Coherent two-dimensional infrared spectroscopy: Quantitative analysis of protein secondary structure in solution
Carlos R.
Baiz
,
Chunte Sam
Peng
,
Mike E.
Reppert
,
Kevin C.
Jones
and
Andrei
Tokmakoff
Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139
First published on 8th March 2012
Abstract
We present a method to quantitatively determine the secondary structure composition of globular proteins using coherent two-dimensional infrared (2DIR) spectroscopy of backbone amide I vibrations (1550–1720 cm−1). Sixteen proteins with known crystal structures were used to construct a library of 2DIR spectra, and the fraction of residues in α-helix, β-sheet, and unassigned conformations was determined by singular value decomposition (SVD) of the measured two-dimensional spectra. The method was benchmarked by removing each individual protein from the set and comparing the composition extracted from 2DIR against the composition determined from the crystal structures. To highlight the increased structural content extracted from 2DIR spectra a similar analysis was also carried out using conventional infrared absorption of the proteins in the library.
1. Motivation
Modern structural biology seeks to establish a molecular understanding of biological processes by exploring structure-function relationships in biomolecules. The mechanisms for protein folding, protein-protein interactions, ligand binding and catalysis are fundamentally linked to structure. Membrane proteins, in particular, drive key biological processes such as cell-signaling, and ion transport through the cell membrane. Modern tools such as cryo-electron-microscopy or X-ray crystallography enable detailed measurements of protein structure on multiple length-scales; however, structures are usually measured under non-biological conditions where key dynamic effects that lead to protein stability and function are not captured,1 and despite the abundance, diversity, and biological importance of membrane proteins, relatively few crystal structures have been solved due to the difficulty in crystallizing the proteins.2 Aside from NMR spectroscopy, most bio-analytical tools in use today are not sensitive to protein structure, heterogeneity, or conformational dynamics.3 Consequently, specific classes of proteins such as fibrous proteins, intrinsically disordered proteins, gels, amyloids, and aggregates, have been particularly difficult to characterize. Therefore, even a simple and reliable analysis to quantify the percentage of amino-acids in α-helix or β-sheet conformations of proteins in solution would constitute a particularly useful bio-structural tool.Infrared spectroscopy in the amide I region has been used to measure protein structure for over four decades. The vibrational modes in these regions involve primarily backbone C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and N–H vibrations that are relatively sensitive to secondary structure and are largely free from the influence of side-chain absorptions. To date, however, most infrared measurements have offered either qualitative information or have required significant efforts to vibrationally isolate individual residues through isotope labeling.4 Attempts to extract structural information from amide-I and II absorption bands in proteins have relied on complex deconvolution and fitting to analyze largely featureless absorption bands.5–8 While these methods offer semi-quantitative agreement with crystal structures, the results are heavily dependent on the specific set of frequencies and fitting functions used, which greatly limits the usefulness of linear infrared spectroscopy as a structural tool.9–12 In this paper we present multidimensional spectroscopy as a new technique to measure structure in solution. The method relies upon the specific signatures of secondary structures in the infrared spectrum which allow for a quantitative decomposition of a protein spectrum into combinations of spectra corresponding to individual secondary structures. Our method assumes that each individual secondary structure has a unique spectrum associated with it, and that the total protein spectrum is the sum of each secondary structure spectrum linearly weighted by its content. Although these assumptions are not entirely correct based on our understanding of vibrational coupling and the overlap between secondary structure spectra, the accuracy of our results justify them. Multidimensional spectroscopy provides more unique spectral information than FTIR, which explains why 2DIR is a more accurate predictor of structure.
O and N–H vibrations that are relatively sensitive to secondary structure and are largely free from the influence of side-chain absorptions. To date, however, most infrared measurements have offered either qualitative information or have required significant efforts to vibrationally isolate individual residues through isotope labeling.4 Attempts to extract structural information from amide-I and II absorption bands in proteins have relied on complex deconvolution and fitting to analyze largely featureless absorption bands.5–8 While these methods offer semi-quantitative agreement with crystal structures, the results are heavily dependent on the specific set of frequencies and fitting functions used, which greatly limits the usefulness of linear infrared spectroscopy as a structural tool.9–12 In this paper we present multidimensional spectroscopy as a new technique to measure structure in solution. The method relies upon the specific signatures of secondary structures in the infrared spectrum which allow for a quantitative decomposition of a protein spectrum into combinations of spectra corresponding to individual secondary structures. Our method assumes that each individual secondary structure has a unique spectrum associated with it, and that the total protein spectrum is the sum of each secondary structure spectrum linearly weighted by its content. Although these assumptions are not entirely correct based on our understanding of vibrational coupling and the overlap between secondary structure spectra, the accuracy of our results justify them. Multidimensional spectroscopy provides more unique spectral information than FTIR, which explains why 2DIR is a more accurate predictor of structure.
