precipitates of the lipid in water are produced. Interestingly, the size and shape of the ordered aggregates are related to a molecular packing parameter,15 which describes the geometric properties of a single amphiphilic molecule in the aggregate. On this basis, amphiphiles with a quasi-cylindrical shape, such as phospholipids, will form bilayer vesicles, while single chain fatty acids with a headgroup considerably bulkier than the hydrophobic tail pack together into spherical micelles (Fig. 3c). In addition, the effective interfacial area per fatty acid headgroup, and thus the shape and form of the resulting aggregate, is strongly influenced by pH. As a consequence, complete deprotonation of the fatty acid at high pH results in a large degree of headgroup repulsion that leads to the formation of micelles, while complete protonation at low pH results in phase separation. Significantly, at intermediate pH values close to the pKa of fatty acid (typically 8–8.5), the amphiphilic molecules self-aggregate into vesicles.18 The closed bilayer membrane is favoured under these conditions because inter-headgroup repulsion is considerably reduced by hydrogen bonding between the protonated and ionized forms of the fatty acid molecules.
The facile entrapment of aqueous micro-volumes by self-ordering amphiphilic molecules acting under simple physiochemical forces has had a profound impact on the modeling of protocellular systems. Indeed, many researchers consider this mechanism of compartmentalization to represent a key step in the formation of early cells.12 In the following sections, we discuss the application of spontaneous aqueous self-assembly of phospholipids or fatty acids in vesicle-based models of protocell organization and function.
4. Amphiphiles for protocell models
Although it seems most likely that primitive cells contained a heterogeneous mixture of amphiphiles generated under prebiotic conditions,18 the majority of experimental studies on vesicle-based protocell models have employed bilayer membranes constructed from one or occasionally two phospholipid species.12 However, phospholipid vesicles are unlikely candidates for protocellular membranes due to their low permeability to many nutrients essential for basic cell function. In addition, phospholipids are relatively complex molecules and require several enzymes for their synthesis in modern cells. In contrast, fatty acids are better suited as components of vesicle-based protocells because their bilayer membranes permit the passive diffusion of ions and small molecules in and out of the vesicle compartment. Moreover, the fast exchange of fatty acids between the vesicle membrane and monomers or micelles in solution results in facile incorporation of new amphiphiles into the bilayer, leading to growth of the membrane and enlargement of the vesicles. Significantly, single-chain amphiphiles, unlike phospholipids, are likely to be of prebiotic origin as fatty acids and related derivatives have been isolated from carbonaceous meteroites,19,20 and synthesised under simulated prebiotic conditions based on artificial interstellar ice environments,21 hydrothermal vents,22 and spark discharges.23 Interestingly, amphiphiles present in meteorites, as well as those obtained synthetically under simulated early Earth conditions, are complex mixtures of compounds with a range of different headgroups and hydrocarbon chain lengths. As mixtures of single-chain amphiphiles produce vesicles that are stable over a wider pH range and have superior permeability properties than their single-component counterparts,24 it seems reasonable to propose that the membranes of primitive cells were compositionally heterogeneous.5. Experimental models of protocells
A number of pioneering studies have been undertaken using phospholipid or fatty acid vesicles as the basis of orchestrating aspects of cellular function in synthetic systems. In each case, the protocell models aim to mimic just a few of the key characteristics of living cells; typically these include the demonstration of simple metabolic, replication, or growth and division processes (Fig. 4).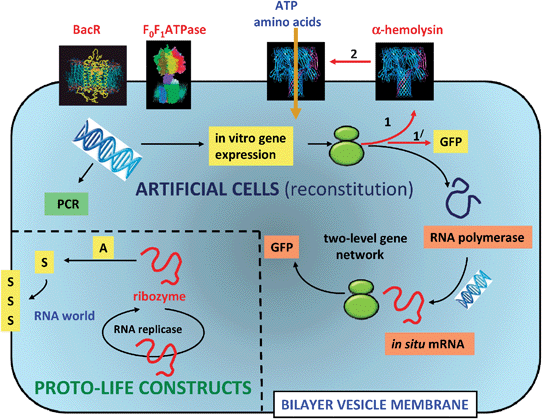 | ||
