 Open Access Article
Open Access ArticleCharacterization of structural changes in aptamer films for controlled release nanodevices†
M. Belén
Serrano-Santos
*a,
Eduard
Llobet
a,
Veli C.
Özalp
b and
Thomas
Schäfer
bc
aEMaS, Department of Electronic, Electric and Automation Engineering, University Rovira i Virgili, Avda. Països Catalans 26, 43007 Tarragona, Spain. E-mail: mariabelen.serrano@urv.cat
bNanoBioSeparations Group, POLYMAT, University of the Basque Country UPV/EHU, Avda. Tolosa 72, 20018 Donostia-San Sebastián, Spain. E-mail: thomas_schafer@ehu.es; Fax: +34 943015347; Tel: +34 943018266
cBasque Foundation for Science, Ikerbasque, Bilbao, Spain
First published on 21st August 2012
Abstract
The dimension of the conformational changes of DNA-aptamers which can be used as stimulus-responsive gate-keepers in controlled delivery nanodevices has been determined by acoustic wave-based sensors upon molecular recognition of a small-sized target, adenosine-5′-monophosphate (AMP).
Supramolecular structures and DNA-based nanosystems have recently been used as molecular tools for amplifying signals, guiding chemical coupling reactions, and creating stimulus-responsive nanomaterials.1 Aptamers, short oligonucleotides with specific affinities to their ligands, are particularly suitable structure-changing molecules for combining highly selective biorecognition and a desired signal-transduction.2 They can be selected toward virtually any ligand molecules through a combinatorial method called SELEX3 and be readily incorporated into functional devices boosting new applications ranging from DNA-machines in sensors to bioseparations and controlled delivery devices where DNA aptamers have been recently employed successfully as nanovalves or so-called “gate-keepers” in nanopores4 (Fig. S1, ESI†). The bottleneck during the systematic design of such DNA-based nanodevices, however, is the knowledge on the quantitative dimensional and viscoelastic property changes which occur in immobilized aptamers upon target binding.
Quartz crystal microbalance with dissipation monitoring (QCM-D) is a surface-sensitive acoustic technique for studying a wide range of interfacial adsorption reactions,5 amongst which there are ligand–receptor interactions, and the resulting change of the viscoelastic properties of the surface layer. Layer-by-layer DNA films,6 DNA hybridization7 and Holliday junctions8 were recently followed through dissipation measurement and a sensor based on dissipation changes occurring during aptamer–protein interactions was reported.9 We here show that using acoustic wave-based sensors we can go beyond these studies by quantifying in situ and in real-time overall structural changes that occur in the DNA aptamer receptor molecule upon binding to a small target molecule and use these data for a rationale design of nanodevices.
As a model system for a small molecule binding aptamer, we investigated the interaction of adenosine-5′-monophosphate (AMP) with AMP-binding aptamer films. The AMP-binding DNA aptamer is a 27 base sequence selected by Huizenga et al.10 and has been used in numerous nanostructures as a model system.
It recognizes specifically adenosine-containing small molecules such as AMP with Mw = 382 Da while not interacting with homologues containing guanosine (Fig. S3, ESI†). The tertiary structure of this aptamer is well-reported and there exist different structure-changing functional designs.11 We compared (i) the AMP-binding aptamer sequence itself and (ii) a hairpin structure of the same aptamer sequence which was created by adding seven additional nucleotides at the 3′-end. It was previously found qualitatively12 that both forms undergo strikingly different structural rearrangements upon binding to the ligand, as illustrated in Fig. 1. The AMP-binding aptamer (Fig. 1A) forms a helix structure with mismatched nucleotides and changes only slightly its overall molecular conformation13 upon binding to two molecules of AMP which intercalate into the distorted minor groove of a zippered-up internal loop segment.14 In contrast, the hairpin structure based on the same aptamer sequence goes through an extensive molecular rearrangement (Fig. 1B). The hairpin structure is used to disrupt the aptamer's original shape which is essential in specific recognition of its ligand and in order to obtain a double stranded duplex neck region bringing both ends of the molecule together. As a consequence, a closed structure referred to as a “hairpin shape” is obtained, which changes to an open structure when interacting with two molecules of AMP owing to the disruption of the neck region. Such a hairpin design has frequently been used to develop fluorescent15 or electrochemical probes based on aptamers.16
 | ||
| Fig. 1 (A) Scheme of the AMP-binding aptamer (blue line), where the overall structure is not significantly disturbed by specific intercalation of the AMP ligand; (B) Scheme of the hairpin form of the AMP-binding aptamer (blue lines) which was created by adding 7 nucleotides at 3′ end (yellow lines) forming a stem-loop structure. | ||
During our experiments, we first created an avidin layer on a biotin-functionalized sensor on top of which either the biotinylated ATP-binding aptamer molecules or the biotinylated AMP-binding aptamer hairpin structures were deposited. Upon immobilization of the AMP-binding aptamer during eight independent experiments, average resulting frequency (ΔF) and dissipation changes (ΔD, dimensionless) were observed to yield around −28 ± 5 Hz and (3.0 ± 0.8)·× 10−6, respectively (Fig. 2A).
