 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The effect of intermolecular hydrogen bonding on the planarity of amides†
James A.
Platts
*,
Hasmerya
Maarof
,
Kenneth D. M.
Harris
*,
Gin Keat
Lim
and
David J.
Willock
School of Chemistry, Cardiff University, Park Place, Cardiff CF10 3AT, Wales, UK. E-mail: Platts@Cardiff.ac.uk; HarrisKDM@Cardiff.ac.uk; Fax: +44 (0)2920-874030; Tel: +44 (0)2920-874950 Tel: +44 (0)2920-870133
First published on 12th July 2012
Abstract
Ab initio and density functional theory (DFT) calculations on some model systems are presented to assess the extent to which intermolecular hydrogen bonding can affect the planarity of amide groups. Formamide and urea are examined as archetypes of planar and non-planar amides, respectively. DFT optimisations suggest that appropriately disposed hydrogen-bond donor or acceptor molecules can induce non-planarity in formamide, with OCNH dihedral angles deviating by up to ca. 20° from planarity. Ab initio energy calculations demonstrate that the energy required to deform an amide molecule from the preferred geometry of the isolated molecule is more than compensated by the stabilisation due to hydrogen bonding. Similarly, the NH2 group in urea can be made effectively planar by the presence of appropriately positioned hydrogen-bond acceptors, whereas hydrogen-bond donors increase the non-planarity of the NH2 group. Small clusters (a dimer, two trimers and a pentamer) extracted from the crystal structure of urea indicate that the crystal field acts to force planarity of the urea molecule; however, the interaction with nearest neighbours alone is insufficient to induce the molecule to become completely planar, and longer-range effects are required. Finally, the potential for intermolecular hydrogen bonding to induce non-planarity in a model of a peptide is explored. Inter alia, the insights obtained in the present work on the extent to which the geometry of amide groups may be deformed under the influence of intermolecular hydrogen bonding provide structural guidelines that can assist the interpretation of the geometries of such groups in structure determination from powder X-ray diffraction data.
Introduction
The geometry of amide CONH2 groups, particularly concerning the planarity at the N atom, has been the subject of a range of experimental and computational studies, revealing that the ground-state geometry of the isolated molecule in the gas phase is planar in some cases (e.g. formamide) and non-planar in other cases (e.g. urea).1 In contrast, in the crystal structure of “pure” urea2 and the crystal structures of the widely studied urea inclusion compounds3 (which contain a tunnel host structure constructed from a hydrogen-bonded arrangement of urea molecules), the urea molecule is completely planar. The potential energy involved in the perturbation of amides from planarity has been studied in detail, and many studies of the origin of planarity versus non-planarity have been reported.1 These calculations generally support the conventional resonance view of the bonding in amides, showing charge transfer from N to C and O giving some double bond character to the C–N bond. However, it is also pointed out that there is accompanying C to N σ-donation so that the calculated atomic charges are smaller than expected from transfer of the N lone pair density to C and O. The hydrogen bonding interactions of amides, most notably with water, have also been the focus of many reports.4Clearly, as observed for urea in the crystalline state, the energy required to deform the amide group from the ground-state geometry of the isolated molecule can be compensated by the formation of an appropriate arrangement of intermolecular hydrogen bonds. Although such observations are already well known, there is nevertheless the need for systematic studies to establish, on a more quantitative basis, the extent to which the degree of planarity of amide groups may be modulated by the formation of intermolecular hydrogen bonds. This may involve one or both of the N–H bonds of the NH2 group as the donor in an N–H⋯X hydrogen bond to a neighbouring hydrogen-bond acceptor X and/or may involve the N atom of the NH2 group as the acceptor in a Y–H⋯N hydrogen bond to a neighbouring hydrogen-bond donor Y–H. A recent example comes from the anticonvulsant drug carbamazepine, for which accurate predictions of observed crystal structures were only possible if the amide group was allowed to deform in order to optimise hydrogen bonding.5 Thus, one of the main motivations underlying the present study is to establish reasonable geometric guidelines for the types of deformation from the ground-state geometry of isolated amide molecules that might occur in crystal structures involving hydrogen bonding of the amide group of the same molecules.
Although neutron diffraction is the most definitive technique for accurate determination of the positions of H (or deuterium) atoms in crystal structures,6 single-crystal X-ray diffraction (XRD) can also establish the positions of H atoms in organic crystal structures with a reasonable degree of accuracy (particularly by exploiting modern instrumentation for high-quality data collection), allowing the degree of planarity of amide groups to be established reliably from analysis of such data.6 However, although single-crystal XRD is a powerful experimental technique for determining crystal structures, the requirement for a suitable single-crystal specimen imposes a limitation on the applicability of such methods. When a suitable single crystal of the material of interest cannot be prepared, structure determination must be tackled instead from powder XRD data.7 The task of carrying out structure determination directly from powder XRD data is considerably more challenging than from single-crystal XRD data, particularly in the case of organic materials, and the accuracy of the structural information obtained from analysis of powder XRD data is significantly lower. Thus, in general, crystal structure determination from powder XRD data cannot establish unambiguously the positions of H atoms in organic crystal structures, and hence the geometry of an amide group may be difficult to establish reliably by this technique. In favourable cases, the locations of the H atoms might be inferred from the positions that give the most reasonable N–H⋯X hydrogen bonds to neighbouring hydrogen-bond acceptor groups, but if optimization of the hydrogen-bond geometry would entail a significant deformation of the amide group from its geometry in the isolated molecule, then caution must be exercised in making such assignments. The positions of H atoms are the consequence of a balance between intra-molecular forces and inter-molecular forces, so that relying on geometric considerations alone without insights into the true balance between these two effects is unlikely to be reliable. Thus, another aim of this work is to provide quantitative insights on the extent to which the planarity of amide groups may be deformed under the influence of intermolecular hydrogen bonding, in order to assist the interpretation of the geometries of such groups when carrying out structure determination from powder XRD data.
