Resonant Mie scattering (RMieS) correction applied to FTIR images of biological tissue samples
Keith R.
Bambery
,
Bayden R.
Wood
and
Don
McNaughton
*
Centre for Biospectroscopy and School of Chemistry, Monash University, Clayton, Victoria 3800, Australia
First published on 10th November 2011
Abstract
Recently a resonant Mie scattering (RMieS) correction approach has been developed and demonstrated to be effective for removing the baseline distortions that compromise the raw data in individual spectra. In this paper RMieS correction is extended to FTIR images of a tissue section from biopsy of the human cervical transformation zone and a coronal tissue section of a Wistar rat brain and compared to the uncorrected images. It is shown that applying RMieS correction to FTIR images a) removes baseline distortions from the image spectra and thus reveals previously hidden information on spatial variation of chemical contents within the tissue and b) can lead to improved automatic tissue feature classification through multivariate cluster analysis.
1. Introduction
Fourier transform infrared (FTIR) micro-spectroscopy coupled with focal plane array (FPA) infrared detectors have been extensively applied for the study of molecular composition in biological tissues. FPA detector based FTIR images of tissues provide simultaneous acquisition of both detailed morphological structure and chemistry. FTIR images generated through both univariate and multivariate processing methods from FTIR microscope data have demonstrated a high spatial correlation with anatomical and histopathalogical features for many tissue types including, for example, cervical,1 brain,2 breast,3 liver,4 heart5 and prostate6 tissues. FTIR micro-spectroscopy is not only useful for investigating biochemical changes that accompany the progress of diseases, such as cancer, but has also attracted interest as a diagnostic tool. Progress in further developing FTIR micro-spectroscopy for disease research and diagnosis has been somewhat hampered by the fact that when mid-infrared radiation penetrates tissue samples, Mie scattering7,8 occurs, as the cells and organelles within the tissue have sizes in the same range as the radiation wavelengths (2.5–12 μm). This light scattering results in distorted spectra with band shapes, band absorbance intensities and band peak wavenumber values altered from those which would be obtained in pure absorption spectra, unadulterated by scattering contributions. The very strong amide I band is particularly susceptible to distortion from Mie scattering and often exhibits both reduced absorbance and a shifted peak wavenumber value. The amide I band is important for determining both protein content and conformation and therefore, removal of light scattering induced distortions from the spectra is vital to facilitate correct interpretation of protein secondary structure composition within the sample.In 2003, Martens et al.9 proposed that an extended multiplicative signal correction (EMSC) approach could be employed to provide a correction model that includes the wavelength dependence of light scattering. Further refinement to this method came in 2008, when Kohler et al.8 described an EMSC based scheme for estimating and correcting for the Mie scattering contribution to FTIR spectra. In this scheme (Mie-EMSC) the Mie scatter component of the recorded spectra is corrected for by utilizing a Principal Component Analysis (PCA) approach to summarize the large number of theoretically expected Mie contributions as a computationally manageable number of principal components. While the Mie-EMSC scheme proved to be successful at removing simple Mie scatter effects from FTIR spectra of biological samples it was of limited effectiveness when the spectra exhibited stronger distortions from scattering, the so-called “dispersion artefact”. In recent work on human prostate cells by Bassan et al.10,11 an improved correction algorithm (RMieS-EMSC) was described which recognizes that this “dispersion artefact” principally arises due to resonant Mie scattering.12
To date, RMieS correction has only been reported for collections of individual FTIR spectra obtained from isolated cells. Indeed, while it has been proposed that RMieS correction may remove baseline contaminations more precisely than other baseline correction methods,11,13,14 further validation from independent laboratories is required until its effectiveness can be proved. This work evaluates the effectiveness of applying the RMieS-EMSC algorithm to FTIR FPA micro-spectroscopic image data obtained from two different tissue sections affected badly by baseline distortions that have been the subject of past research in our laboratory, namely, a tissue section from biopsy of the human cervical transformation zone1 and a coronal tissue section of a Wistar rat brain.2 To facilitate direct comparison with our past research, multivariate analysis of the RMieS corrected FTIR images was performed using unsupervised hierarchical cluster analysis (UHCA), which was the same method of analysis applied to these samples previously.1,2
2. Materials and methods
2.1 Tissue sample preparation
Details of the preparation and collection of the tissue samples have already been published elsewhere, for the cervical biopsy tissue in Bambery et al.1 and for the rat brain tissue in Bambery et al.2 In summary, the tissues were fixed in 10% formalin, embedded in paraffin and sectioned by microtome into 4 μm tissue sections. The sections were mounted on IR reflecting slides (MirrIR, Kevley Technologies, OH, USA) and deparaffinised by washing three times in clean xylene and then air dried.2.2 FTIR micro-spectroscopy
FTIR spectra were acquired in transflection mode using a “Stingray” FTIR microscope system (Agilent Technologies) equipped with a 64 × 64 pixel FPA (mercury-cadmium-telluride) detector using a 15 x Cassegrain objective. Spectra were recorded at 6 cm−1 spectral resolution over the ranges 4000–900 cm−1 and 4000–950 cm−1 for the cervical and rat brain tissues respectively.(a) The cervical tissue section was imaged as a sixteen-tile mosaic (4 × 4) image with a pixel spatial resolution of 5.5 μm. The final mosaic image has dimensions of 256 × 256 pixels and covers an area of 1.4 × 1.4 mm. This FTIR image comprises 65,536 individual spectra and to date is the largest FTIR image data set analysed by multivariate methods in our laboratory. Our earlier studies on cervical tissue sections were limited to a maximum of 16,384 spectra. This 4-fold increase in the size of the spectral data sets that can be analysed is a direct result of both the larger addressable memory available to 64 bit operating systems and improvement in processor speeds.