2. Background: Amide-I multidimensional infrared spectroscopy
Over the last ten years, coherent multidimensional spectroscopy has been developed as a technique to study the structure and dynamics of complex systems in solution.13–18 In particular, two-dimensional infrared (2DIR) spectroscopy has the ability to directly measure the structure, static disorder, conformational flexibility, and solvent exposure of residues within a protein.19–21 To date, 2DIR spectroscopy has offered new insights into protein structure and heterogeneity,22 protein dynamics,23 protein-protein interactions,24 and ligand binding.25 However, the amount of structural information extracted from 2DIR has remained largely qualitative, and in most cases the equilibrium structure of the protein under investigation is known a priori. In this work, we seek to exploit the structural-sensitivity of 2DIR in order to develop a quantitative method for measuring secondary structure in solution.Amide-I vibrations involve primarily combinations of backbone carbonyl stretches.26,27 The ‘local’ carbonyl stretches, corresponding to individual amino acids within the protein, form the basis for the delocalized normal modes.28 The frequencies of the normal modes are given by the frequencies of the local C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretches as well as the magnitude of the couplings between these local oscillators. Coupling patterns report on the secondary structure and conformational disorder of the residues: structured regions exhibit highly regular coupling patterns whereas disordered regions exhibit a random coupling pattern.29 Each oscillator within a particular secondary structure is coupled strongly to other oscillators in the secondary structure, giving rise delocalized normal modes. The frequency of the delocalized vibrations tends to separate into particular ranges: β-sheets exhibit two peaks near 1630 and 1680 cm−1 whereas α-helices and unstructured regions appear near 1650 cm−1.30,31 Despite the secondary structure-sensitivity of the peak frequencies, conformational disorder and solvent exposure render the absorption bands broad and largely featureless.
O stretches as well as the magnitude of the couplings between these local oscillators. Coupling patterns report on the secondary structure and conformational disorder of the residues: structured regions exhibit highly regular coupling patterns whereas disordered regions exhibit a random coupling pattern.29 Each oscillator within a particular secondary structure is coupled strongly to other oscillators in the secondary structure, giving rise delocalized normal modes. The frequency of the delocalized vibrations tends to separate into particular ranges: β-sheets exhibit two peaks near 1630 and 1680 cm−1 whereas α-helices and unstructured regions appear near 1650 cm−1.30,31 Despite the secondary structure-sensitivity of the peak frequencies, conformational disorder and solvent exposure render the absorption bands broad and largely featureless.
Analogous to NMR, a 2DIR spectrum can be interpreted as follows: the vibrations are first excited, or labeled, by two infrared pulses, and after a certain waiting time, a third pulse either induces absorption to higher excited states or stimulates emission down to the ground state. The excitation frequency (horizontal axis) and a detection frequency (vertical axis) are then combined in a two-dimensional plot. Diagonal peaks are due to excitation and detection at the same frequency, and cross peaks arise from excitation of one mode, and detection of a different mode. Cross peaks are observed only if two vibrations share common atoms, or in this context, common residues. For proteins, 2DIR spectroscopy offers enhanced structure-sensitivity by spreading the spectral content onto two frequency axes. For example, in the case of β-sheets, off-diagonal features are observed between the two main peaks at 1630 and 1680 cm−1, while no cross peaks are observed between α-helix and β-sheet vibrations since the two modes primarily involve different residues. Further analysis of the main spectral features associated with each secondary structure is provided in the discussion section below.