For this study, we focus on formamide and urea as model amide molecules, recognizing (as stated above) that the degree of planarity of the amide group in the isolated molecule is significantly different in the case of formamide and urea. Furthermore, we also consider glycyl glycine as an example of an amide group in a model peptide. We consider complexes comprising these molecules together with one or more hydrogen-bond acceptor or hydrogen-bond donor molecules, focusing on hydrogen cyanide (NCH) as a model hydrogen-bond acceptor and hydrogen fluoride (HF) as a model hydrogen-bond donor. Rather than searching for minima on the potential energy surface, we take the approach of fixing the position of the hydrogen-bond donor or acceptor to explore the response of the amide in a systematic and controlled manner. In each case, the linear geometry of these molecules ensures that the results are not influenced by the formation of secondary interactions. In the case of glycyl glycine, water is also considered as a hydrogen-bond donor.
Computational methods
All geometries were optimised using tight convergence criteria at the B3LYP/aug-cc-pVnZ (n = D or T) level8,9 using Gaussian03.10 B3LYP was chosen due to its proven ability to describe the geometry and strength of hydrogen bonds.11 In the calculations on individual molecules, point group symmetry was conserved, i.e. Cs for formamide, C2 for urea and C∞v for HF and NCH. For the calculations on the hydrogen-bonded complexes, no such symmetry constraints were applied. Glycyl glycine was constructed in its extended form, i.e. ϕ = ψ = 180°, but no symmetry constraints were applied. Local minima and transition states were confirmed as such from the number of negative eigenvalues of the Hessian matrix following harmonic frequency calculation. The effect of the orientation of hydrogen bonding was explored by fixing relevant dihedral angles that define the geometry of interaction of the donor and acceptor, and possible complications due to secondary interactions were avoided by enforcing linearity of all hydrogen bonds. All other geometric parameters, including donor–acceptor distances, were fully optimized in all cases.Single-point energies were evaluated at these constrained optimized geometries using the density-fitting, local 2nd order Møller–Plesset (DF-LMP2) method,12 again with the aug-cc-pVTZ basis set.9 This method has been shown to closely reproduce conventional MP2 energies at a fraction of the computational cost, and also renders basis set superposition error negligible by construction, and hence has been tested extensively for a range of non-covalent interactions.13 Auxiliary fitting basis sets appropriate to aug-cc-pVTZ were taken from ref. 14, and orbitals were localized by the Pipek–Mezey scheme15 with orbital domains selected using the criteria of Broughton and Pulay.16 All such calculations were performed using Molpro version 2010.1.17
In this study, we use Ebind to denote the binding energy of the complex, representing the total energy associated with formation of the geometry-optimized complex starting from the individual (isolated) molecules in their ground-state geometries. The binding energy Ebind can be broken down into two separate contributions denoted Edef (deformation energy) and Eint (interaction energy). Thus, Edef is the energy associated with deformation of the isolated molecules from their ground-state geometry to the geometry that they adopt in the geometry-optimized complex, and Eint is the energy associated with formation of the (geometry-optimized) complex starting from the isolated molecules already in the deformed geometry adopted in the complex, such that Ebind = Edef + Eint.
Results and discussion
1. Formamide
The geometry of formamide optimized in Cs symmetry is in excellent agreement with the results of previous experiments and high-level ab initio calculations,1a and harmonic frequency calculation confirms that the planar structure is a true minimum. In this geometry, the amide N atom may be expected to act only as a weak hydrogen-bond acceptor due to delocalization of its lone pair. This view is supported by the electrostatic potential (plotted on a 0.001 a.u. electron density isosurface; Fig. 1), which shows effectively zero potential directly above and below the N atom. For comparison, the analogous value for NH3 constrained to a planar (D3d) geometry is −0.037 a.u., confirming that delocalisation across the amide affects the electrostatic potential.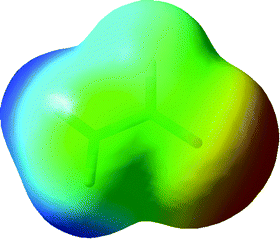 | ||
| Fig. 1 Electrostatic potential of formamide plotted on a 0.001 a.u. electron density isosurface: values run from +0.077 a.u. (blue) to −0.077 a.u. (red), and green represents zero. | ||
Introducing HF as a hydrogen-bond donor at a fixed ∠(CN⋯H) = 109.471°, with linearity of the hydrogen bond enforced but all other geometrical parameters relaxed, leads to a stable complex with r(N⋯H) = 1.863 Å.‡ Geometric and energetic properties of this complex are summarized in Table 1 and illustrated in Fig. 2 (full geometric details of all complexes are available as ESI†, as are results obtained from other DFT methods18). The most striking geometric feature is the non-planarity of the NH2 group, which adopts a slightly asymmetric pyramidal form, evident from the dihedral angles involving the H atoms in the syn and anti positions. The degree of non-planarity can be further quantified by the sum of the angles around the N atom, which equals 345.4° in formamide⋯HF compared to the extremes of 360° for planar and 328.4° for tetrahedral forms. The formation of the hydrogen bond with HF also leads to a decrease in r(C–O) and an increase in r(C–N), suggesting that the delocalization across the amide bond is disrupted. While B3LYP performs well for hydrogen bonding, we felt it necessary to examine the dependence of these properties on the method used. Tables S2 and S3 (ESI†) show that a range of modern DFT methods, as well as MP2, give essentially identical information to that reported for formamide⋯HF in Table 1.