(b) The rat brain tissue section was imaged as a 1024-tile mosaic (32 × 32) image at a pixel spatial resolution of 88 μm. The mosaic image has pixel dimensions of 128 × 128 (16384 individual spectra) and covers an area of 11.26 × 11.26 mm.
2.3 FTIR image pre-processing and RMieS-EMSC correction
The spectra contained in both these FTIR images were subjected to quality testing in CytoSpec 64 bit (www.cytospec.com). Two copies of each of the quality tested FTIR images were saved, one copy of the image data was further processed with the RMieS-EMSC algorithm (version 2) to test the effectiveness of resonant Mie scattering correction and the other copy was left uncorrected to serve as the control. The FTIR image data (full resolution, 4000–950 cm−1) was passed as input to the RMieS-EMSC algorithm by a MATLAB® routine written by us. This routine preserved the x,y spatial coordinates for each spectrum for later image reconstruction and ensured that only those spectra which had passed the quality testing were processed by the RMieS-EMSC algorithm. The RMieS-EMSC algorithm options were set such that the spectral resolution of the data sets was left unchanged and resonant Mie scattering correction was applied for a modelled range of scattering particles radii from 2 to 8 μm. Lower and upper ranges for average refractive indices were set at the default values of 1.1 and 1.5 respectively and 10 equidistant values were used for constructing the Mie scattering curve matrix from the above scattering parameters which was compressed to 7 orthogonal principal components. (See Bassan et al.11). All computational analyses were performed on a desktop PC with 2.93 GHz Intel® Core™ i7 CPU, 16 GB RAM and Windows 7 Enterprise 64 bit operating system.For preliminary tests of the applicability of RMieS corrections to FTIR image data only a single iteration of the algorithm was performed. Single iteration estimations for resonant Mie scatter correction were able to be computed in just a few minutes. However, all final RMieS corrected images and spectra presented in this paper were obtained by iterating the algorithm 8 times. The full 8 iteration correction took approximately 53 h to complete for the larger cervical tissue FTIR images and 10 h for the rat brain images. Bassan et al.,13 have adapted some of the RMieS-EMSC algorithm to be parallel processed on a graphics processing unit (GPU) to reduce computation time. The version of the RMieS-EMSC algorithm used in this work did not make use of parallel processing.