3. Methods
a. Protein selection and sample preparation
The set of proteins used in this study (Fig. 1) was selected to span a wide variety of structures ranging from mainly α-helical proteins (Myoglobin) to mainly β-sheet proteins (γ-globulins) while minimizing structural redundancies. The solubility, robustness, and commercial availability of each protein were also considered. All samples were purchased from Sigma-Aldrich (St. Louis, MO) and were used without further purification. The samples were hydrogen/deuterium exchanged in D2O (Cambridge Isotopes, Andover MA) at 60 °C for 1 h, and lyophilized after freezing in liquid nitrogen. Solid protein samples were stored at −20 °C to prevent degradation. The proteins were then re-dissolved in pure D2O at neutral pH* to a final concentration of 20 mg ml−1. Due to low solubility at neutral pH*, Fibrigonen, Immunoglobulin G and Insulin samples were dissolved in pH* = 2 DCl solution. The solutions were placed in a temperature-controlled sample cell equipped with two CaF2windows and a 50 μm Teflon spacer. To prevent degradation of the samples, spectra were acquired within approximately one hour after preparing each solution, during this period the samples were kept at room temperature under a dry air atmosphere. All experiments were carried out at 25 °C.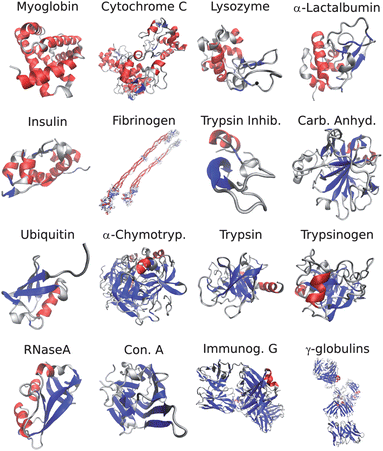 | ||
| Fig. 1 PDB Structures of the proteins used for SVD analysis. α-helix and β-sheet structures are colored in red and blue respectively. The proteins are arranged in order of increasing β-sheet contents as calculated by the DSSP program. | ||
b. Infrared spectroscopy
The two-dimensional spectrometer has been described in detail elsewhere.32,33 Briefly, 90-fs mid-infrared pulses are generated by an optical parametric amplifier/difference frequency generation setup pumped by a regeneratively amplified Ti:Sapphire laser. Three IR pulses are overlapped at the sample in a box geometry, and the signal electric field is interferometrically measured in a phase-matched direction by overlapping the emitted signal with a reference pulse. The relative polarization of the excitation and detection pulses is set to 90° so as to enhance off-diagonal features in the spectrum. The excitation frequency is measured by scanning the first two IR pulses and Fourier-transforming the resulting oscillations along the first time delay (t1); the detection frequency is measured using a spectrometer equipped with a 64 × 2-pixel mercury-cadmium-telluride (MCT) array. The excitation time axis (t1) is scanned from 0 to 3 ps in steps of 4 fs; the frequency resolution along the detection axis is approximately 4.5 cm−1 per pixel. Dispersed pump–probe spectra, collected in the parallel polarization condition, were used to independently phase the rephasing and non-rephasing spectra. Following Fourier-transformation and phasing, all correlation spectra were interpolated to 2 cm−1 uniformly-spaced frequency axes for subsequent SVD analysis. An absorption spectrum of each sample was collected immediately before each 2DIR measurement using a Nicolet 380 Fourier-transform infrared spectrometer. D2O baseline subtraction of FTIR spectra was carried out by fitting a line between two frequency points at 1550 and 1720 cm−1. Two-dimensional spectra were not corrected for HOD absorption. Since the 2DIR signal depends on the fourth order of the transition dipoles, the small transition dipole of the HOD bend vibration makes its 2DIR peak negligible relative to the amide-I C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrations.
O vibrations.
c. Structure assignment and singular value decomposition
The X-ray structure of each protein was downloaded from the Brookhaven Protein Databank34 (PDB) and the secondary structure assignment was calculated with the DSSP program.35 DSSP uses the atomic coordinates and hydrogen-bonding patterns to assign each residue to the following structural elements: β-bridge, extended β-strand, 3–10 helix, α-helix, hydrogen bonded turns, and bends. For SVD analysis we combined the β-bridge and extended β-strand into a single structural element β-sheet, and combined the α-helix and 3–10 helix residues into an effective α-helix, all other residues were labeled unassigned.Singular value decomposition (SVD) is a matrix-diagonalization technique used to describe the data in terms of a set of principal component spectra. Within this method a matrix of spectra M, is decomposed into three matrices such that,
| M = USVT, | (1) |
Some of the U columns, V columns, and S diagonal values are zero or near zero. We discard these components and only keep the nonzero parts. As a result of this rejection, S becomes a square, invertible matrix.
If we assume that the protein spectra, M, are the linear combination of the secondary structure spectra weighted by their content, then we can decompose the spectra into:
| ΣPT = M | (2) |
To transform the SVD results from eqn (1) into the spectra and fractions in eqn (2), we insert XX−1 = I into eqn (1):
| M = USXX−1VT. | (3) |
| X−1VT = PT. | (4) |
X −1 can then be inverted and multiplied to give Σ:
| USX = Σ. | (5) |
To assign the secondary structure P(u) = {p(β)(u), p(α)(u), p(una)(u)} of an “unknown” spectrum m(u), we solve the equation:
| Σ(P(u))T = m(u). | (6) |
Substitution of previous equations allows us to solve for P(u) with:
| P(u) = PTVS−1UTm(u) | (7) |
The SVD procedure is cross-validated for each sample by removing a specific spectrum from the initial set, building an SVD basis with the remaining spectra, and using the removed spectrum as the unknown. Finally the predicted percentage of secondary structure is compared with the percentage computed from the PDB structure.
