A highly effective polymerase chain reaction enhancer based on dendrimer-entrapped gold nanoparticles†
Jingjing
Chen
ab,
Xueyan
Cao
b,
Rui
Guo
b,
Mingwu
Shen
b,
Chen
Peng
a,
Tongyu
Xiao
b and
Xiangyang
Shi
*abc
aState Key Laboratory for Modification of Chemical Fibers and Polymer Materials, Donghua University, Shanghai, 201620, People's Republic of China
bCollege of Chemistry, Chemical Engineering and Biotechnology, Donghua University, Shanghai, 201620, People's Republic of China. E-mail: xshi@dhu.edu.cn; Fax: +86-21-67792306-804; Tel: +86-21-67792656
cCQM—Centro de Química da Madeira, Universidade da Madeira, Campus da Penteada, 9000-390, Funchal, Portugal
First published on 7th November 2011
Abstract
In molecular biology, polymerase chain reaction (PCR) has played an important role but suffers a general problem of low efficiency and specificity. Development of suitable PCR additives to improve the specificity and efficiency still remains a great challenge. Here we report the use of dendrimer-entrapped gold nanoparticles (Au DENPs) as a novel class of enhancers to improve the specificity and efficiency of PCR. We show that the Au DENPs prepared using amine-terminated generation 5 poly(amidoamine) dendrimers (G5.NH2) as templates are much more effective than the same dendrimers without AuNPs entrapped in improving the specificity and efficiency of an error-prone two-round PCR system. With the increase of the molar ratio between Au atom and G5.NH2 dendrimer in the Au DENPs, the optimum concentration of Au DENPs used to improve the PCR specificity and efficiency is decreased and can be as low as 0.37 nM when the Au atom/G5.NH2 dendrimer molar ratio reaches 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Our PCR results along with the dynamic light scattering data suggest that unlike the flexible soft dendrimers without NPs entrapped that may display a non-spherical shape when interacting with the PCR components, the Au DENPs with increasing Au atom/dendrimer molar ratio are able to reserve the spherical shape of dendrimers, enabling much more efficient interaction with the PCR components. Therefore, as a NP-based PCR enhancer, both the surface charge and the shape of the particles should be responsible for effective interaction with the PCR components for improving the PCR specificity and efficiency. Furthermore, the used Au DENPs were proved to be stable after the PCR process, enabling them to be potentially used for enhancing different PCR systems.
1. Our PCR results along with the dynamic light scattering data suggest that unlike the flexible soft dendrimers without NPs entrapped that may display a non-spherical shape when interacting with the PCR components, the Au DENPs with increasing Au atom/dendrimer molar ratio are able to reserve the spherical shape of dendrimers, enabling much more efficient interaction with the PCR components. Therefore, as a NP-based PCR enhancer, both the surface charge and the shape of the particles should be responsible for effective interaction with the PCR components for improving the PCR specificity and efficiency. Furthermore, the used Au DENPs were proved to be stable after the PCR process, enabling them to be potentially used for enhancing different PCR systems.