| Fig. 4 Experimental design for protocell models based on phospholipid or fatty acid vesicles. Two scenarios are illustrated; the reconstitution and operation of functional components inside the vesicles (artificial cells), and the construction of vesicles with prebiotically relevant components (proto-life constructs). In the former, single genes or simple gene networks derived from modern cells can be used to express proteins and enzymes that have functional relevance as fluorescent markers (GFP), membrane porins (α-hemolysin) and catalysts for mRNA synthesis (RNA polymerase). The translation of mRNA into proteins occurs via entrapped ribosomes (green structures). Entrapped DNA strands can be amplified by temperature cycling using polymerase chain reaction (PCR) procedures. Proton transport proteins such as bacteriorhodopsin (BacR) and F0F1ATPase can be incorporated into the vesicle membrane by direct addition of the biomolecules. Alternatively, prebiotically plausible components such as catalytically active strands of RNA (ribozymes), or nucleic acid templates that participate in non-enzymatic extension to produce double stranded informational polymers, are incorporated into the vesicles. Ideally, a single RNA molecule that could self-replicate (RNA replicase) and act as a ribozyme for the synthesis of new membrane components, could represent the simplest protocell model comprising primitive aspects of metabolism, replication and compartmentalization within a RNA world.31 | ||
5.1 Protocells based on lipid vesicles
Despite the above limitations in terms of their prebiotic relevance, phospholipid vesicles have been used extensively for the preparation of model cell-like systems,3 principally due to their similarity with respect to modern cell membranes, the existence of established techniques for their preparation as cell-sized compartments,25,26 and their relatively high stability with respect to variations in pH and temperature.18 A striking example involves the incorporation of a totally cell-free functional gene expression system within the intravesicular aqueous space.27 Typically, all the necessary components—approximately 100 or so in number—including an engineered plasmid DNA coded specifically for the expressed protein (green fluorescent protein, GFP), an enzyme (RNA polymerase) for the synthesis of messenger RNA (mRNA) from the DNA template, the cellular machinery (ribosomes) required to translate the as-produced mRNA into the amino acid sequence of the designated protein, and small molecule building blocks such as amino acids and activated mononucleotides, as well as an energy source (ATP), are trapped simultaneously within the vesicle.27 The results show that encapsulation of the GFP gene in the phospholipid vesicles prolongs the expression of the protein compared to the same system monitored in bulk solution. However, due to the low membrane permeability, the system grinds to a halt once the entrapped amino acids and nucleotides are used up. To circumvent this, a second gene that expresses the protein α-hemolysin was encapsulated along with the GFP gene and cell-free expression system. The expressed α-hemolysin assembles spontaneously into the vesicle membrane to produce molecular pores that facilitate the influx of new nutrient molecules from the external environment. Using this strategy, GFP expression is continued for up to 4 days.The above example illustrates a semi-synthetic approach to protocell modeling, in which genes and enzymes extracted from living organisms are used to test whether biochemical transformations can be carried out and sustained within membrane-bounded aqueous compartments (see ref. 28 for other examples). Although the components are clearly not prebiotically relevant, the approach has considerable merit in providing a step towards the construction of primitive artificial cells or semi-synthetic bioreactors. Another limitation of the lipid vesicle system is that it is difficult to conceive how this protocell model can be developed to incorporate notions of self-reproduction, which is a key feature of all living cells. To be truly self-reproducing, the cell-mimicking system needs to be capable of membrane growth and division, and replicate genetic material. In contemporary cells, membrane growth is associated with the synthesis of phospholipids by localised enzymes, and advanced biochemical machinery mediates cell division.3 Thus, one possibility is to entrap within the vesicles genes or enzymes responsible for phospholipid synthesis such that new amphiphilic building blocks can be generated internally and then integrated into the existing bilayer membrane to promote vesicle growth and division. A first step towards this goal has been recently published.29