| r(C–O) | r(C–N) | OCNHsyn | OCNHanti | E int | E def | E bind | |
|---|---|---|---|---|---|---|---|
| a ∠(CN⋯H) was fixed at 109.471°. b HF molecule was constrained as in a, and the other HF molecule was engaged in hydrogen bonding to the O atom of formamide. c The (O)CN⋯NC(H) dihedral angle was fixed at the value indicated. | |||||||
| Formamide | 1.211 | 1.357 | 0.00 | 180.00 | |||
| Formamide⋯HFa | 1.202 | 1.389 | 19.72 | 156.99 | −5.67 | +1.14 | −4.53 |
| Formamide⋯(HF)2b | 1.213 | 1.370 | 17.58 | 160.02 | −3.27 | +1.09 | −2.18 |
| NCH syn 0c | 1.213 | 1.354 | 0.00 | 180.00 | −3.99 | +0.12 | −3.88 |
| NCH syn 15c | 1.213 | 1.355 | 8.37 | 174.67 | −3.95 | +0.16 | −3.79 |
| NCH syn 30c | 1.212 | 1.357 | 14.62 | 171.46 | −3.76 | +0.28 | −3.48 |
| NCH anti 180c | 1.214 | 1.352 | 0.00 | 0.00 | −4.80 | +0.11 | −4.70 |
| NCH anti 165c | 1.214 | 1.353 | 4.52 | 170.77 | −4.72 | +0.15 | −4.57 |
| NCH anti 150c | 1.214 | 1.356 | 7.24 | 163.64 | −4.45 | +0.27 | −4.18 |
| 2 NCH 0/180c | 1.216 | 1.349 | 0.00 | 180.00 | −7.79 | +0.21 | −7.58 |
| 2 NCH 30/150c | 1.215 | 1.355 | 13.89 | 164.16 | −7.06 | +0.38 | −6.68 |
| 2 NCH −30/150c | 1.216 | 1.350 | −7.62 | 170.41 | −6.67 | +0.58 | −6.09 |
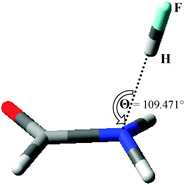 | ||
| Fig. 2 Optimized geometry of the formamide⋯HF complex. | ||
The calculated energies, also reported in Table 1, indicate that the interaction between formamide and HF is reasonably strong with an overall binding energy of −4.5 kcal mol−1. The components of this binding energy give further insights into the nature of this interaction: the deformation energy Edef is 1.14 kcal mol−1, with contributions of 0.90 kcal mol−1 for formamide and 0.24 kcal mol−1 for HF, and the “frozen” interaction energy Eint is −5.7 kcal mol−1. Thus, it is clear that the energy required to deform the individual molecules into the geometries adopted in the hydrogen-bonded complex is more than compensated by the stabilising energy of the hydrogen bond. To investigate whether the formation of a hydrogen bond to the O atom of formamide affects the hydrogen bonding and planarity at the N atom, we have also examined a formamide⋯(HF)2 complex. From the results in Table 1, the geometry of the NH2 group in this complex is less pyramidal than that in the formamide⋯HF complex; both the r(C–O) and r(C–N) distances are closer to those in the isolated formamide molecule, and the hydrogen-bond strength is reduced. Possible underlying reasons are explored in more detail below.
In addition to acting as a hydrogen-bond acceptor through the N atom, formamide can also act as a hydrogen-bond donor involving one or both of the N–H bonds, and we have studied this type of interaction using hydrogen cyanide (NCH) as a model acceptor. Specifically, the geometry and energy of such N–H⋯N hydrogen bonds has been probed as a function of the extent to which the NCH molecule lies out of the plane of the formamide molecule (Table 1). Unsurprisingly, the strongest N–H⋯N hydrogen bonds are formed when the NCH molecule lies in the same plane as the formamide molecule; in this case, the hydrogen bonds do not perturb the planarity of formamide, and the hydrogen bond strength is similar to that in the formamide⋯HF complex discussed above, with very small deformation energies. Moving the NCH molecule out of the plane of formamide by 15° or 30° is observed to induce non-planarity of the NH2 group. With a single NCH molecule, the H atom involved in the hydrogen bond moves out of the plane rather more than the non-interacting H atom, which also moves out of the plane (even though it is not close to the NCH molecule). For instance, with NCH placed close to Hanti at 30° out of the mean plane of the non-H atoms, the N–Hanti bond is oriented more than 16° from the OCN plane and the N–Hsyn bond is oriented more than 7° from this plane. As the NCH molecule moves further out of the plane of the formamide molecule, the strength of the N–H⋯N hydrogen bonds is reduced, and the deformation energy increases slightly; nevertheless, it is apparent that the hydrogen bonds are more than strong enough to overcome the innate preference for planarity of formamide.
It is also possible to form stable complexes of formamide with two NCH molecules, in which each N–H bond of formamide forms a hydrogen bond with an NCH molecule (denoted “2 NCH” in Table 1). The configuration in which both NCH molecules are in the same plane as formamide is confirmed as a true energy minimum by harmonic frequency calculation. This complex has slightly shorter r(C–N) than the isolated formamide molecule, but is otherwise similar to that in the formamide⋯NCH complexes. Moving both NCH molecules out of the OCN plane by 30° brings the syn and anti H atoms out of the plane, again to a similar extent as observed in the formamide⋯HF complexes.
We have also considered another type of formamide⋯(NCH)2 complex, in which one NCH molecule lies above the formamide plane and the other NCH molecule lies below this plane, which results in a “twisted” orientation of the NH2 group. The deviations from the mean plane of formamide are less than 10°, which is smaller than the deviations observed in the pyramidal cases discussed above, while the C–NH2 group remains approximately planar (the HCNH improper dihedral angle is 178.06°). The binding energy of this complex is rather more negative than the sum of the individual binding energies of the two formamide⋯NCH complexes involving the syn and anti N–H bonds as the hydrogen-bond donor, and the deformation energy is higher than for either of the formamide⋯NCH complexes.
It is apparent from Table 1 that, as the NCH molecule moves out of the plane of the formamide molecule, the H atoms of the NH2 group “follow” it out of the plane, although the dihedral angle that defines the deformation of the NH2 group from planar geometry is rather smaller than that used to fix the orientation of the NCH molecule. To further probe this behaviour, the results obtained with the orientation of a single NCH molecule fixed at dihedral angles ranging from 0 to 60° are shown in Fig. 3. On increasing this angle from 0 to 30°, an approximately linear increase of the OCNH dihedral angle is observed, beyond which the value of the OCNH dihedral angle levels off at a maximum of around 20°. Throughout the linear regime, the OCNH dihedral angle is approximately 50% of the value used to fix the orientation of the NCH molecule, indicating that even for small deviations from planarity, the hydrogen bonds are not perfectly linear and that the deformation of the NH2 group “lags behind” the hydrogen-bond acceptor molecule.