4. Results
a. Interpretation of amide-I spectra
Experimental FTIR and 2DIR spectra are shown in Fig. 2 and 3 respectively. Qualitatively, absorption spectra associated with primarily α-helical proteins are characterized by a single band centered near 1650 cm−1 with an approximate diagonal width of 50 cm−1, while proteins composed of primarily β-sheet show two peaks centered near 1620 and 1680 cm−1 resulting from vibrations whose main transition dipoles lie perpendicular and parallel to the β-strands respectively. The amplitude ratio of νparallel to νperp reports on the size of the β-sheet.36 In this paper we limit the analysis to the number of residues in α-helix and β-sheet conformations. In principle, 2DIR spectroscopy is sensitive to the size and shape of secondary structures as well, but a larger and more diverse protein library would be required for such analysis. Mixed α/β proteins show a combination of the features associated with helix, sheet and unstructured regions where the relative contributions of the individual features depends on the fraction of residues that compose each secondary structure. Peaks assigned to unstructured regions are centered near 1640 cm−1 and thus tend to overlap with the α-helix peaks. This overlap increases the difficulty of unambiguously assigning α-helix and unstructured conformations in proteins. In general, diagonal linewidths report on the static heterogeneity (disorder) of the amide-I frequencies whereas the anti-diagonal linewidths report on the sub-picosecond dynamic fluctuations that are a result of protein-solvent interactions; and the ratio of diagonal to anti-diagonal linewidth serves as a measure of structural rigidity and solvent exposure.23 In this context it should be pointed out that since vibrations are highly delocalized; residue-level resolution can be achieved through isotope labeling.4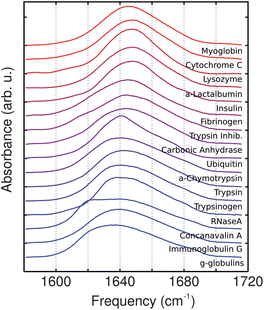 | ||
| Fig. 2 Amide-I linear absorption spectra. Peaks are normalized to the area in the 1580 to 1720 cm−1 and vertically offset for clarity. | ||
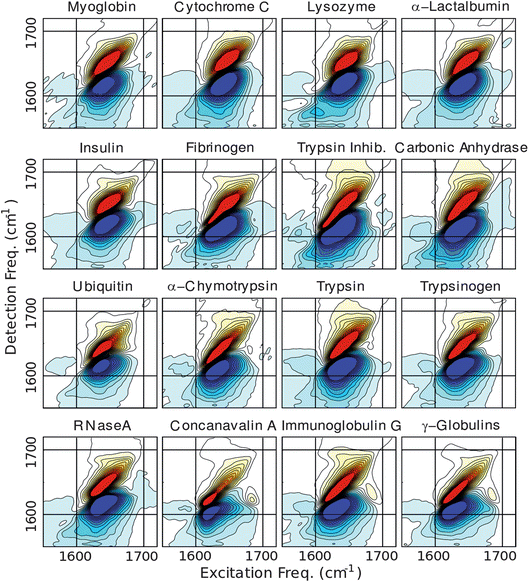 | ||
| Fig. 3 Correlation 2DIR spectra of the proteins acquired in the perpendicular polarization geometry. Contours are plotted from ±50% of the maximum amplitude in 4% intervals and spectra are arranged in order of increasing β-sheet content (see Fig. 1). | ||
The amide-I vibrations shift approximately 5–10 cm−1 upon deuteration; while this is not a large shift relative to the peak width, it could likely contribute to the errors in the assigned structure, it is therefore important to ensure that proteins are fully deuterated. The Amide-II modes, centered near 1550 cm−1 and involving primarily C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretch and N–H bend, tend to shift by approximately 100 cm−1 to 1450 cm−1 upon deuteration (Amide-II′), therefore it is relatively straightforward to check for partial deuteration by comparing the intensity of the 1550 cm−1 band to the 1450 cm−1 band.31 All the samples used here showed negligible amide-II peak intensities compared to amide-II′ following H/D exchange. It is also important to note that certain side-chain absorptions overlap the amide-I peaks. Different protonation states will shift these absorption bands and may thus interfere with the structural assignment of spectra.