Introduction
Polymerase chain reaction (PCR) has been identified as a fundamental technique in contemporary molecular biology research and clinical medicine. This gene amplification technique can increase the number of copies of target genes by 6 orders of magnitude.1–3 Since its initial use in the early 1980s, this technique has always suffered a problem of low specificity and efficiency, and therefore research efforts have been continuously made to improve the PCR specificity and efficiency.4 Besides the development of various small molecular additives, a great deal of attention has been paid to use various nanoparticle (NP)-based enhancers in recent years. For example, gold NPs,5–8 quantum dots,9,10 carbon nanotubes,11–13 and carbon nanopowders14 have been utilized to improve the PCR specificity and efficiency. It is generally believed that there are 3 different mechanisms involved in PCR optimization with NPs: (1) the NPs can somehow mimic the function of single-stranded DNA-binding protein (SSB), which selectively binds to single-stranded DNA rather than double-stranded DNA and then minimizes the mispairing between the primers and the templates in the PCR system;5 (2) the NPs with high surface area are able to strongly interact or bind with the DNA template or DNA polymerase, which would significantly increase the local concentration of the polymerase or DNA template, greatly improving the specificity and efficiency of PCR;10,12,15 and (3) the NPs with high thermal conductivity in the PCR mixture are helpful for PCR to increase the specific annealing of primers with templates, lowering the chance of forming nonspecific or smear products.13 These 3 mechanisms may occur simultaneously, or with 1 or 2 mechanisms playing a major role in PCR optimization.In our previous studies,11,15 we show that the electrostatic interaction between the PCR components and the positively charged dendrimers15 or polyethyleneimine (PEI)-modified multiwalled carbon nanotubes (MWCNTs)11 plays a major role in optimizing an error-prone two-round PCR system. Our studies show that amine-terminated dendrimers have a higher PCR enhancing effect than those with terminal acetyl and carboxyl groups.15 Similarly, in the presence of PEI or PEI-modified MWCNTs, the specificity and efficiency of the PCR can be significantly improved, suggesting that the PCR enhancing mechanism is not necessarily based only on the thermal conductivity of MWCNTs.11 The strong interaction, especially the electrostatic interaction between NPs and PCR components is believed to be dependent not only on the surface charge density of the particles but also on the particle morphology upon interaction with the PCR components. However, the latter concern has never been addressed before in the literature.
Dendrimers are a class of highly branched, monodispersed, synthetic macromolecules with well-defined structure, composition, geometry, and abundant terminal functional groups.16,17 The unique features of dendrimers allow one to use them as templates or stabilizers to prepare inorganic NPs.18–25 Our previous study has shown that using amine-terminated poly(amidoamine) (PAMAM) dendrimers of generation 5 (G5.NH2) as templates, gold NPs with a size ranging from 2 to 4 nm can be synthesized within the dendrimer interior.26 Therefore, the formed dendrimer-entrapped gold NPs (Au DENPs) may serve as an ideal NP model which has similar size and surface characteristics as G5.NH2 dendrimers, but have a different morphology from that of G5.NH2 dendrimers, especially when interacting with other substances. Amine-terminated PAMAM dendrimers of higher generation (e.g., generation 5, G5.NH2) are soft and flexible, forming a “pancake” shape when interacting with other solid surfaces or interfaces.27,28 The “soft” dendrimer NPs would have limited sites for interaction with PCR components, thereby impacting the efficiency to enhance the PCR specificity and efficiency. Our hypothesis is that the use of Au DENPs with decreased flexibility and close to spherical shape even after being interacted with solid surfaces or interfaces would overcome the drawbacks of pure dendrimers that can easily lose their intact 3-dimensional (3D) spherical shape with limited binding sites. In this case, Au DENPs would have much more surface sites to be interacted with PCR components, effectively enhancing the PCR specificity and efficiency.
To prove our hypothesis, in this present study, we synthesized Au DENPs using G5.NH2 dendrimers as templates according to our previous work.26 With the molar ratio between gold salt and G5.NH2, the spherical shape of dendrimers would be attained more and more, which is beneficial for their effective interactions with the PCR components. We systematically investigated the effect of Au DENPs with different compositions on the PCR specificity and efficiency. Possible molecular mechanisms were discussed and verified by dynamic light scattering studies. The stability of the Au DENPs during the PCR process was tested using UV-Vis spectrometry. To our knowledge, this is the first report attempting to address the effect of NP morphology (with either “soft” or “hard” particles) on the PCR optimization.
Experimental section
Materials
An ethylenediamine core G5.NH2 dendrimer with a polydispersity index less than 1.08 was purchased from Dendritech (Midland, MI). The Au DENPs with gold salt (HAuCl4)/dendrimer molar ratio at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50
1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 75
1, 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 100
1, and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were synthesized and characterized according to our previous work.26 The structures of both G5.NH2 dendrimers and Au DENPs formed using G5.NH2 dendrimers as templates are shown in Fig. 1. The water used in all the experiments was purified using a Milli-Q Plus 185 water purification system (Millipore, Bedford, MA) with a resistivity higher than 18 MΩ cm.