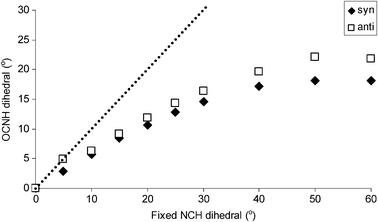 | ||
| Fig. 3 Response of the OCNH dihedral angle to changes in the NCH dihedral angle (which defines the orientation of the NCH molecule out of the plane of the formamide molecule). The dotted line denotes an idealised case of intercept = 0 and slope = 1, which would correspond to the OCNH dihedral angle being equal to the fixed NCH dihedral. | ||
2. Urea
In contrast to formamide, the equilibrium geometry of urea in the gas-phase is non-planar. For urea, B3LYP optimization in C2 symmetry again gives a geometry in close agreement with previous experimental and theoretical results,1 and harmonic frequency calculation confirms that this geometry represents a true minimum. In this respect, the behaviour of urea and formamide contrast significantly with each other, leading to a significant change in the electrostatic potential, which for urea is negative around the “lone pair” region of each N atom (Fig. 4).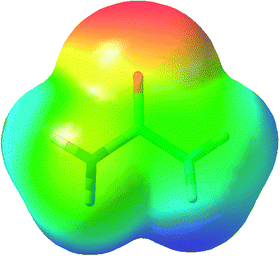 | ||
| Fig. 4 Electrostatic potential of urea plotted on a 0.001 a.u. electron density isosurface (colours as in Fig. 1). A region of slightly negative (yellow) potential is evident on the left-hand N atom, but this region is obscured for the right-hand N atom due to the C2 symmetry of the molecule. | ||
A urea⋯HF complex in which HF acts as the donor in an N⋯H–F hydrogen bond to a single N atom of urea [with ∠(CN⋯H) fixed at 109.471°] is a stable complex, with r(N⋯H) = 1.773 Å. Geometric and energetic aspects of this complex are given in Table 2. The results show that formation of this N⋯H–F hydrogen bond induces the NH2 group to become more non-planar than in the isolated urea molecule, with the OCNH dihedral angles changing by more than 5°. As observed for formamide, the r(C–O) and r(C–N) distances also change substantially; in particular, r(C–O) decreases and r(C–N) increases, which may indicate that the hydrogen bonding leads to a reduction in the delocalization over the OCNH unit. Interestingly, the NH2 group that is not involved in hydrogen bonding is perturbed by the hydrogen-bonding interaction of the other NH2 group with HF. Thus, for the NH2 group that is not engaged in the N⋯H–F hydrogen bond, r(C–N) is shorter and the geometry around the N atom is less pyramidal than in the isolated urea molecule (discussed in more detail below). The N⋯H–F hydrogen bond to urea is rather stronger than the corresponding hydrogen bond to formamide, and the deformation energy is slightly lower, presumably as a consequence of the inherently pyramidal geometry of urea itself.
| r(C–O) | r(C–N)a | OCNHsyna | OCNHantia | E int | E def | E bind | |
|---|---|---|---|---|---|---|---|
| a Where relevant, the first line reports data for the atoms closest to the hydrogen bond, and the second line reports data for the more distant atoms. b ∠(CN⋯H) was fixed at 109.471°. c The (O)CN⋯NC(H) dihedral angle was fixed at the value indicated. | |||||||
| Urea | 1.216 | 1.382 | 12.46 | 153.38 | |||
| Urea⋯HFb | 1.209 | 1.416 | 17.85 | 145.05 | −8.35 | +0.98 | −7.37 |
| 1.370 | 10.68 | 156.44 | |||||
| NCH syn 0c | 1.218 | 1.373 | 6.36 | 156.54 | −3.38 | +0.29 | −3.10 |
| 1.390 | 13.00 | 151.14 | |||||
| NCH syn 30c | 1.217 | 1.379 | 18.19 | 156.40 | −3.34 | +0.14 | −3.20 |
| 1.388 | 13.91 | 152.91 | |||||
| NCH anti 180c | 1.220 | 1.368 | 8.81 | 170.77 | −5.08 | +0.69 | −4.39 |
| 1.384 | 15.83 | 159.07 | |||||
| NCH anti 150c | 1.219 | 1.378 | 11.78 | 152.85 | −4.52 | +0.14 | −4.39 |
| 1.383 | 14.19 | 156.98 | |||||
| 2 NCH 0/180c | 1.221 | 1.363 | 2.59 | 174.09 | −7.76 | +0.94 | −6.83 |
| 1.390 | 16.64 | 157.50 | |||||
| 2 NCH 30/150c | 1.220 | 1.376 | 17.22 | 154.57 | −6.78 | +0.22 | −6.56 |
| 1.387 | 15.49 | 156.47 | |||||
| 2 NCH −30/150c | 1.221 | 1.367 | −3.41 | 156.09 | −6.26 | +0.86 | −5.40 |
| 1.390 | 13.97 | 153.83 | |||||
Complexes of urea with NCH as a hydrogen-bond acceptor were also studied, and the results are reported in Table 2. As observed for formamide, the geometry of the NH2 group of urea is sensitive to the position of the NCH molecule. Thus, forcing the NCH molecule to remain in the plane of the non-H atoms of urea increases the planarity of the NH2 group by between 3° and 6° in comparison to the isolated urea molecule, with the H atom involved in the N–H⋯N hydrogen bond lying closer to this plane than the other H atom. Formation of this N–H⋯N hydrogen bond to one NH2 group also affects the remote NH2 group of the urea molecule, leading to a slight decrease in planarity. In contrast, forcing the NCH molecule to lie 30° out of the plane of the non-H atoms of urea results in the geometry of the NH2 group becoming even more pyramidal than in the isolated urea molecule.
These effects are enhanced when each N–H bond of a single NH2 group forms an N–H⋯N interaction with an NCH molecule (denoted “2 NCH” in Table 2, and illustrated in Fig. 5). In this case, with two NCH molecules in the plane of the non-H atoms of urea, the OCNH dihedral angles become 2.6° and 174.1°, while with both NCH molecules lying out of the plane by 30°, the OCNH dihedral angles are larger than in the complexes with a single NCH molecule. Table 2 also reports results for a twisted complex, with one NCH molecule above the plane of the non-H atoms of urea and one NCH molecule below this plane. Again, the NH2 group deforms in order to establish hydrogen bonding, resulting in one negative dihedral angle and one positive dihedral angle. However, in this case, the C–NH2 group remains pyramidal (the HCNH improper dihedral angle is 159.50°).