N stretch and N–H bend, tend to shift by approximately 100 cm−1 to 1450 cm−1 upon deuteration (Amide-II′), therefore it is relatively straightforward to check for partial deuteration by comparing the intensity of the 1550 cm−1 band to the 1450 cm−1 band.31 All the samples used here showed negligible amide-II peak intensities compared to amide-II′ following H/D exchange. It is also important to note that certain side-chain absorptions overlap the amide-I peaks. Different protonation states will shift these absorption bands and may thus interfere with the structural assignment of spectra.
b. Singular value decomposition
Once spectra are decomposed and structure projections are computed (eqn (1) and 4), it is informative to reconstruct the 2DIR spectra corresponding to purely α-helix, β-sheet components (eqn (3)). Note that since the unassigned structure percentages are not linearly independent from the helix and sheet, the number of components in the analysis is effectively reduced to two. Fig. 4 below shows the reconstructed SVD spectra; the α-helix spectrum shows a single peak centered at 1650 cm−1 whereas the β-sheet spectrum exhibits two peaks near 1630 and 1670 cm−1 with the corresponding cross-peaks, giving the spectrum the characteristic “Z-shape” associated with primarily β-sheet proteins. SVD spectra are in agreement with our spectral assignment described in the previous section as well as with experimental and simulated spectra of idealized structures published previously.37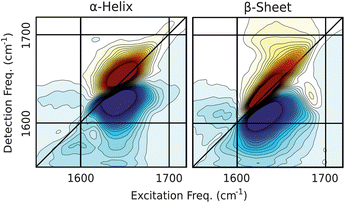 | ||
| Fig. 4 Spectra reconstructed from the SVD decomposition. Contours are plotted from ±50% of the maximum amplitude in 4% intervals and spectra are arranged in order of increasing β-sheet content (see Fig. 1). | ||
c. Secondary structure prediction
The SVD method is validated by removing each protein from the initial set, creating an SVD basis with the remaining structures, and using the new basis to analyze the “unknown” structure (see Methods). Fig. 5 shows the fraction of each secondary structure predicted from SVD analysis along with the fractions obtained from the X-ray structure. Overall there is an excellent correlation between the predicted structures in solution with those derived from the crystal structure. The largest error is observed in Myoglobin, perhaps due to the fact that this protein lies at the edge of the conformational space sampled by the protein library. Alternatively, the large error may suggest that this sample was somehow anomalous. The root-mean-squared errors in comparing the SVD prediction with the X-ray structure are 12.5, 9.2 and 9.1% for α-helix, β-sheet, and unstructured components respectively; however removing Myoglobin from the set reduces the values to 6.7, 7.7, and 8.2% respectively. Naturally, the β-sheet component is the most distinct and thus the errors in predicting the percentage of residues in this conformation tend to be smaller. Distinguishing between α-helices and unstructured regions has remained challenging for amide-I infrared spectroscopy as both peaks appear in the same region of the spectrum. For comparison, we performed the same SVD decomposition on the measured FTIR spectra. The RMS errors obtained for an FTIR-derived SVD basis are: 19.5, 8.3 and 21.5% respectively, consistent with published benchmarks.38–40 This simple comparison highlights the structural-sensitivity gain associated with projecting the spectral information onto two frequency axes and measuring the diagonal as well as the anti-diagonal linewidths. Incomplete background subtraction is one of the largest sources of error in FTIR spectroscopy. Differences in the H2O background, however do not affect the 2DIR data since the non-linear interactions largely suppress the broad background signal, and the spectra are corrected for any distortions due to absorption of the IR pulses by H2O.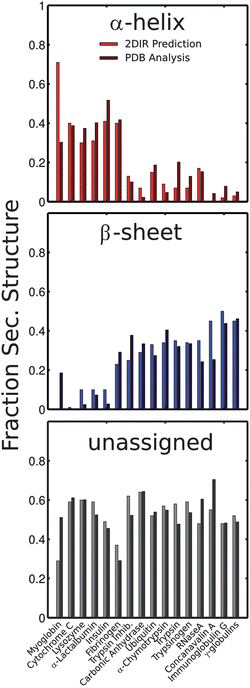 | ||
| Fig. 5 Comparison between amount of secondary structure predicted by SVD analysis (light red, blue, and gray bars) of the 2DIR spectra and the amount extracted from the X-ray structures (dark bars, respectively) using the DSSP program. See text for additional details. | ||
Discussion
Circular dichroism (CD) spectroscopy is a standard method to measure protein conformation in solution. Although the light-matter interactions of amide-I IR spectroscopy and ultraviolet CD spectroscopy are quite different, both methods measure the structure of the backbone of proteins by exciting transitions that are delocalized over multiple amide-I units and where the local structure of the units affects the absorption frequencies and intensities.41 The errors associated with secondary structure determination by 2DIR are comparable to those obtained from circular dichroism. The accepted RMSE values for CD are: 9% α-helix, 12% anti-parallel β-sheet, 8% parallel β-sheet, 7% β-turn, and 9% unassigned.42 These results show that 2DIR spectroscopy is a viable alternative to CD spectroscopy with the added advantage of ultrafast time resolution and the ability to isotope-label individual residues for increased structural resolution.The observed errors between structure determination techniques in solution compared to crystal structure reflect, in part, on the structural heterogeneity and the inherent differences in structure that are induced upon crystallization: it is well-known that water plays a central role in maintaining the balance between entropic and enthalpic contributions that determine the structure and conformational flexibility needed for protein function.43