1 were synthesized and characterized according to our previous work.26 The structures of both G5.NH2 dendrimers and Au DENPs formed using G5.NH2 dendrimers as templates are shown in Fig. 1. The water used in all the experiments was purified using a Milli-Q Plus 185 water purification system (Millipore, Bedford, MA) with a resistivity higher than 18 MΩ cm.
 | ||
| Fig. 1 Schematic illustration of the structure of G5.NH2 dendrimers and Au DENPs formed using G5.NH2 dendrimers as templates. | ||
PCR optimization and evaluation
In the error-prone two-round PCR system, primer 1 (5′-GGCTTCGGTCCCTTCTGT-3′) and primer 2 (5′-CACCACCTGTTCAAACTCTGC-3′) were designed to amplify a 283-bp fragment from λDNA (Takara Bio Inc.). PCR reagents were mixed in a final volume of 25 μL according to the following conditions: 10 mM Tris–HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.2 μM primers (Shanghai Sangon Biological Engineering and Technology and Service Co. Ltd), 0.25 mM each dNTP (Takara Bio Inc.), and 0.025 U μL−1 Ex Taq DNA polymerase (Takara Bio Inc.). The PCR procedure was: 2 min at 95 °C for pre-denaturation, followed by 35 cycles of: 20 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C. Then cycling was terminated after 5 min incubation at 72 °C. Amplifications were carried out in a S1000™ Thermal Cycler (Bio-Rad Inc.).PCR products (3 μL) were examined with 1% agarose gel electrophoresis with 1 μL loading buffer. The effectiveness of Au DENPs was described through the assignment of two densitometric quantities, termed specificity and efficiency. From electropherograms, the specificity of amplification was calculated as a ratio of the densitometric value of the specific band and that of all bands amplified by PCR, including undesired nonspecific bands. By definition, the maximal value of specificity, in the absence of non-specific bands, equals to 1.0. The efficiency of an additive is defined as a ratio of the densitometric value of the target DNA band determined after PCR to 500 bp of DL2000 DNA marker, which is assigned a value of 1.0. If the obtained efficiency data are close to 1.0 or higher than 1.0, we can consider that the PCR additive is highly effective in PCR optimization. The concentration of each additive that made the PCR produce the most specific and the brightest target band (maximal efficiency) on the gel was identified to be the optimum concentration.
Dynamic light scattering (DLS) measurements
DLS measurements were carried out using a Zetasizer Nano ZS system (Malvern, UK) equipped with a standard 633 nm laser. Solutions of G5.NH2 dendrimers and Au DENPs with their optimum concentrations used to optimize the PCR in the absence and presence of PCR components were prepared before measurement. The particle sizes were measured in triplicate at room temperature of 25 °C.UV-Vis spectrometry
UV-Vis spectrometry was used to test the stability of Au DENPs before and after the PCR process. The spectra were collected using a Lambda 25 UV-Vis Spectrometer (Perkin Elmer, U.S.A.). All Au DENP samples were dissolved in water at a concentration of 0.1 mg mL−1. For comparison, G5.NH2 in the absence of AuNPs at a concentration of 0.1 mg mL−1 was also measured.Results and discussion
The two-round PCR system5 works on the similar principle as the nested PCR. However, due to the use of the same primers, even though the specific band is observed in the first round (Fig. S1†, Lanes 1 and 2, ESI†), the second round PCR always fails to amplify any meaningful band, showing an “error-prone” phenomenon including broad molecular size distribution or smear in agarose gel electrophoresis (Fig. S1†, Lanes 3 and 4). These “error-prone” non-specific amplifications could not be improved by only optimizing the annealing temperature or the concentration of Mg2+ ions based on a previous work reported by Hu and coworkers.5 Therefore, it is an appropriate model to detect if the additives could improve the specificity of DNA amplification.To determine whether the specificity and efficiency of PCR could be improved, four kinds of different Au DENPs with the gold atom/dendrimer molar ratio at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50
1, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 75
1, 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 100
1, and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (denoted as {(Au0)25-G5.NH2}, {(Au0)50-G5.NH2}, {(Au0)75-G5.NH2}, and {(Au0)100-G5.NH2}, respectively) with an Au NP size of 1.9, 2.2, 2.5, and 2.6 nm, respectively were tested in this PCR system (Fig. 2). G5.NH2 dendrimers without the AuNPs entrapped were tested as a control (Fig. 3). Each additive with different concentrations was added into the reaction mixture to optimize the enhancing effect, and the PCR products were analyzed after electrophoretic fractionation on agarose gels and staining with ethidium bromide. It is clear that all the Au DENPs can improve the efficiency and specificity of the error-prone two-round PCR, similar to the control G5.NH2 dendrimers without AuNPs entrapped. When all the Au DENPs were added into the PCR mixture, the brightness or intensity of the target DNA 283-bp band was strengthened, while the non-specific bands or smear tailed bands were decreased and disappeared. However, when the concentration of these additives exceeded their corresponding optimum concentrations, the amplification of the target band and the non-specific products was significantly inhibited. This phenomenon is similar to other conventional PCR additives, in agreement with our previous reports.11,15
1 (denoted as {(Au0)25-G5.NH2}, {(Au0)50-G5.NH2}, {(Au0)75-G5.NH2}, and {(Au0)100-G5.NH2}, respectively) with an Au NP size of 1.9, 2.2, 2.5, and 2.6 nm, respectively were tested in this PCR system (Fig. 2). G5.NH2 dendrimers without the AuNPs entrapped were tested as a control (Fig. 3). Each additive with different concentrations was added into the reaction mixture to optimize the enhancing effect, and the PCR products were analyzed after electrophoretic fractionation on agarose gels and staining with ethidium bromide. It is clear that all the Au DENPs can improve the efficiency and specificity of the error-prone two-round PCR, similar to the control G5.NH2 dendrimers without AuNPs entrapped. When all the Au DENPs were added into the PCR mixture, the brightness or intensity of the target DNA 283-bp band was strengthened, while the non-specific bands or smear tailed bands were decreased and disappeared. However, when the concentration of these additives exceeded their corresponding optimum concentrations, the amplification of the target band and the non-specific products was significantly inhibited. This phenomenon is similar to other conventional PCR additives, in agreement with our previous reports.11,15
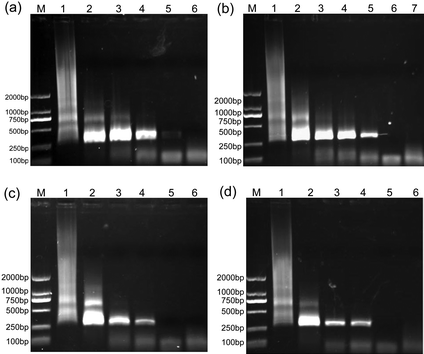 | ||
| Fig. 2 The effect of Au DENPs on the specificity of PCR. Lane M is for marker, the last lane is the negative control in every image. (a) {(Au0)25-G5.NH2} was added into the PCR mixture, and for Lane 1 to 5, its final concentration was 0, 0.40, 0.45, 0.51, and 0.57 nM, respectively. (b) {(Au0)50-G5.NH2} was added into the PCR mixture, and for Lane 1 to 6, its final concentration was 0, 0.35, 0.40, 0.45, 0.49, and 0.54 nM, respectively. (c) {(Au0)75-G5.NH2} was added into the PCR mixture, and for Lane 1 to 5, its final concentration was 0, 0.39, 0.45, 0.51, and 0.57 nM, respectively. (d) {(Au0)100-G5.NH2} was added into the PCR mixture, and for Lane 1 to 5, its final concentration was 0, 0.31, 0.37, 0.43, and 0.49 nM, respectively. | ||