2.4 Multivariate analysis
UHCA was performed in CytoSpec on both the RMieS corrected FTIR images and the control FTIR images. Quality tested raw spectra and second derivative spectra were both trialled as input for UHCA across the spectral window 1800–950 cm−1. UHCA was computed using the D-values distance method and Ward's algorithm as the clustering method.3. Results
3.1 Preliminary assessment of RMieS correction for FTIR images
To our knowledge this work is the first report on RMieS correction of FTIR images and also the first report on correction of FTIR spectra from tissues rather than from cells and so, no published work or guidance was available on how to proceed. For this reason, preliminary tests were performed on using the RMieS-EMSC algorithm simply to assess RMieS correction's general applicability for FTIR image data from tissues. For these preliminary tests a single pass of the algorithm was used (i.e. no iteration) so as to rapidly obtain a first estimation of the resonant Mie scatter corrected images. FTIR micro-spectroscopic images are hyperspectral data sets and consequently they contain both spatial and spectral information. Thus, when assessing RMieS correction on FTIR image data the effect of the correction process must be considered in terms of both image contrast (spatial dimensions) and chemical information content (spectral dimension). An image or map can be generated from the FTIR micro-spectroscopic data set by either univariate or multivariate approaches. Univariate (chemical) images have image contrast based on peak height, integrated areas under specific bands or band ratios. These images can provide information on the distribution and relative content of the macromolecules within the tissue section. In multivariate imaging, the information content of the entire spectrum can be used to generate pseudo-coloured cluster maps. In this imaging approach a statistical measure of similarity is established between all the spectra and sufficiently similar spectra are then collected together into a common cluster. All spectra in a cluster are assigned the same colour. To produce multivariate cluster maps the assigned colour for each spectral cluster is displayed at the image coordinates at which the cluster member spectra were collected. Of course in practice, the spatial and spectral dimensions are not separable since the image contrast is, by one means or another, always generated from the spectral information content.3.2 Assessment of RMieS correction on spatial information content in FTIR images
The RMieS-EMSC algorithm normalizes all the spectra using the un-weighted EMSC approach. Spectral pre-processing with normalization has little impact for multivariate FTIR imaging methods where the image maps are constructed from cluster colour coded pixels corresponding to statistically similar spectral profiles. In fact, multivariate methods algorithms generally incorporate some normalization or mean-centring processing of the spectral data set. Univariate images however, are normally generated from FTIR data without any prior normalization. Consider the case where RMieS correction is applied as pre-processing before generating univariate FTIR images. Poor quality spectra from overly thin sections of the tissue, spectra from holes in the tissue section and spectra from blank areas on the substrate would also be “corrected” and normalized. This is undesirable and indeed it was found that careful quality testing of the image spectra was a necessary prior step to obtain useful RMieS corrected univariate FTIR images. The normalization process might also be expected to result in loss of variation amongst the spectra contained in the image and consequently a loss of image contrast might result in the corrected univariate FTIR image. In these tissue samples the EMSC normalization was not found to cause loss of FTIR univariate image contrast, i.e. the univariate images were not featureless. RMieS-EMSC correction models each raw spectrum as the sum of a constant value offset, a sloping baseline, a resonant Mie scattering spectrum, the reference spectrum amplified by a multiplicative factor and an un-modelled residual spectrum. The corrected spectrum is then the reference spectrum plus the un-modelled residual spectrum divided by the multiplicative factor. It is presumed that the un-modelled residual spectrum preserves the chemical variation important for providing image contrast in univariate FTIR images. Alternative normalization methods (min-max and vector) were tested and the resultant FTIR images were completely lacking in contrast. Normalization by EMSC is relatively conservative. EMSC based baseline correction is valued for preservation of the chemical (spectral dimension) information as well as preserving the spectral variation important for providing image contrast in univariate FTIR images.3.3 Assessment of RMieS correction on spectral information content in FTIR images
While the use of only a single iteration of the RMieS-EMSC algorithm gave results in very rapid time, the resultant spectra retain some characteristics of the reference spectrum and further iterations are required to remove these. For studies where the aim is only to separate spectra from different sample groups (e.g. diagnostic; normal versus diseased), the single iteration result may very well be sufficient to distinguish the groups using automated methods such as artificial neural networking. However, in this study we wished to interpret the spectra within the RMieS corrected images to obtain detailed information on the chemical composition of the sample. Iterating the algorithm 8 times gave resultant spectral profiles with significantly improved correspondence (in terms of relative peak heights and band widths) to the individual original spectra in the image (minus baseline distortion).Similarly, the resultant RMieS corrected spectra had even better correspondence with the original spectra when the average spectrum from the sample set was used as the reference spectrum rather than use of the routine's in-built Matrigel spectrum as the reference. Using the average spectrum from a set of spectra as the reference spectrum is a long accepted approach for EMSC and it appears to be a good choice for RMieS-EMSC as well. The average spectrum was itself corrected for resonant Mie scattering before it was used as the reference spectrum for correcting the FTIR image spectra. This was achieved by first averaging together all the spectra in the image which had passed quality testing followed by RMieS correction with 8 iterations using the Matrigel standard spectrum as the reference.