Another source of error in our measurements is the protein library itself. To minimize errors, the SVD basis should reflect the region of conformational space of the unknown protein to be analyzed. For example, constructing a basis set of purely β-sheet proteins and using such basis to analyze an α-helical protein would likely result in a large error. A particularly large basis set is unlikely to capture the structural details of an individual proteins and therefore likely to produce an “averaged-out” measure of structure. To circumvent this limitation, it may be possible to perform a multi-level SVD analysis where the protein is initially screened using a diverse basis set, and based on the initial results the SVD basis is then further tailored to include only proteins that are similar in structure to the unknown protein. Such analysis would, however, be susceptible to errors in biasing the basis set towards a particular structure that may not accurately reflect the structure of the unknown protein.
Computational models and symmetry considerations for idealized β-sheets suggest that 2DIR spectroscopy is sensitive to the size and type of β-structure.28,36 To date a systematic measurement of pure β-sheet proteins has not been carried out. More generally, quantifying the spectral signatures of various structural motifs, with particular attention to heterogeneity and site disorder, will further enhance the capabilities of 2DIR spectroscopy as a structural technique. Similarly, the ∼12 cm−1 shift that occurs upon deuteration, also suggests that additional information can be derived from the 2DIR of partially deuterated proteins, analogous to amide-I/II 2DIR hydrogen/deuterium exchange experiments,31 which report directly on the solvent exposure of secondary structures within a protein. Finally, an underlying assumption of the analysis presented here is that the spectra of individual secondary structures are additive. However, the delocalized nature of amide-I vibrations coupled with the inherent nonlinearity of 2DIR spectroscopy, renders the peaks sensitive to tertiary contacts. Preliminary simulations suggest that the couplings between secondary structures are relatively weak (∼2 cm−1), and thus peak distortions due to coupling between secondary structures is only a small source of error within the SVD analysis. Future studies will benefit from the tertiary structure information present in the spectra but will require a significant effort in developing a large basis set as well as computational modeling to understand the spectral signatures of particular tertiary contacts.
5. Summary and outlook
We have presented a general method to measure protein structure in solution based on coherent two-dimensional infrared spectroscopy. Spectra are analyzed by singular-value decomposition, a well-known analysis method which remains free from the inherent subjectivity and bias of phenomenological fitting models. The model was tested on a set of sixteen commercially-available globular proteins with well-defined structures. The results presented here show that 2DIR spectroscopy is capable of quantitatively determining the structure of stock proteins without the need for labeling. One key advantage, however, of 2DIR spectroscopy lies in its picosecond time resolution and ability to spread the spectral information over two axes by correlating pump and probe frequencies. Combined with a laser-induced temperature-jump44–46 the method can be used to determine protein folding/unfolding pathways in solution and expand the current understanding of protein folding and function.Acknowledgements
Funding for this project was provided by the National Science Foundation (CHE-0616575 and CHE-0911107) as well as a grant from Agilent Technologies.References
- J. Frank, Single-particle imaging of macromolecules by cryo-electron microscopy, Annu. Rev. Biophys. Biomol. Struct., 2002, 31, 303–319 CrossRef CAS
.
- S. H. White, Biophysical dissection of membrane proteins, Nature, 2009, 459(7245), 344–346 CrossRef CAS
.
- W. Braun, Distance Geometry and Related Methods for Protein-Structure Determination from Nmr Data, Q. Rev. Biophys., 1987, 19(3–4), 115–157 CrossRef CAS
.
- C. T. Middleton,
et al., Residue-specific structural kinetics of proteins through the union of isotope labeling, mid-IR pulse shaping, and coherent 2D IR spectroscopy, Methods, 2010, 52(1), 12–22 CrossRef CAS
.