 | ||
| Fig. 2 Frequency (black lines) and dissipation (green lines) changes during binding of AMP to the (A) AMP-binding aptamer and (B) AMP-binding aptamer hairpin as monitored by the QCM-D, with dotted vertical lines delimiting the aptamer and the hairpin immobilization, respectively, prior to target injection at concentrations up to 750 μM. | ||
This level of deposition is comparable with literature data for similar-sized oligonucleotides.17 The increase in dissipation upon aptamer deposition indicates the formation of an overall softer layer than with avidin alone due to the less dense packing of aptamer molecules on top of the tightly packed avidin. We then contacted first the aptamer film with AMP in binding buffer at a series of concentrations (10 up to 750 μM). A clear decrease in frequency was detected during subsequent AMP injections and a corresponding increase in dissipation energy was recorded (Fig. 2A). This response could unambiguously be attributed to specific aptamer–target interactions rather than bulk changes in medium composition given that the measurements without the aptamer film did not cause any significant response in the acoustic wave-based sensor. Average values of ΔD/ΔF from independent experiments were plotted against AMP concentrations to obtain a dissociation constant (KD) of 43 ± 12 μM (Fig. 3A), which is in good agreement with the literature citing the affinity constants of the AMP-binding DNA aptamer to be in the range of 6 to 30 μM depending on the method used in the respective assessments.11,18
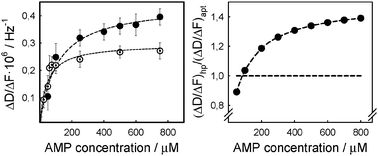 | ||
| Fig. 3 Left (A): dissipation changes normalized by the corresponding frequency change as a function of the respective AMP concentrations, and the resulting ligand binding curve (dashed line) for the hairpin structure (solid circles) and the aptamer (open circles). Right (B): comparison of the normalized dissipation changes using as reference those of the aptamer (horizontal line) indicating significantly larger conformational rearrangement of the hairpin structure (solid circles). | ||
When titrating successive concentrations of AMP to the immobilized aptamer hairpin structure, responses resembled those observed during the aptamer experiments (Fig. 2B). However, the binding plot (Fig. 3A) yielded a slightly higher dissociation constant, namely 107 ± 22 μM for ΔD/ΔF, which nevertheless is in line with data reported on a similarly designed AMP–aptamer sequence18b and can be explained by a distortion of the original aptamer structure when incorporated in the hairpin form. In fact we could confirm this by measuring the dissociation constant of an AMP-binding aptamer hairpin structure in solution, maintaining the same buffer conditions, and where a binding constant KD of about 350 μM was obtained (Fig. S2, ESI†).
Given the good agreement between the dissociation constants derived from QCM-D measurements and those reported previously in the literature, we further investigated into how far the structural changes occurring during binding to the small target molecule, AMP, could be quantified. For this purpose, we compared the aptamer and the aptamer hairpin structure at a practically saturating AMP concentration of 750 μM where maximum conformational changes were expected according to the experimental data (Fig. 3B). We observed that changes in the dissipation energy were almost twice as high in the hairpin structure with 0.38·10−6 ± 0.06·10−6 than in the aptamer with (0.22 ± 0.04) × 10−6 (Fig. 4A and B). This proved that larger conformational arrangements took place in the hairpin structure upon interaction with AMP, compared to those occurring in the aptamer. With regard to the hairpin structure (Fig. 1B and 4B), this can be explained by a loss of the double stranded neck region upon AMP binding, while at the same time the single stranded loop region is stabilizing the mismatched double helix structure of the sequence which interacts with the ligand. “Softer” films may then be conceived as an extension of the hairpin film in the vertical direction resulting from the more open structure of the hairpin compared to that of the aptamer. Concerning the aptamer, NMR studies do not suggest any major molecular conformational change upon binding to AMP,19 but rather a relatively small rearrangement in the internal binding pocket leading to a slightly more upright position of the molecule. The latter would result in a minor change in the film density which is indeed confirmed by an only slight increase in dissipation (Fig. 4A and 1A).