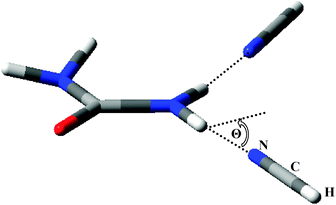 | ||
| Fig. 5 Geometry of urea⋯(NCH)2, illustrating the dihedral angle used to constrain the complex. | ||
The DF-LMP2 energies indicate that the urea⋯HF complex is rather more strongly bound than the formamide⋯HF complex, whereas the urea⋯NCH complex is slightly more weakly bound than the formamide⋯NCH complex. Thus, urea is a better hydrogen-bond acceptor but a worse hydrogen-bond donor than formamide. The deformation energies for urea are generally slightly smaller than for formamide. The overall value of 0.98 kcal mol−1 for the urea⋯HF complex stems equally from urea and from HF, such that less than 0.5 kcal mol−1 is required to deform the urea molecule to the more pyramidal structure adopted in this complex. The largest deformation energies are found for the urea⋯(NCH)2 complexes, for which the values approach 1 kcal mol−1 for the 0/180 case and the twisted complexes.
Several geometric parameters reported in Tables 1 and 2 suggest that hydrogen bonding affects the electronic delocalization across the amide bond. To probe this issue in more detail, Table 3 shows the atomic charges and bond orders calculated using the Natural Bond Orbital (NBO) approach19 from B3LYP/aug-cc-pVTZ orbitals. In the isolated formamide molecule, the C–O and C–N bond orders support the delocalized picture of bonding. For the isolated urea molecule, the charges on the O and N atoms are more negative, and the C–O and C–N bond orders are lower, than in formamide. We propose that these differences are responsible for the contrasting degrees of planarity of the isolated formamide (planar) and urea (non-planar) molecules; thus, each N atom in urea donates some electron density to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group, but the capacity of the C
O group, but the capacity of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group to accept this electron density is limited. As a consequence, each N atom in urea retains more electron density (q = −0.83) than the N atom in formamide (q = −0.80) and cannot achieve full delocalization.
O group to accept this electron density is limited. As a consequence, each N atom in urea retains more electron density (q = −0.83) than the N atom in formamide (q = −0.80) and cannot achieve full delocalization.
| q(O) | q(N) | C–O | C–N | |
|---|---|---|---|---|
| Formamide | −0.60 | −0.80 | 1.73 | 1.22 |
| Formamide⋯HF | −0.55 | −0.85 | 1.80 | 1.12 |
| Formamide⋯NCH syn 30 | −0.60 | −0.80 | 1.72 | 1.23 |
| Formamide⋯NCH anti 30 | −0.61 | −0.80 | 1.72 | 1.23 |
| Urea | −0.64 | −0.83 | 1.65 | 1.12 |
| Urea⋯HF | −0.61 | −0.88 | 1.69 | 1.03 |
| −0.82 | 1.16 | |||
| Urea⋯NCH syn 0 | −0.66 | −0.84 | 1.64 | 1.10 |
| −0.84 | 1.15 | |||
| Urea⋯NCH anti 180 | −0.66 | −0.83 | 1.62 | 1.11 |
| 1.15 |
Formation of the urea⋯HF and formamide⋯HF complexes results in less negative charge at the O atom and more negative charge at the N atom, as well as increasing the C–O bond order and decreasing the C–N bond order, and indicating that pyramidalization of NH2 diminishes the delocalization of the lone pair of the N atom into the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group. In the urea⋯HF complex, there are also changes in the charge and bond order of the NH2 group that is not involved in hydrogen bonding to HF, resulting in less negative charge on the N atom and increased C–N bond order. These changes may be interpreted as evidence that hydrogen bonding to one N atom allows the other N atom to participate in greater delocalization with C
O group. In the urea⋯HF complex, there are also changes in the charge and bond order of the NH2 group that is not involved in hydrogen bonding to HF, resulting in less negative charge on the N atom and increased C–N bond order. These changes may be interpreted as evidence that hydrogen bonding to one N atom allows the other N atom to participate in greater delocalization with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, in general agreement with the geometric changes noted in Table 2. In contrast to the complexes with HF, the atomic charges and bond orders for the urea⋯NCH and formamide⋯NCH complexes are almost identical to those for the isolated urea and formamide molecules respectively, in spite of the substantial geometric re-arrangements that take place on forming these complexes. The reason underlying this difference is not immediately clear, but may indicate that the H atoms in an amide group are relatively free to move to maximise their strength as hydrogen bond donors without overall disruption of electronic structure, whereas for the N atom to act as a hydrogen bond acceptor inevitably requires substantial electronic rearrangement throughout the group.
O, in general agreement with the geometric changes noted in Table 2. In contrast to the complexes with HF, the atomic charges and bond orders for the urea⋯NCH and formamide⋯NCH complexes are almost identical to those for the isolated urea and formamide molecules respectively, in spite of the substantial geometric re-arrangements that take place on forming these complexes. The reason underlying this difference is not immediately clear, but may indicate that the H atoms in an amide group are relatively free to move to maximise their strength as hydrogen bond donors without overall disruption of electronic structure, whereas for the N atom to act as a hydrogen bond acceptor inevitably requires substantial electronic rearrangement throughout the group.
Of course, the physical significance of the non-planarity of the urea molecule depends on the relative magnitudes of the barrier associated with adopting a planar structure and the zero-point/thermal energy of the relevant vibrational mode(s). We estimate that this barrier is 289 cm−1 at the DF-LMP2 level,§ while the harmonic vibrational frequency for planarization of NH2 is 525 cm−1, corresponding to a zero-point energy level of 263 cm−1. Thus, the zero-point energy is insufficient to overcome the barrier, but at sufficiently high temperature, thermal motion may be expected to lead to the averaged structure being observed. Hydrogen bonding to HF alters this picture drastically. From our results, the barrier is estimated to be 2233 cm−1,§ while relevant vibrational modes are observed at 921 and 932 cm−1. In the urea⋯(NCH)2 complex with the two NCH molecules fixed at 30° and 150°, we estimate that the barrier to planarity is 883 cm−1, with relevant vibrational normal modes at 589 and 636 cm−1. Thus, even at elevated temperatures, we predict that hydrogen bonding should promote the non-planar form of urea.