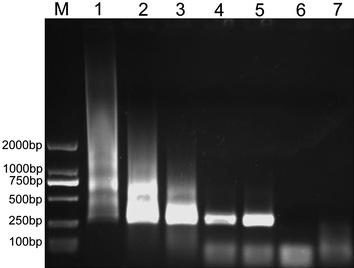 | ||
| Fig. 3 The effect of G5.NH2 dendrimers on the specificity of PCR. Lane 7 is the negative control, and for Lane 1 to 6, the final concentration of G5.NH2 dendrimers was 0, 0.91, 0.95, 0.98, 1.05, and 1.08 nM, respectively. | ||
The specificity and efficiency of each additive were calculated by semi-quantitative analysis (Table 1). We show that all of the Au DENPs exhibit the same performance as that of G5.NH2 without AuNPs, but with much lower optimum concentrations. The optimum concentrations of Au DENPs are all below 0.51 nM, which is about 2 times lower than that of pure G5.NH2 dendrimers (1.05 nM) without AuNPs. The slight deviation of the determined optimum concentration of G5.NH2 dendrimers from that (1.35 nM) determined in our previous study11,15 is probably due to the use of different PCR instruments. It is believed that under similar PCR experimental conditions, the obtained data should be comparable and reliable. The optimum concentration of Au DENPs decreases with the Au atom/G5.NH2 molar ratio. The optimum concentrations of {(Au0)25-G5.NH2}, {(Au0)50-G5.NH2}, {(Au0)75-G5.NH2}, and {(Au0)100-G5.NH2} were estimated to be 0.51 nM, 0.49 nM, 0.45 nM, and 0.37 nM, respectively. We note that for the improvement of the PCR specificity and efficiency, both G5.NH2 dendrimers and Au DENPs have similar performance. However, with the decreased optimum concentration required to optimize PCR, we can say that Au DENPs are more effective in enhancing the PCR than G5.NH2 dendrimers without AuNPs entrapped. Similarly, when compared with other PCR additives such as acid-treated multiwalled carbon nanotubes (MWCNTs) under similar experimental conditions,11,15 the calculated specificity (1.0) and efficiency (1.2) are also satisfactory although the required optimum concentration to enhance PCR is quite different. For enhanced PCR optimization with satisfactory specificity and efficiency, the key parameter determining if the added PCR additive is highly efficient is the required optimum concentration used for PCR optimization: the lower the optimum concentration, the higher the PCR enhancing effect.
| Additives | Optimum concentrations | Maximal efficiency | Maximal specificity |
|---|---|---|---|
| G5.NH2 | 1.05 nM | 1.34 | 1 |
| {(Au0)25-G5.NH2} | 0.51 nM | 1.34 | 1 |
| {(Au0)50-G5.NH2} | 0.49 nM | 1.34 | 1 |
| {(Au0)75-G5.NH2} | 0.45 nM | 1.12 | 1 |
| {(Au0)100-G5.NH2} | 0.37 nM | 0.98 | 1 |
Data presented in this study indicate that Au DENPs are effective enhancers to improve the PCR specificity and efficiency. With the increase of the Au atom/G5.NH2 molar ratio, the optimum concentrations of Au DENPs could be decreased to as low as 0.37 nM, which is presumably due to the much more intact spherical morphology of the G5.NH2 dendrimers after entrapment of AuNPs with increased Au loading content, allowing for effective interaction with the PCR components (Fig. 4b). In contrast, the flexible “soft” G5.NH2 dendrimer particles without AuNPs likely lose their 3D spherical morphology when interacting with PCR components, consequently the interaction sites are limited, leading to limited interaction with the PCR components (Fig. 4a). Accordingly, the concentration required to optimize the PCR is increased when compared with that of Au DENPs. To prove the proposed mechanism, the hydrodynamic sizes of both G5.NH2 dendrimers and Au DENPs under their corresponding optimum concentrations used for enhancing PCR before and after addition of PCR components were measured by DLS (Table 2). The PCR mixture has a size of 800.2 nm