 | ||
| Fig. 4 Dissipation changes observed upon binding at 750 μM AMP in buffer solution (arrow) in the (A) aptamer and (B) aptamer hairpin structure, as well as the corresponding thickness changes observed upon binding at 750 μM AMP in buffer solution in the (C) aptamer and (D) aptamer hairpin structure; vertical lines indicate reference points from buffer solution and maximum response to the target. | ||
Using a Voigt-based viscoelastic model20 (Fig. S4, ESI†), we derived the quantitative thickness changes of the aptamer and aptamer hairpin layer, as well as the respective viscoelastic properties. These parameters are related to the efficiency of aptamer-functionalized nanopores with gating or controlled release characteristics (for more details, see ESI†). In a first step, the avidin layer deposited was determined to have an average thickness of about 5 nm which agreed well with previous observations validating the model chosen. Subsequently, the thickness of both the aptamer film and the hairpin structure during ligand binding was modelled and is depicted in Fig. 4C and D, respectively. The aptamer film increased only slightly in thickness upon AMP binding, namely about 0.1 nm which was in line with the fact that the ATP binding aptamer has a structure stabilized by hybridizations at two ends and an asymmetric internal bubble such that the binding of AMP molecules involves widening of the minor groove in the binding pocket into which two AMP molecules intercalate, however, without significantly changing the overall dimension of the aptamer sequence.22 The thickness increase was dramatically more conspicuous in the case of the hairpin form of the aptamer, namely 1.6 nm. This difference certainly is related to the seven nucleotide long single-stranded linker added to the aptamer sequence in order to form the hairpin, extending the hairpin structure vertically away from the surface as it opens upon AMP binding. However, with recent measurements reporting the length of one nucleotide to be 6.3 Å21 for ssDNA in solution, seven nucleotides would yield a length of 4.4 nm when elongated most, which is about three times of what we measured in our experiments during AMP binding. It is here that it must be recalled that an aptamer hairpin film is measured during our experiments rather than a single molecule with the consequence that ssDNA can stay partially tilted on surfaces depending on the surface density of the film and the interactions with the surface material.22 While this appears to be a limitation of the measurement protocol presented as for the characterization of single-molecule conformational changes, it certainly represents best functionalized mesoporous structures with aptamer hairpins as nanovalves where an overall response owing to an average conformational change would be observed, rather than the action of one single aptamer molecule. The modelling revealed furthermore that the observed increase in layer thickness at increasing concentrations of AMP was accompanied by a progressive decrease in shear viscosity and shear modulus in both structures for which the data obtained (see ESI†) were in the same range of magnitude as viscoelastic parameters reported previously for DNA hybridization.8 This needs to be considered in applications where the shear stress of fluid flow may affect the structural arrangement of DNA-based gate-keepers immobilized in or at the mouth of nanopores.
In conclusion, acoustic wave-based sensors yield reliable information in real time and in situ on the overall dimensional changes of aptamer and aptamer hairpin thin-films when specifically binding to a small target molecule and thus help in the systematic design of nanovalves or gating architectures in DNA-based controlled release devices.
This work was supported by the European Research Council through ERC Grant 209842-MATRIX and by the European Commission through the Marie Curie Grant PIEF-GA-2009-236628 (M. B. Serrano Santos). T. Schäfer would like to thank Prof. P. M. Echenique for being hosted in the Donostia International Physics Centre (DIPC).
Notes and references
- (a) C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef CAS; (b) T. D. Nguyen, H.-R. Tseng, P. C. Celeste, A. H. Flood, Y. Liu, J. F. Stoddart and J. I. Zink, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10029–10034 CrossRef CAS; (c) M. Barboiu, Chem. Commun., 2010, 46, 7466 RSC; (d) J. Wu, X. Zhan and B. J. Hinds, Chem. Commun., 2012, 48, 7979–7981 RSC.