As discussed above, the crystal structure of urea has a perfectly planar C2v structure of the urea molecule, with H atom positions determined accurately from neutron diffraction data.2Table 4 reports the results of calculations on a set of clusters extracted from the crystal structure determined at 12 K. The clusters are constructed to introduce the local crystal environment for a central urea molecule in a systematic manner. Optimisation of the larger clusters is unfeasible with the basis set used above, so the smaller 6-311++G(2d,2p) was chosen as a compromise between speed and accuracy for these studies (see ESI†). The clusters were constructed to introduce the local crystal environment for a central urea molecule in a systematic manner. Two approaches were taken: in one approach, all atoms of the central urea molecule were free to move without any symmetry or other constraint (denoted “free” in Table 4), whereas in the second approach, the non-H atoms of the central urea molecule were constrained to their positions in the crystal structure and only H atoms were fully optimized (denoted “constrained”). In both cases, the other urea molecules that represent the crystal environment were held fixed at their geometries in the crystal structure at 12 K.
| Free | Constrained | |||
|---|---|---|---|---|
| OCNHsyn | OCNHanti | OCNHsyn | OCNHanti | |
| a Results for the hydrogen-bond donor molecule. b Results for the central molecule. | ||||
| Monomer | 13.4 | 151.0 | 10.9 | 156.4 |
| Dimera | 12.5 | 163.5 | 12.0 | 171.3 |
| Trimer – linearb | 15.5 | 164.6 | 11.3 | 173.7 |
| Trimer – H-shapedb | 15.6 | 157.4 | 1.5 | 175.8 |
| Pentamerb | 5.0 | 169.7 | 0.2 | 179.8 |
These data show that constraining non-H atoms to their positions in the crystal structure affects even the planarity of a single urea molecule, with the fully optimised molecule exhibiting slightly larger dihedral angles. The presence of a single neighbour is sufficient to reduce the non-planarity of the urea molecule. In the free optimisation of the dimer, the free urea molecule moves out of the plane of the dimer and the hydrogen bonds between the molecules are elongated relative to the crystal structure (see Fig. S1, ESI†). Even with this flexibility, the dihedral angle of the anti H atoms (involved in hydrogen bonding) increases by more than 12°, whereas that of the syn H atoms (remote from the hydrogen bonding) changes by less than 1°. The constrained dimer retains the overall pattern of hydrogen bonding observed in the crystal structure, and exhibits larger changes towards planarity in the dihedral angles for both the syn and anti H atoms.
Extending the cluster to a “linear” trimer has only a small effect on the planarity of the central urea molecule compared to that for the dimer. In both optimisations, the syn H atoms are slightly further out-of-plane than in the monomer or dimer, whereas the anti H atoms lie slightly more in the plane than in the dimer. In contrast, an “H-shaped” trimer, in which the central molecule is hydrogen-bonded through its syn H atoms, shows large differences in the geometries obtained from the free and constrained optimisations. In the former case, all H atoms are significantly out-of-plane, whereas in the latter case, the syn and anti H atoms are within 2° and 5° of the idealised planar values, respectively. This trend is continued in the pentamer, which is the only structure considered that includes hydrogen bonds to both sets of H atoms of the central molecule. In the free optimisation, the dihedral angles of the central molecule are closer to planarity than in the monomer, but are still far from 0° and 180°, whereas for the constrained optimization, the central molecule is effectively planar. This observation suggests that the local hydrogen-bonding interactions in the crystal structure affect the planarity around the N atoms, but that longer range crystal packing forces are also required to hold the molecule as a whole at the geometry observed in the crystal structure (Fig. 6).
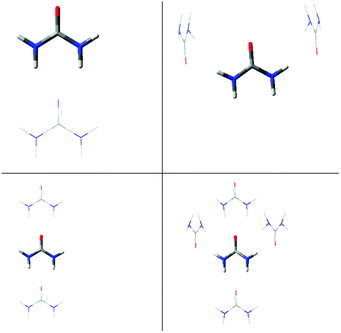 | ||
| Fig. 6 Constrained optimised structures of urea clusters. The “central” molecule for which data are reported in Table 4 is shown in bold, with all other molecules shown as wireframe. | ||
Further evidence of the important effect that intermolecular hydrogen bonding may exert on the planarity of the urea molecule stems from the crystal structures (determined from neutron diffraction) of two co-crystals of urea and oxalic acid,20 with stoichiometries (urea)2(oxalic acid)1 and (urea)1(oxalic acid)1 (CCDC references UROXAL01 and UROXAM01 respectively). For the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 co-crystal, the NH2 groups are significantly non-planar (OCNHsyn = −5.41° and −6.08°; OCNHanti = −176.2° and +178.4°), apparently due to the disposition of the hydrogen-bond acceptors (torsion angles: −3.2° and −7.3°; −176.8° and +173.6°). For the 1
1 co-crystal, the NH2 groups are significantly non-planar (OCNHsyn = −5.41° and −6.08°; OCNHanti = −176.2° and +178.4°), apparently due to the disposition of the hydrogen-bond acceptors (torsion angles: −3.2° and −7.3°; −176.8° and +173.6°). For the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 co-crystal, only the H atoms in the anti positions deviate significantly from planarity (−174.3°), and are again close to the orientation of the hydrogen-bond acceptor (−173.7°).
1 co-crystal, only the H atoms in the anti positions deviate significantly from planarity (−174.3°), and are again close to the orientation of the hydrogen-bond acceptor (−173.7°).
3. Glycyl-glycine
The importance of amide bonds in biology stems from their central role in peptides and proteins. Such bonds are widely assumed to be completely planar, for instance in fitting molecular structures to diffraction data or in developing atomistic force fields for simulations. However, the results presented above suggest that hydrogen bonding may distort peptide bonds from their preferred planarity. In order to probe the hydrogen bonding properties of such peptide bonds, we have examined the simplest possible model, glycyl-glycine (GlyGly).The B3LYP/aug-cc-pVTZ optimized geometry of GlyGly is essentially planar about the peptide bond. The formation of N⋯H–F hydrogen bonding to an HF molecule placed in the proximity of the amide N atom (with geometry optimization) does indeed induce a distortion of planarity, as summarized in Table 5 and shown in Fig. 7. The distortions from planarity of GlyGly induced by hydrogen bonding with HF are not as large as those observed for formamide, but are nonetheless significant at 15 to 20°. Moreover, the interaction of GlyGly with HF leads to a decrease of r(C–O) and an increase of r(C–N), as also observed for formamide. Table 5 also shows that the hydrogen bond in GlyGly⋯HF is rather stronger than that in formamide⋯HF, but not as strong as the hydrogen bond in urea⋯HF. The deformation energy (almost 2 kcal mol−1) associated with the formation of the hydrogen bond in GlyGly⋯HF is substantial, most of which (1.4 kcal mol−1) originates from deformation of the GlyGly molecule.