5.2 Protocells based on fatty acid vesicles
As discussed above, there is a growing consensus that fatty acid vesicles are plausible models of primitive cellular compartments. An early example of a protocell model based on oleic acid/oleate vesicles involved the enzymatic polycondensation of ADP into the homo-polyribonucleotide, poly(A),30 which can be considered as a prototype of RNA. For this, the enzyme polynucleotide phosphorylase (PPNase), which is too large to permeate the fatty acid membrane, is encapsulated in the vesicles, and ADP then added to the external medium. Slow diffusion of ADP across the vesicle membrane results in an increase in the intra-vesicular substrate concentration, and as a consequence poly(A) is produced specifically within the enclosed compartment. As poly(A) does not leak out of the vesicles, the enzyme-driven polymerization reaction stops as the concentration and osmotic pressure rise. Recently, similar experiments involving the non-enzymatic extension of a vesicle-entrapped DNA template have been demonstrated as a more a plausible prebiotic scenario.31 For this, fatty acid vesicles prepared from a mixture of decanoic acid, decanol and decanoic acid glycerol monoester are used to entrap a single-stranded polycytosine template with an attached DNA primer that acts as an initiation point for guanosine polymerization along the template to produce a double-stranded product. The components of the vesicle membrane are optimised to maximise permeability to imidazole-activated nucleotides, and to minimise the leakage of the encapsulated DNA template. In this way, efficient uptake of activated guanosine molecules required for the slow non-enzymatic extension of the attached primer sequence along the DNA template is achieved within the vesicle interior. However, the replication is terminated when the entire population of template molecules has been converted into double stranded DNA, indicating that some mechanism of separating the duplex is required if successive cycles of replication are to occur. One possibility might be to heat the vesicles at this stage to disassemble the double strands. Indeed, temperature-dependent studies indicate that vesicles composed of various mixtures of single-chain amphiphiles are stable to increases in temperature, and retain encapsulated DNA and RNA oligonucleotides over a temperature range of 0 to 100 °C.32 Moreover, double strands of encapsulated DNA are separated by heating and subsequently re-anneal when the temperature is lowered, suggesting that thermal cycling could mediate multiple rounds of replication of genetic polymers inside fatty acid vesicles provided that self-combination of the separated DNA strands occurs at a slower rate than primer extension.One major problem with the above studies is that to keep the system going, some means of generating multiple cycles of vesicle growth and division—that is, self-reproduction—is required. An early model for fatty acid vesicle division involved extrusion of the vesicles through small pores under pressure to drive their division.33 However, it seems unlikely that analogous extrusion process could have occurred on early Earth. Other key studies have shown that vesicle growth can be achieved by adding an amphiphile feedstock in the form of monomeric fatty acids, a micellar solution at high pH, or water-insoluble fatty acid anhydrides to the external aqueous medium.34 Berclaz et al.35 first demonstrated that the addition of fatty acid micelles at basic pH to a buffered suspension of fatty acid vesicles results in destabilization of the micelles and concomitant growth of the pre-existing vesicles. In contrast, addition of phospholipid molecules to a suspension of phospholipid vesicles results in the formation of new vesicles, rather than growth of the preformed vesicles. More detailed studies on the mechanism of fatty acid vesicle growth and division have been recently reported.36,37