3.4 RMieS correction of FTIR images of cervical transformation zone tissue sections
Fig. 1 shows two FTIR images of a cervical tissue section together with a photomicrograph of an adjacent H&E stained section. The H&E section in Fig. 1a) shows the location of cellular tissue types. The FTIR image in Fig. 1b) was constructed from the raw FTIR image (without normalization) by colour coding the relative intensity of the band arising from amide I (1700–1575 cm−1) before RMieS correction. Because the background arises from light scattering from the sample, this image is essentially a physico-chemical map. There is little correspondence between high and low amide I absorbance regions of the tissue compared with the tissue types. The basal layer can be seen in this image but examination of the spectra reveal this is likely due to strong scattering in this optically dense layer (compared to surroundings). Fig. 1c) shows the same data acquisition as in Fig. 1b) but after RMieS correction. The latter image is extremely close to a purely chemical map because the physical light scattering contributions have been removed or almost completely removed by RMieS correction. In this image the regions of differing amide I intensity show good correspondence with the tissue types in the sample section. In particular, relatively lower absorbance is observed for the epithelial tissue (regions 1,2 and 3), intermediate absorbance in the stromal (connective) tissue (region 5) and relatively high absorbance in the region of the basal layer and basement membrane (region 4), attributable to increasing protein content in these tissues in that order.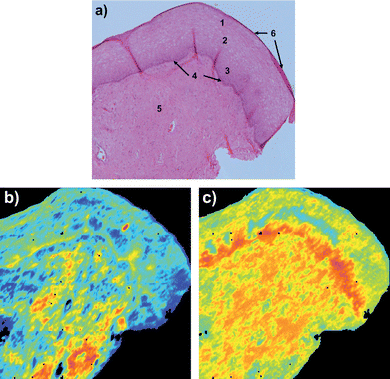 | ||
| Fig. 1 Cervical tissue section a) H&E stained section showing cellular tissue types numbered as, 1 superficial epithelial, 2 intermediate epithelial, 3 parabasal epithelial, 4 bas-al layer, 5 stroma (connective tissue) and 6 exfoliating cells; b) before RMieS correction image of amide I (1700–1575 cm−1) band intensity (colours red to blue corresponding to high to low absorbance respectively); c) after RMieS correction image of amide I band (same scale as b)). | ||
Fig. 2 shows UHCA multivariate maps for the same cervical tissue section both before (fig. 2a)) and after (fig. 2b)) RMieS correction. In both these images UHCA was performed on second derivative spectra to classify the image into 8 clusters. The choice of 8 clusters for UHCA was based on this being the minimum number of clusters required to obtain at least one cluster corresponding to each of the major tissue features. The UHCA map produced from the uncorrected FTIR image data shows good agreement between the clusters and the tissue features, as has been previously reported.1,15 The UHCA map produced from the RMieS corrected FTIR image is very similar to the uncorrected result but superior in that the epithelium shows improved layer demarcation into the exfoliating cell layer (gold), superficial layer of squamous epithelium (dark green), the intermediate layer (pink and some glycogen rich areas in blue) and the parabasal layer (orange). The stromal tissue is described by 3 clusters in the uncorrected UHCA map but only 2 clusters in the corrected UHCA map. The stroma is reasonably homogeneous in chemical content so the stroma being described by a lesser number of clusters correlates well with the known properties of stromal tissue and is an improved result. Inspection of the original uncorrected spectra from the stromal region showed that they exhibited significant distortion from light scattering effects in their baselines. Despite the use of second derivative spectra for minimization of the baseline influence on the analysis, the UHCA still ascribed more clusters to the stroma in the uncorrected FTIR UHCA image than in the RMieS corrected UHCA image. All the cluster average spectra from both the corrected and the uncorrected images exhibited spectral features distinct from the other clusters except for the additional stroma cluster (light green) in the uncorrected FTIR image which appeared different from the adjacent stroma cluster spectra (pale blue) mainly in baseline offset (i.e. tissue “thickness” or optical density).