| r(C–O) | r(C–N) | OCNC | OCNH | E int | E def | E bind | |
|---|---|---|---|---|---|---|---|
| GlyGly | 1.221 | 1.356 | 1.05 | −176.77 | |||
| GlyGly⋯HF | 1.211 | 1.395 | −13.73 | −151.24 | −7.09 | +1.77 | −5.33 |
| GlyGly⋯H2O | 1.217 | 1.368 | −8.04 | −161.02 | −3.34 | +0.33 | −3.00 |
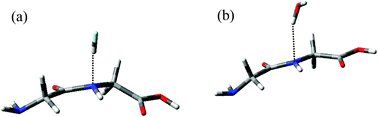 | ||
| Fig. 7 Optimized geometries of (a) GlyGly⋯HF and (b) GlyGly⋯H2O. | ||
Using HF as a model hydrogen-bond donor is convenient due to its small size and lack of secondary interactions, although HF would not be found in living systems. For this reason, a complex of GlyGly with water, the universal solvent of life, was also studied, and the results are reported in Table 5. Our results indicate that the N⋯H–O hydrogen bond formed between the N atom of GlyGly and water is weaker than that formed between the N atom of GlyGly and HF, and the corresponding distortions in molecular geometry of GlyGly are somewhat smaller for water than for HF, but with the same general pattern of bond lengths and dihedral angles observed in each case.
Concluding remarks
In conclusion, we have employed theoretical methods to explore the extent to which intermolecular hydrogen bonding can modify the degree of planarity of amide groups, motivated in part by the need to establish quantitative guidelines on the extent to which the geometry of amide groups may be deformed under the influence of intermolecular hydrogen bonding to assist the interpretation of the geometries of such groups in structure determination from powder XRD data, as well as by the interest in understanding the disparity in the structure of the urea molecule between gas-phase and crystal environments.Formamide has been used as a simple example for which the amide group is completely planar in the isolated molecule, as demonstrated previously by microwave spectroscopy and high-level calculations. Although the isolated formamide molecule is planar, the N atom can act as a hydrogen-bond acceptor (with HF selected in the present work as a model hydrogen-bond donor) to form a complex in which the amide becomes non-planar at the N atom. Our results indicate that the deformation energy required to form the non-planar structure (ca. 1 kcal mol−1) is more than compensated by the stabilisation due to hydrogen bonding (ca. 4.5 kcal mol−1). Moreover, delocalisation is disrupted by such hydrogen bonding, leading to a shorter C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond with greater double bond character, and a longer C–N bond with reduced multiple bond character, compared to the isolated formamide molecule. Interaction with hydrogen-bond acceptors can also affect the planarity of formamide (with NCH employed as a model acceptor). In the most stable geometry, all atoms are exactly in the plane of the formamide molecule, but if the acceptors are constrained to lie out of this plane, then the H atoms of the NH2 group “follow” them to produce non-planar structures. Progressively increasing the angle between the acceptor and the mean plane of the formamide molecule causes the non-planarity of the NH2 group also to increase, up to a maximum of approximately 20°, beyond which no significant increase in the deformation of the NH2 group is observed.
O bond with greater double bond character, and a longer C–N bond with reduced multiple bond character, compared to the isolated formamide molecule. Interaction with hydrogen-bond acceptors can also affect the planarity of formamide (with NCH employed as a model acceptor). In the most stable geometry, all atoms are exactly in the plane of the formamide molecule, but if the acceptors are constrained to lie out of this plane, then the H atoms of the NH2 group “follow” them to produce non-planar structures. Progressively increasing the angle between the acceptor and the mean plane of the formamide molecule causes the non-planarity of the NH2 group also to increase, up to a maximum of approximately 20°, beyond which no significant increase in the deformation of the NH2 group is observed.
A similar picture emerges for urea, which is well known to have a non-planar, C2 structure in the gas-phase, but is planar in the crystal structure. The N atoms of the NH2 groups in urea are stronger hydrogen-bond acceptors than the N atom of the NH2 group in formamide: a complex formed with HF as hydrogen-bond donor to this N atom induces greater non-planarity than the isolated urea molecule, with the deformation energy (ca. 1 kcal mol−1) readily outweighed by the hydrogen-bond stabilisation (ca. 7 kcal mol−1). The planarity of the urea molecule is also affected when the NH2 group forms one or two N–H⋯N hydrogen bonds with NCH molecule(s) as hydrogen-bond acceptor(s). If the acceptors are constrained to lie in the plane of the non-H atoms, then the H atoms respond by approaching a planar geometry for the NH2 group, whereas if the acceptors are constrained to lie out of this plane, then the urea molecule becomes less planar than the isolated molecule. In clusters ranging from a dimer to a pentamer extracted from the crystal structure of urea, the planarity of the amide group increases as the size of the cluster is increased. However, even for the pentamer, in which the central molecule forms all hydrogen-bond contacts to nearest neighbours formed by a molecule in the crystal structure, the central urea molecule is not exactly planar, indicating that longer-range effects play a role in controlling the planarity of the urea molecule in the crystal structure.
Amide groups play a particularly important role in biomolecules, forming the peptide bonds that make up all proteins. As a model of such molecules, we have examined the potential for intermolecular hydrogen bonding to induce non-planarity in glycyl-glycine, which is approximately planar as an isolated molecule (at the DFT level). Hydrogen bonds formed by the central N atom of this peptide with HF or H2O as donors show similar behaviour to the N atoms in formamide and urea discussed above, namely substantial non-planarity and small deformation energies that are easily overcome by the overall stabilisation due to hydrogen bonding.
Acknowledgements
JAP is grateful to the Leverhulme Trust for a Research Fellowship. GKL and HM are grateful to the Government of Malaysia for studentships.References
- (a) J. Demaison, A. G. Csázár, I. Kleiner and H. Møllendal, J. Phys. Chem. A, 2007, 111, 2574 CrossRef CAS; (b) B. E. Mannfors, N. G. Mirkin, K. Palmo and S. Krimm, J. Phys. Chem. A, 2003, 107, 1825 CrossRef CAS , and references cited therein; (c) K. B. Wiberg and K. E. Laidig, J. Am. Chem. Soc., 1987, 109, 5935 CrossRef CAS; (d) J. I. Mujika, J. M. Matxain, L. A. Eriksson and X. Lopez, Chem.–Eur. J., 2006, 12, 7215 CrossRef CAS; (e) Y. R. Mo, P. V. Schleyer, W. Wu, M. H. Lin, Q. Zhang and J. L. Gao, J. Phys. Chem. A, 2003, 107, 10011 CrossRef CAS; (f) H. Basch and S. Hoz, Chem. Phys. Lett., 1998, 294, 117 CrossRef CAS; (g) G. Fogarasi and P. G. Szalay, J. Phys. Chem. A, 1997, 101, 1400 CrossRef CAS.