The use of oleic anhydride as a potential feedstock for fatty acid vesicle growth and self-reproduction is particularly interesting. Oleic anhydride sequesters into the membrane bilayer where it undergoes slow hydrolysis into oleic acid, which results in growth and multiplication of the vesicles. This approach has been adopted to establish enzymatic RNA replication within multiplying oleic acid/oleate vesicles.38 The enzyme, Qβ replicase, which replicates RNA by catalysing the polymerization of a RNA template, is entrapped along with a suitable RNA template, substrate mononucleotides (ATP, CTP, GTP and UTP) and Mg2+ ions inside the fatty acid vesicles. Replication of RNA is observed to proceed for a few generations, and reproduction of the vesicles is achieved by external addition of oleic anhydride. These studies illustrate how the internalized synthesis/replication of an informational molecule and growth of the compartment boundary can be achieved in a protocell model of combined self-reproduction. However, the biochemistry taking place in the vesicle interior and the process of membrane self-reproduction are mutually independent. Indeed, the ability to couple the self-replication of an internal component to the self-reproduction of the fatty acid vesicle membrane—for example, by genetic expression of an enzyme capable of catalysing the intra-vesicular synthesis of new membrane molecules—would represent a major advance in protocell modelling. Furthermore, the continuous increase in the number of vesicles in a system capable of self-reproduction has to be commensurate with a regeneration mechanism for continued production of the entrapped enzymes and DNA/RNA templates. Otherwise, the daughter vesicles contain decreasing numbers of entrapped molecules and so become non-functional after only a few generations—a process referred to in reference 3 as “death by dilution”! Thus, replication inside the vesicles must be coupled not only to the synthesis of new membrane constituents via enzyme/protein expression, but also to the self-copying of the template molecules if a primitive level of systems autonomy is to be achieved within a model protocell.
6. Conclusions
Compartmentalization of prebiotic reactions within primitive vesicles is considered a necessary step in the transition of inanimate matter to life. By maintaining the identity of distinct self-replicating systems, compartmentalization facilitates competition and natural selection in early cells, and hence opens the door towards the onset of Darwinian evolution. Membrane boundaries are critical to maintain sufficient concentrations of nutrients essential for primitive metabolism, generate gradients required for energy capture and transfer, and retain genetically derived polymeric products within specific microscale compartments. Single-chain amphiphiles such as fatty acids are plausible prebiotic compounds, and can spontaneously self-assemble into vesicles exhibiting encapsulation properties, selective permeability and dynamical behaviour that together comprise a suitable system for development as a feasible protocell model. In recent years, although significant progress has been made towards the synthesis of cell-mimicking systems in the laboratory, these advances only partially fulfil the requirements necessary for the realization of a fully developed protocell model. For example, most experiments to date have investigated only one or two component subsystems in isolation, such as vesicle properties, confinement of biochemical transformations, or template copying of encapsulated genetic molecules. Clearly, a protocell model that could successfully combine these elements in one integrated system would represent a major breakthrough in this emerging field of exciting interdisciplinary research.Acknowledgements
We thank Avinash Patil and Adam Perriman for reading the manuscript and helpful comments.Notes and references
- P. Mazzarello, Nat. Cell Biol., 1999, 1, E13–E15 CrossRef CAS.
- A. I. Oparin, The Origin of Life, Macmillan, New York, 1938 Search PubMed.
- P. L. Luisi, The Emergence of Life, Cambridge University Press, 2006 Search PubMed.
- W. Gilbert, Nature, 1986, 319, 618 CrossRef.
- J. C. Lacey, G. W. Cook and D. W. Mullins, CHEMTRACTS—Biochemistry and Molecular Biology, 1999, 12, 398–418 CAS.
- F. J. Dyson, Origins of Life, Cambridge University Press, 1985 Search PubMed.
- S. Mann, Angew. Chem., Int. Ed., 2008, 47, 5307–5320 CrossRef.
- C. M. Fraser, et al. , Science, 1995, 270, 397–403 CAS.
- B. Alberts, et al., Molecular Biology of the Cell, Garland Sciences, New York, 5th edn, 2008 Search PubMed.