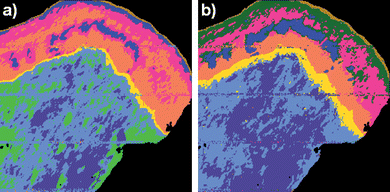 | ||
| Fig. 2 UHCA map of cervical tissue section produced from a) raw uncorrected FTIR micro-spectroscopic image and b) RMieS corrected image data. In the RMieS corrected image (b)) the exfoliating cell layer appears in gold, the superficial layer of squamous epithelium is dark green, the intermediate layer is pink and blue, the parabasal layer is orange, the basal layer is yellow and the stroma is light blue and slate blue. In the uncorrected image (a)) the superficial layer of the epithelium has not been separately identified by the UHCA and an additional (light green) cluster has been assigned to the stroma. | ||
It was also noted that successful demarcation of tissue features could not be achieved from UHCA of uncorrected “raw” (i.e. non-derivative) spectra whereas; for RMieS corrected raw spectra all the major tissue features were eventually found for cluster maps containing 15 or more clusters. In this last test involving non-derivative spectra the uncorrected raw spectra were compared with normalized and baseline corrected spectra. In our experience, pre-processing FTIR spectra with some form of baseline correction and normalization will generally result in a significant improvement in multivariate classification. This work has only compared the UHCA of spectra baseline corrected with combined RMieS-EMSC and differentiation versus differentiation only. Classification of FTIR spectra (expressing biochemical variation) to specific tissue features has been successfully demonstrated many times in the past using alternative baseline and normalization methods (e.g. basic EMSC or “rubberband” baseline correction). This ability to correctly classify FTIR spectra to specific tissue features is not unique to RMieS-EMSC. For studies where it is perhaps only necessary to identify the presence or absence of a specific spectral marker (e.g. diseased state markers) then the relatively computationally intensive RMieS-EMSC may not offer significant advantage over simpler baseline correction techniques.
However, these observations do demonstrate that pre-processing FTIR image spectra with RMieS correction prior to performing UHCA can improve the clustering correspondence with tissue features as well as achieve clustering parsimony. Derivative spectra are often used as inputs for various multivariate analyses4,16,17 based on the argument that derivative pre-processing suppresses baseline influence on the results. Even after baseline correction with RMieS-EMSC, the UHCA performed on second derivative spectra still gave best demarcation of tissue features in fewer clusters. Presumably this is because second derivative pre-processing also enhances the separate resolution of overlapping band features in the spectra.
In FTIR spectra of isolated proteins the positions of the underlying amide I band features may be used to estimate protein secondary structure. However, in tissues, the complex mixture of the proteins present makes interpretation of the protein secondary structure information contained in the spectra difficult. Furthermore, absorptions in the amide I region of the spectrum arise in tissue samples from non-proteins such as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups in nucleic acids and the O–H bending vibration of water. These limitations preclude the use of FTIR spectroscopy to quantitatively assess protein secondary structure content in tissue samples. Despite difficulties in achieving quantitative analysis in tissue samples, FTIR is nevertheless a good tool for qualitative monitoring of relative changes in secondary conformation across the mixture of proteins present. Fig. 3 shows the same FTIR micro-spectroscopic image acquisition but colour coded based on the wavenumber value of the amide I band peak (i.e. a frequency image) rather than absorbance intensity, both before a) and after b) RMieS correction. Comparing the before and after RMieS correction wavenumber images it can be readily seen that the amide I band in areas of stroma have changed from being predominantly characterized by a relatively red shifted amide I band to a relatively blue shifted amide I band.
O groups in nucleic acids and the O–H bending vibration of water. These limitations preclude the use of FTIR spectroscopy to quantitatively assess protein secondary structure content in tissue samples. Despite difficulties in achieving quantitative analysis in tissue samples, FTIR is nevertheless a good tool for qualitative monitoring of relative changes in secondary conformation across the mixture of proteins present. Fig. 3 shows the same FTIR micro-spectroscopic image acquisition but colour coded based on the wavenumber value of the amide I band peak (i.e. a frequency image) rather than absorbance intensity, both before a) and after b) RMieS correction. Comparing the before and after RMieS correction wavenumber images it can be readily seen that the amide I band in areas of stroma have changed from being predominantly characterized by a relatively red shifted amide I band to a relatively blue shifted amide I band.