- S. Swaminathan, B. M. Craven and R. K. McMullan, Acta Crystallogr., Sect. B: Struct. Sci., 1984, 40, 300 CrossRef.
- (a) K. D. M. Harris and J. M. Thomas, J. Chem. Soc., Faraday Trans., 1990, 86, 2985 RSC; (b) K. D. M. Harris, Supramol. Chem., 2007, 19, 47 CrossRef CAS.
- (a) R. L. T. Parreira, G. F. Caramori, N. H. Morgon and S. E. Galembeck, Int. J. Quantum. Chem., 2012, 112, 1401 CrossRef CAS; (b) M. Nagaraju and G. N. Sastry, Int. J. Quantum. Chem., 2010, 110, 1994 CAS; (c) A. A. Samchenko, A. V. Kabanov and V. M. Komarov, Biofizika, 2010, 55, 197 CAS; (d) R. L. T. Parreira, H. Valdes and S. E. Galembeck, Chem. Phys., 2006, 331, 96 CrossRef CAS; (e) J. Binoy, N. B. Prathima, C. M. Krishna, C. Santhosh, I. H. Joe and V. S. Jayakumar, Laser Phys., 2006, 16, 1253 CrossRef CAS; (f) S. Chalmet and M. F. Ruiz-Lopez, J. Chem. Phys., 1999, 111, 1117 CrossRef CAS; (g) T. Ishida, P. J. Rossky and E. W. Castner, J. Phys. Chem. B, 2004, 108, 17483 CrossRef.
- A. J. Cruz Cabeza, G. M. Day, W. D. S. Motherwell and W. Jones, Cryst. Growth Des., 2006, 6, 1858 CAS.
- (a) J. D. Dunitz, X-ray Analysis and the Structures of Organic Molecules, Verlag Helvetica Chimica Acta, Basel, 1995 Search PubMed; (b) J. P. Glusker and K. N. Trueblood, Crystal Structure Analysis – A Primer, Oxford University Press, Oxford, 1985 Search PubMed.
- (a) K. D. M. Harris and M. Tremayne, Chem. Mater., 1996, 8, 2554 CrossRef CAS; (b) ed. W. I. F. David, K. Shankland, L. B. McCusker and C. Baerlocher, Structure Determination from Powder Diffraction Data, OUP/IUCr, 2002 Search PubMed; (c) K. D. M. Harris, Cryst. Growth Des., 2003, 3, 887 CrossRef CAS.
- (a) A. D. Becke, J. Chem. Phys., 1993, 98, 5648 CrossRef CAS; (b) C. T. Lee, W. T. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785 CrossRef CAS.
- R. A. Kendall, T. H. Dunning Jr. and R. J. Harrison, J. Chem. Phys., 1992, 96, 6796 CrossRef CAS.
- M. J. Frisch, et al., Gaussian03 Rev. E.01, Gaussian Inc, Wallingford CT, 2004 Search PubMed.
- See for example (a) M. Lozynski, D. Rusinska-Roszak and H.-G. Mack, J. Phys. Chem. A, 1998, 102, 2899 CrossRef CAS; (b) I. V. Alabugin, M. Manoharan, S. Peabody and F. Weinhold, J. Am. Chem. Soc., 2003, 125, 5973 CrossRef CAS; (c) A. D. Rabuck and G. E. Scuseria, Theor. Chem. Acc., 2000, 104, 439 CrossRef CAS.
- H.-J. Werner, F. R. Manby and P. J. Knowles, J. Chem. Phys., 2003, 118, 8149 CrossRef CAS.
- (a) M. Schütz, G. Rauhut and H.-J. Werner, J. Phys. Chem. A, 1998, 102, 5997 CrossRef; (b) J. G. Hill, J. A. Platts and H.-J. Werner, Phys. Chem. Chem. Phys., 2006, 8, 4072 RSC.
- F. Weigend, A. Köhn and C. Hättig, J. Chem. Phys., 2002, 116, 3175 CrossRef CAS.
- J. Pipek and P. G. Mezey, J. Chem. Phys., 1989, 90, 4916 CrossRef CAS.
- J. W. Boughton and P. J. Pulay, J. Comput. Chem., 1993, 14, 736 CrossRef CAS.
- H.-J. Werner, P. J. Knowles, R. Lindh, F. R. Manby and M. Schütz, Molpro, version, 2010.1, a package of ab initio programs; see: http://www.molpro.net Search PubMed.
- Bond lengths differ from B3LYP values by less than 0.01 Å and dihedral angles by less than 1° with four different DFT methods. See Table S2 (ESI†) for more information.
- (a) A. E. Reed, R. B. Weinstock and F. Weinhold, J. Chem. Phys., 1985, 83, 735 CrossRef CAS.
- (a) G. J. van Hummel and R. B. Helmholdt, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1991, 47, 213 CrossRef; (b) S. Harkema, J. H. ter Brake and R. B. Helmholdt, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1984, 40, 1733 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Optimised coordinates of all molecules and complexes reported, basis set dependence of the binding energy of the urea dimer, and method dependence of formamide⋯HF geometry and binding energy. See DOI: 10.1039/c2cp41716b |
| ‡ Full unconstrained optimization of formamide⋯HF starting from ∠(CN⋯H) = 109.471° gives a true minimum with r(N⋯H) = 1.865 Å and ∠(CN⋯H) = 109.661°, but with non-linear N–H–F indicating possible secondary interactions. For this reason, we prefer to analyze the constrained complex. |
| § This barrier is estimated as the difference in electronic energy between the fully optimised B3LYP geometry and a form with OCNH dihedral angles fixed at 0 and 180°. While the latter is not a true transition state, this approach avoids complications due to constraints of HF/NCH position and motion of the non-hydrogen bonded NH2 group. |
| This journal is © the Owner Societies 2012 |