- D. M. Byler and H. Susi, Examination of the Secondary Structure of Proteins by Deconvolved Ftir Spectra, Biopolymers, 1986, 25(3), 469–487 CrossRef CAS
.
- M. Ruegg, V. Metzger and H. Susi, Computer Analyses of Characteristic Infrared Bands of Globular Proteins, Biopolymers, 1975, 14(7), 1465–1471 CrossRef CAS
.
- G. D. Fasman, Protein Conformational Prediction, Trends Biochem. Sci., 1989, 14(7), 295–299 CrossRef CAS
.
- E. Goormaghtigh, J. M. Ruysschaert and V. Raussens, Evaluation of the information content in infrared spectra for protein secondary structure determination, Biophys. J., 2006, 90(8), 2946–2957 CrossRef CAS
.
- M. Jackson and H. H. Mantsch, The Use and Misuse of Ftir Spectroscopy in the Determination of Protein-Structure, Crit. Rev. Biochem. Mol. Biol., 1995, 30(2), 95–120 CrossRef CAS
.
- M. Jackson and H. H. Mantsch, Artifacts Associated with the Determination of Protein Secondary Structure by Atr-Ir Spectroscopy, Appl. Spectrosc., 1992, 46(4), 699–701 CrossRef CAS
.
- W. Hubner, H. H. Mantsch and H. L. Casal, Beware of Frequency-Shifts, Appl. Spectrosc., 1990, 44(4), 732–734 CrossRef
.
- W. K. Surewicz, H. H. Mantsch and D. Chapman, Determination of Protein Secondary Structure by Fourier-Transform Infrared-Spectroscopy - a Critical-Assessment, Biochemistry, 1993, 32(2), 389–394 CrossRef CAS
.
- P. Hamm, M. H. Lim and R. M. Hochstrasser, Structure of the amide I band of peptides measured by femtosecond nonlinear-infrared spectroscopy, J. Phys. Chem. B, 1998, 102(31), 6123–6138 CrossRef CAS
.
- M. Khalil, N. Demirdoven and A. Tokmakoff, Coherent 2D IR spectroscopy: Molecular structure and dynamics in solution, J. Phys. Chem. A, 2003, 107(27), 5258–5279 CrossRef CAS
.
- M. D. Fayer, Dynamics of Liquids, Molecules, and Proteins Measured with Ultrafast 2D IR Vibrational Echo Chemical Exchange Spectroscopy, Annu. Rev. Phys. Chem., 2009, 60(1), 21 CrossRef CAS
.
- Z. Ganim,
et al., Amide I two-dimensional infrared Spectroscopy of proteins, Acc. Chem. Res., 2008, 41(3), 432–441 CrossRef CAS
.
- C. R. Baiz,
et al., Two-Dimensional Infrared Spectroscopy of Metal Carbonyls, Accounts of Chemical Research, 2008 Search PubMed
. In Press.
- S. T. Roberts, K. Ramasesha and A. Tokmakoff, Structural Rearrangements in Water Viewed Through Two-Dimensional Infrared Spectroscopy, Acc. Chem. Res., 2009, 42(9), 1239–1249 CrossRef CAS
.
- M. D. Fayer, Fast protein dynamics probed with infrared vibrational echo experiments, Annu. Rev. Phys. Chem., 2001, 52, 315–356 CrossRef CAS
.
- Y. S. Kim,
et al., 2D IR provides evidence for mobile water molecules in beta-amyloid fibrils, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(42), 17751–17756 CrossRef CAS
.
- S. H. Shim,
et al., Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(16), 6614–6619 CrossRef CAS
.
- A. Ghosh,
et al., Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(15), 6115–6120 CrossRef CAS
.
- A. W. Smith,
et al., Melting of a beta-Hairpin Peptide Using Isotope-Edited 2D IR Spectroscopy and Simulations, J. Phys. Chem. B, 2010, 114(34), 10913–10924 CrossRef CAS
.
- Z. Ganim, K. C. Jones and A. Tokmakoff, Insulin dimer dissociation and unfolding revealed by amide I two-dimensional infrared spectroscopy, Phys. Chem. Chem. Phys., 2010, 12(14), 3579–3588 RSC
.
- J. Bredenbeck,
et al., Protein ligand migration mapped by nonequilibrium 2D-IR exchange spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(36), 14243–14248 CrossRef CAS
.
- Y. Kim,
et al., Cyanide binding and active site structure in heme-copper oxidases: Normal coordinate analysis of iron-cyanide vibrations of a(3)(2+)CN(−) complexes of cytochromes ba(3) and aa(3), Biospectroscopy, 1998, 4(1), 1–15 CrossRef CAS
.