- We have not included the evolutionary capacity of cells in the above list even though this is clearly a fundamental property of life. Not all cells exhibit Darwinian evolution—for example, sterile cells are metabolically active but do not have the ability to evolve, and a male mule is infertile. However, such examples are dependent on living, evolving progenitors.
- P. L. Luisi, P. Walde and T. Oberholzer, Curr. Opin. Colloid Interface Sci., 1999, 4, 33–39 CrossRef CAS.
- I. A. Chen and P. Walde, Cold Spring Harbor Perspect. Biol., 2010, 2, 13 CrossRef.
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS.
- J. P. Schrum, T. F. Zhu and J. W. Szostak, Cold Spring Harbor Perspect. Biol., 2010, 2, 15 CrossRef.
- J. N. Israelachvili, D. J. Mitchell and B. W. Ninham, Biochim. Biophys. Acta, Biomembr., 1977, 470, 185–201 CrossRef CAS.
- I. Budin, R. J. Bruckner and J. W. Szostak, J. Am. Chem. Soc., 2009, 131, 9628–9629 CrossRef CAS.
- R. Zana, Encyclopedia of Supramolecular Chemistry, Taylor & Francis, 2004 Search PubMed.
- P. A. Monnard and D. W. Deamer, Anat. Rec., 2002, 268, 196–207 CrossRef CAS.
- D. W. Deamer, Nature, 1985, 317, 792–794 CrossRef CAS.
- J. G. Lawless and G. U. Yuen, Nature, 1979, 282, 396–398 CrossRef CAS.
- L. P. Dworkin, D. W. Deamer, S. A. Sandford and L. J. Allamandola, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 815–819 CrossRef.
- T. M. McCollom, G. Ritter and B. R. T. Simoneit, Origins Life Evol. Biospheres, 1999, 29, 153–166 CrossRef CAS.
- G. U. Yuen, J. G. Lawless and E. H. Edelson, J. Mol. Evol., 1981, 17, 43–47 CrossRef CAS.
- C. L. Apel, D. W. Deamer and M. N. Mautner, Biochim. Biophys. Acta, Biomembr., 2002, 1559, 1–9 CrossRef CAS.
- P. Walde, BioEssays, 2010, 32, 296–303 CrossRef CAS.
- J. P. Reeves and R. M. Dowben, J. Cell. Physiol., 1969, 73, 49–60 CrossRef CAS.
- V. Noireaux and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17669–17674 CrossRef CAS.
- P. L. Luisi, F. Ferri and P. Stano, Naturwissenschaften, 2005, 93, 1–13 CrossRef.
- Y. Kuruma, P. Stano, T. Ueda and P. Luisi, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 567–574 CrossRef CAS.
- P. Walde, A. Goto, P. A. Monnard, M. Wessicken and P. L. Luisi, J. Am. Chem. Soc., 1994, 116, 7541–7547 CrossRef CAS.
- S. S. Mansy, J. P. Schrum, M. Krishnamurthy, S. Tobe, D. A. Treco and J. W. Szostak, Nature, 2008, 454, 122–U110 CrossRef CAS.
- S. S. Mansy and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 13351–13355 CrossRef CAS.
- M. M. Hanczyc, S. M. Fujikawa and J. W. Szostak, Science, 2003, 302, 618–622 CrossRef CAS.
- P. Stano and P. L. Luisi, Chem. Commun., 2010, 46, 3639–3653 RSC.
- N. Berclaz, M. Muller, P. Walde and P. L. Luisi, J. Phys. Chem. B, 2001, 105, 1056–1064 CrossRef CAS.
- I. A. Chen and J. W. Szostak, Biophys. J., 2004, 87, 988–998 CrossRef CAS.
- T. F. Zhu and J. W. Szostak, J. Am. Chem. Soc., 2009, 131, 5705–5713 CrossRef CAS.
- T. Oberholzer, R. Wick, P. L. Luisi and C. K. Biebricher, Biochem. Biophys. Res. Commun., 1995, 207, 250–257 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