- (a) Y. Huang, S. Zhao, Z. F. Chen, M. Shi and H. Liang, Chem. Commun., 2012, 48, 9400–9402 RSC; (b) F. Luo, L. Zheng, S. Chen, Q. Cai, Z. Lin, B. Qiu and G. Chen, Chem. Commun., 2012, 48, 6387–6389 RSC.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS.
- V. C. Özalp and T. Schäfer, Chem.–Eur. J., 2011, 17, 9893–9896 CrossRef.
- F. Höök, B. Kasemo, T. Nylander, C. Fant, K. Sott and H. Elwing, Anal. Chem., 2001, 73, 5796–5804 CrossRef.
- A. Singh, S. Snyder, L. Lee, A. P. R. Johnston, F. Caruso and Y. G. Yingling, Langmuir, 2010, 26, 17339–17347 CrossRef CAS.
- C. Larsson, M. Rodahl and F. Höök, Anal. Chem., 2003, 75, 5080–5087 CrossRef CAS.
- G. Papadakis, A. Tsortos and E. Gizeli, Nano Lett., 2010, 10, 5093–5097 CrossRef CAS.
- Q. Chen, W. Tang, D. Z. Wang, X. J. Wu, N. Li and F. Liu, Biosens. Bioelectron., 2010, 26, 575–579 CrossRef CAS.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 CrossRef CAS.
- (a) J. W. Liu, Z. H. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS; (b) J. M. Picuri, B. M. Frezza and M. R. Ghadiri, J. Am. Chem. Soc., 2009, 131, 9368–9377 CrossRef CAS; (c) M. Wieland, A. Benz, J. Haar, K. Halder and J. S. Hartig, Chem. Commun., 2010, 46, 1866–1868 RSC; (d) J. Elbaz, R. Tel-Vered, R. Freeman, H. B. Yildiz and I. Willner, Angew. Chem., Int. Ed., 2008, 48, 133–137 CrossRef.
- M. N. Stojanovic and D. M. Kolpashchikov, J. Am. Chem. Soc., 2004, 126, 9266–9270 CrossRef CAS.
- T. Hermann and D. J. Patel, Science, 2000, 287, 820–825 CrossRef CAS.
- S. Nonin-Lecomte, C. H. Lin and D. J. Patel, Biophys. J., 2001, 81, 3422–3431 CrossRef CAS.
- Z. W. Tang, P. Mallikaratchy, R. H. Yang, Y. M. Kim, Z. Zhu, H. Wang and W. H. Tan, J. Am. Chem. Soc., 2008, 130, 11268–11269 CrossRef CAS.
- R. J. White, A. A. Rowe and K. W. Plaxco, Analyst, 2010, 135, 589–594 RSC.
- (a) A. Gunnarsson, P. Jonsson, R. Marie, J. Tegenfeldt and F. Höok, Nano Lett., 2008, 8, 183–188 CrossRef CAS; (b) K. M. M. Aung, X. N. Ho and X. D. Su, Sens. Actuators, B, 2008, 131, 371–378 CrossRef.
- (a) Q. Deng, I. German, D. Buchanan and R. T. Kennedy, Anal. Chem., 2001, 73, 5415–5421 CrossRef CAS; (b) L. J. Nielsen, L. F. Olsen and V. C. Özalp, ACS Nano, 2010, 4, 4361–4370 CrossRef CAS.
- C. H. Lin and D. J. Patel, Chem. Biol., 1997, 4, 817–832 CrossRef CAS.
- A. Domack, O. Prucker, J. Rühe and D. Johannsmann, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1997, 56, 680–689 CrossRef CAS.
- M. C. Murphy, I. Rasnik, W. Cheng, T. M. Lohman and T. J. Ha, Biophys. J., 2004, 86, 2530–2537 CrossRef CAS.
- H. Wackerbarth, M. Grubb, J. Zhang, A. G. Hansen and J. Ulstrup, Angew. Chem., Int. Ed., 2004, 43, 198–203 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2cc35683j |
| This journal is © The Royal Society of Chemistry 2012 |