- A. Barth, Infrared spectroscopy of proteins, Biochim. Biophys. Acta, Bioenerg., 2007, 1767(9), 1073–1101 CrossRef CAS
.
- H. S. Chung and A. Tokmakoff, Visualization and characterization of the infrared active amide I vibrations of proteins, J. Phys. Chem. B, 2006, 110(6), 2888–2898 CrossRef CAS
.
- W. H. Moore and S. Krimm, Transition Dipole Coupling in Amide I Modes of Beta Polypeptides, Proc. Natl. Acad. Sci. U. S. A., 1975, 72(12), 4933–4935 CrossRef CAS
.
- Z. Ganim,
et al., Amide I two-dimensional infrared Spectroscopy of proteins, Acc. Chem. Res., 2008, 41(3), 432–441 CrossRef CAS
.
- L. P. DeFlores,
et al., Amide I ′–II ′ 2D IR Spectroscopy Provides Enhanced Protein Secondary Structural Sensitivity, J. Am. Chem. Soc., 2009, 131(9), 3385–3391 CrossRef CAS
.
- K. C. Jones, Z. Ganim and A. Tokmakoff, Heterodyne-Detected Dispersed Vibrational Echo Spectroscopy, J. Phys. Chem. A, 2009, 113(51), 14060–14066 CrossRef CAS
.
- H. S. Chung,
et al., Transient two-dimensional IR spectrometer for probing nanosecond temperature-jump kinetics, Rev. Sci. Instrum., 2007, 78(6) CrossRef CAS
.
- H. Valafar, F. Valafar and J. H. Prestegard, One and two dimensional statistical data-mining of protein databank for crystallization patterns of proteins, Metmbs'01: Proceedings of the International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences, 2001, 67–72 CAS
.
- R. P. Joosten,
et al., A series of PDB related databases for everyday needs, Nucleic Acids Res., 2011, 39, D411–D419 CrossRef
.
- A. W. Smith,
et al., Two-dimensional infrared spectroscopy of beta-sheets and hairpins, Biophysical Journal, 2004, 86(1), 619a–619a Search PubMed
.
- C. M. Cheatum, A. Tokmakoff and J. Knoester, Signatures of beta-sheet secondary structures in linear and two-dimensional infrared spectroscopy, J. Chem. Phys., 2004, 120(17), 8201–8215 CrossRef CAS
.
- R. Pribic, Principal Component Analysis of Fourier-Transform Infrared and/or Circular-Dichroism Spectra of Proteins Applied in a Calibration of Protein Secondary Structure, Anal. Biochem., 1994, 223(1), 26–34 CrossRef CAS
.
- R. Pribic,
et al., Protein Secondary Structure from Fourier-Transform Infrared and or Circular-Dichroism Spectra, Anal. Biochem., 1993, 214(2), 366–378 CrossRef CAS
.
- K. Rahmelow and W. Hubner, Secondary structure determination of proteins in aqueous solution by infrared spectroscopy: A comparison of multivariate data analysis methods, Anal. Biochem., 1996, 241(1), 5–13 CrossRef CAS
.
- J. Jiang,
et al., Simulation of Two-Dimensional Ultraviolet Spectroscopy of Amyloid Fibrils, J. Phys. Chem. B, 2010, 114(37), 12150–12156 CrossRef CAS
.
- I. H. M. Vanstokkum,
et al., Estimation of Protein Secondary Structure and Error Analysis from Circular-Dichroism Spectra, Anal. Biochem., 1990, 191(1), 110–118 CrossRef CAS
.
- Y. Levy and J. N. Onuchic, Water mediation in protein folding and molecular recognition, Annu. Rev. Biophys. Biomol. Struct., 2006, 35, 389–415 CrossRef CAS
.
- H. S. Chung,
et al., Transient 2D IR spectroscopy of ubiquitin unfolding dynamics, Biophysical Journal, 2007, 207a–207a Search PubMed
.
- H. S. Chung, M. Khalil and A. Tokmakoff, Using nonlinear IR spectroscopy to probe early events in the thermal unfolding of proteins, Abstracts of Papers of the American Chemical Society, 2004, 227, U332–U332 Search PubMed
.
- H. S. Chung and A. Tokmakoff, Temperature-dependent downhill unfolding of ubiquitin. I. Nanosecond-to-millisecond resolved nonlinear infrared spectroscopy, Proteins: Struct., Funct., Bioinf., 2008, 72(1), 474–487 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2012 |