and the Au DENPs with larger Au atom/dendrimer molar ratios seem to have smaller sizes when compared with G5.NH2 dendrimers without AuNPs. After addition of the PCR component to the solutions of G5.NH2 dendrimers or Au DENPs, the sizes of the formed complexes are much smaller than those of the corresponding G5.NH2 dendrimers or Au DENPs, indicating the strong interaction between dendrimers/Au DENPs and the PCR components. It is obvious that the size of the formed Au DENPs/PCR component complex becomes smaller with the Au atom/dendrimer molar ratio, which is probably ascribed to the consequence of the more reserved 3D spherical morphology of the dendrimers that have more binding sites for stronger interaction with the PCR components. Although the determined size of the {(Au0)25-G5.NH2}/PCR component complex is slightly bigger than that of the G5.NH2/PCR component complex, which is an exception, we could still deduce that the proposed mechanism is quite rational to explain the enhancing effect of Au DENPs in optimizing PCR. Similar to our previous study related to the PCR enhancing effect using PEI-modified MWCNTs,11 the main PCR optimization mechanism using Au DENPs is still based on the electrostatic interaction between the PCR components and the positively charged Au DENPs in improving the PCR specificity and efficiency. However, for the particles with similar surface characteristics, one has to consider another important factor related to the 3D spherical particle morphology, which may significantly impact the interaction between the particles and the PCR components. It should be noted that the observed enhancement of PCR specificity for Au DENPs versus dendrimer itself may not result from the interaction between the entrapped AuNPs5–8 and the PCR components, since the AuNPs entrapped within the dendrimers can only be accessed by small molecules and cannot be accessed by larger biomacromolecules such as the DNA polymerase, DNA templates, and DNA primers.
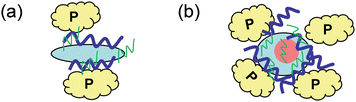 | ||
| Fig. 4 Schematic illustration of the interaction of the PCR mixture with dendrimer (a) and with Au DENPs (b). Yellow pieces: DNA polymerase; thick blue line: template; thin green line: primers; the light blue particle: G5.NH2 dendrimer; red particle: AuNP. | ||
| Sample | Hydrodynamic size/nm | |
|---|---|---|
| Before addition of PCR components | After addition of PCR components | |
| G5.NH2 | 1317.5 | 843.6 |
| {(Au0)25-G5.NH2} | 1269.0 | 862.5 |
| {(Au0)50-G5.NH2} | 1248.8 | 834.6 |
| {(Au0)75-G5.NH2} | 1128.4 | 806.7 |
| {(Au0)100-G5.NH2} | 1039.2 | 767.6 |
There are three major steps in a typical PCR process, which are repeated for 30 or 40 cycles, including denaturation, annealing, and extension. The temperature for denaturation may be as high as 94 °C, which could easily lead to the instability and aggregation of the added Au DENPs, consequently resulting in a decreased PCR enhancing effect. In this context, it is important to explore the thermal stability of Au DENPs during the PCR process. In our study, we used UV-Vis spectrometry to compare the stability of the Au DENPs before and after the PCR process without addition of the PCR components. Fig. 5 shows the UV-Vis spectra of the Au DENP solutions with different compositions. It is clear that the surface plasmon band at around 510 nm becomes more prominent with the increase of the molar ratio of Au atom/dendrimer, indicative of the existence of the AuNPs. After the PCR process, the absorption feature and the surface plasmon band at 510 nm for all Au DENPs do not have any significant changes, indicating that the Au DENPs tested in our study are very stable, which is essential for enhancement of different PCR systems.
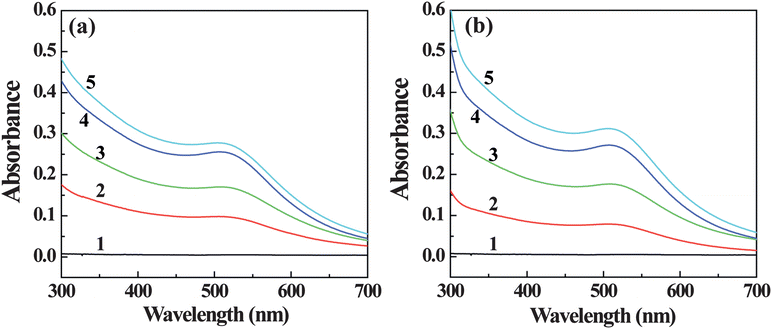 | ||
| Fig. 5 UV-Vis spectra of G5.NH2 dendrimers (1) and {(Au0)25-G5.NH2} (2), {(Au0)50-G5.NH2} (3), {(Au0)75-G5.NH2} (4), and {(Au0)100-G5.NH2} (5) DENPs in PCR buffer solution before (a) and after the PCR process (b). | ||
Conclusions
In summary, we have used Au DENPs synthesized using amine-terminated G5 dendrimers as templates with different compositions as additives to investigate their effects on the specificity and efficiency of PCR amplification. Our results indicate that an addition of Au DENPs to the PCR mixture significantly increases the specificity and efficiency of PCR at their optimal concentrations in the error-prone two-round PCR system. With the increase of the molar ratio between gold atom and G5.NH2, the optimum concentration of Au DENPs could be decreased to as low as 0.37 nM, much lower than that of G5.NH2 dendrimers without AuNPs entrapped. The enhanced PCR optimization effect using Au DENPs when compared with G5.NH2 dendrimers without AuNPs suggests that besides the major role played by the electrostatic interaction between the dendrimers and the PCR components in the PCR optimization mechanism, the entrapment of AuNPs within the dendrimers likely reserves the intact 3D spherical morphology of the dendrimers, making their electrostatic interaction with the PCR components much more effective for improving the PCR specificity and efficiency. With good thermal stability, the developed Au DENPs should be applicable for enhancing various PCR systems.Acknowledgements
Xueyan Cao and Jingjing Chen equally contributed to this work. This research is financially supported by the National Basic Research Program of China (973 Program, 2007CB936000), the National Natural Science Foundation of China (20974019 and 81101150), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, and the Fundamental Research Funds for the Central Universities (for R. G., M. S., and X. S.). C. P. thanks the Innovation Funds of Donghua University Doctorate Dissertation of Excellence (BC201102). X. S. gratefully acknowledges the Fundação para a Ciência e a Tecnologia (FCT) and Santander bank for the Chair in Nanotechnology.Notes and references
- R. J. Chen, Y. G. Zhang, D. W. Wang and H. J. Dai, J. Am. Chem. Soc., 2001, 123, 3838–3839 CrossRef CAS.
- C. Dwyer, M. Guthold, M. Falvo, S. Washburn, R. Superfine and D. Erie, Nanotechnology, 2002, 13, 601–604 CrossRef CAS.
- J. F. Siqueira and I. N. Rocas, Oral Microbiol. Immunol., 2003, 18, 263–265 CrossRef CAS.
- Q. Chou, M. Russell, D. E. Birch, J. Raymond and W. Bloch, Nucleic Acids Res., 1992, 20, 1717–1723 CrossRef CAS.
- H. K. Li, J. H. Huang, J. H. Lv, H. J. An, X. D. Zhang, Z. Z. Zhang, C. H. Fan and J. Hu, Angew. Chem., Int. Ed., 2005, 44, 5100–5103 CrossRef CAS.
- M. Li, Y. C. Lin, C. C. Wu and H. S. Liu, Nucleic Acids Res., 2005, 33, e184 CrossRef.
- L. Mi, Y. Wen, D. Pan, Y. Wang, C. Fan and J. Hu, Small, 2009, 5, 2597–2600 CrossRef CAS.
- W. Yang, L. Mi, X. Cao, X. Zhang, C. Fan and J. Hu, Nanotechnology, 2008, 19, 255101 CrossRef.
- A. Khaliq, P. J. Sonawane, B. K. Sasi, B. S. Sahu, T. Pradeep, S. K. Das and N. R. Mahapatra, Nanotechnology, 2010, 21, 255704 CrossRef.
- L. Wang, Y. Zhu, Y. Jiang, R. Qiao, S. Zhu, W. Chen and C. Xu, J. Phys. Chem. B, 2009, 113, 7637–7641 CrossRef CAS.
- X. Cao, J. Chen, S. Wen, C. Peng, M. Shen and X. Shi, Nanoscale, 2011, 3, 1741–1747 RSC.
- D. X. Cui, F. R. Tian, Y. Kong, I. Titushikin and H. J. Gao, Nanotechnology, 2004, 15, 154–157 CrossRef CAS.
- Z. Z. Zhang, C. C. Shen, M. C. Wang, H. Han and X. H. Cao, BioTechniques, 2008, 44, 537–545 CrossRef CAS.
- Z. Z. Zhang, M. C. Wang and H. J. An, Nanotechnology, 2007, 18, 355706 CrossRef.
- X. Y. Cao, X. Y. Shi, W. C. Yang, X. D. Zhang, C. H. Fan and J. Hu, Analyst, 2009, 134, 87–92 RSC.
- Dendrimers and Other Dendritic Polymers, ed. D. A. Tomalia and J. M. J. Frechet, John Wiley & Sons Ltd, New York, 2001 Search PubMed.
- D. A. Tomalia, A. M. Naylor and W. A. Goddard, III, Angew. Chem., Int. Ed., 1990, 29, 138–175 CrossRef.
- X. Shi, K. Sun and J. R. Baker, Jr, J. Phys. Chem. C, 2008, 112, 8251–8258 CAS.
- X. Shi, S. Wang, S. Meshinchi, M. E. Van Antwerp, X. Bi, I. Lee and J. R. Baker, Jr, Small, 2007, 3, 1245–1252 CrossRef CAS.
- X. Shi, S. Wang, H. Sun and J. R. Baker, Jr, Soft Matter, 2007, 3, 71–74 RSC.
- X. Shi, S. H. Wang, I. Lee, M. Shen and J. R. Baker, Jr, Biopolymers, 2009, 91, 936–942 CrossRef CAS.
- X. Shi, S. H. Wang, M. E. Van Antwerp, X. Chen and J. R. Baker, Jr, Analyst, 2009, 134, 1373–1379 RSC.
- X. Y. Shi, I. Lee and J. R. Baker, Jr, J. Mater. Chem., 2008, 18, 586–593 RSC.
- X. Y. Shi, K. Sun, L. P. Balogh and J. R. Baker, Jr, Nanotechnology, 2006, 17, 4554–4560 CrossRef CAS.
- S. H. Wang, X. Y. Shi, M. Van Antwerp, Z. Y. Cao, S. D. Swanson, X. D. Bi and J. R. Baker, Jr, Adv. Funct. Mater., 2007, 17, 3043–3050 CrossRef CAS.
- R. Guo, H. Wang, C. Peng, M. W. Shen, M. J. Pan, X. Y. Cao, G. X. Zhang and X. Y. Shi, J. Phys. Chem. C, 2010, 114, 50–56 CAS.
- A. W. Bosman, H. M. Janssen and E. W. Meijer, Chem. Rev., 1999, 99, 1665–1688 CrossRef CAS.
- T. Imae, K. Funayama, K. Aoi, K. Tsutsumiuchi, M. Okada and M. Furusaka, Langmuir, 1999, 15, 4076–4084 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c1an15816c |
| This journal is © The Royal Society of Chemistry 2012 |