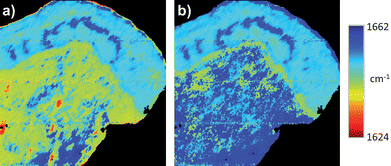 | ||
| Fig. 3 Univariate wavenumber images of cervical tissue section based on the wavenumber value of the amide I band peak of the same FTIR micro-spectroscopic image acquisition both a) before and b) after RMieS correction. Colour coding is blue to red corresponding to spectra with relatively blue shifted amide I band peaks to relatively red shifted amide I band peaks, respectively. | ||
Fig. 4 shows the cluster average spectrum from the stroma both before (light blue) and after RMieS correction (dark blue) derived from the UHCA in Fig. 2. Before RMieS correction, the FTIR spectra in the region of stromal tissue exhibited the amide I peak at 1644 cm−1, which is not unreasonable if a mixed content of α-helical, unordered structure and β-sheet protein conformations was present.18 However, considering that a significant collagen contribution is expected in connective tissues this value appears too low. After RMieS correction the stroma spectra exhibited the amide I band with a peak at 1659 cm−1 consistent with that observed for type I collagen.19 Unlike the other tissues present, the stroma spectra also exhibited strong bands at 1337, 1282, 1236 and 1204 cm−1, all of which are characteristic of extra-cellular matrix collagen.20 Typical collagen fibre diameters in human skin are reported in the range 2–15 μm,21,22 a size range very likely to produce significant Mie scattering. The red shift of the amide I band in the uncorrected stroma spectra can be attributed to significant light scattering distortion in their baselines and arising from the collagen fibres. The RMieS-EMSC algorithm has successfully corrected for this Mie scatter. Also shown in Fig. 4 are RMieS corrected cluster average spectra representative of the basal layer (yellow) and epithelial tissue (pink) which exhibit the amide I band with peak wavenumber values of 1643 cm−1 and 1648 cm−1 respectively (both are amide I intermediate values consistent with a mixed population of α-helical, unordered turns and β-sheet protein conformations18).
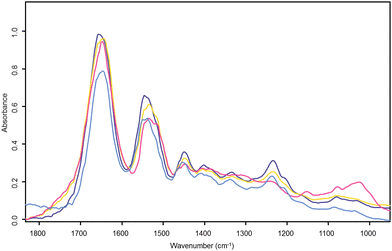 | ||
| Fig. 4 Cervical tissue section UHC analysis cluster average spectra. The light blue spectrum is the UHC analysis cluster average spectrum from raw uncorrected stroma found in the light blue cluster of Fig. 2a). The other three spectra correspond to their like coloured clusters in the RMieS corrected UHC analysis map of Fig. 2b). The dark blue spectrum is from the corrected stroma cluster, yellow is the corrected basal layer cluster and pink the corrected superficial layer in the epithelium. | ||
3.5 RMieS correction of FTIR images of coronal rat brain tissue section
Amide I absorbance images from the same FTIR micro-spectroscopy data set of the tissue features in the rat brain, before and after RMieS correction are shown in Fig. 5. In Fig. 5b) before RMieS correction (raw spectra without normalization) the caudate-putamen and the cerebral cortex appear in light blue and yellow indicating a relatively low amide I absorbance when compared with the corpus callosum seen in red colours of relatively higher amide I absorbance. Following RMieS correction (fig. 5c)), the corpus callosum appears in light blue and yellow while the caudate-putamen and the cerebral cortex are seen in orange and red. The RMieS correction has apparently reversed the order of relative absorbances in these univariate images based on the amide I band. Inspection of the raw spectra in the various brain regions reveals there to be greater baseline distortion from scattering effects in the cerebral cortex than in the corpus callosum. The RMieS algorithm is attributing different amounts of the spectral baseline absorbance to scattering. It is difficult to know if the RMieS corrected image is describing variation of relative protein content amongst the different regions more correctly than the uncorrected images, especially as EMSC normalization of the spectra has occurred as part of the correction process. Banay-Schwartz et al.,23 have reported total protein assay of regions in the rat brain. They found the same protein content in the cerebral cortex and the caudate regions to within the uncertainty of their measurements. This apparent change in relative protein content emphasizes the need to exercise caution in assigning increased or decreased absorbance to higher or lower molecular content between tissues spectra exhibiting differing degrees of scattering effects, especially in the amide I region of the spectra which are most susceptible to light scatter induced baseline distortion.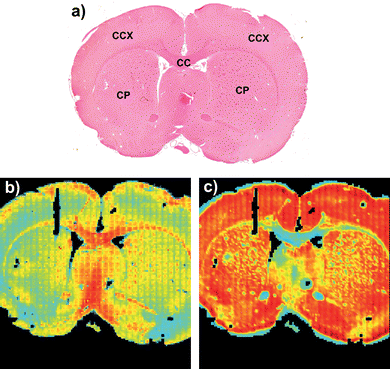 | ||
| Fig. 5 A coronal rat brain tissue section FTIR image of relative amide I (1680–1620 cm−1) band absorbance in a) H&E stained tissue section. CC, corpus callosum; CCX, cerebral cortex; CP, caudate-putamen. b) before RMieS correction and in c) after RMieS correction. Figures a) and b) are adapted from images published in Bambery et al.,.2 Reused with permission from Elsevier. | ||
The seven cluster UHC analysis in Fig. 6 illustrates that the RMieS corrected image data can be cluster analysed equally effectively as the uncorrected image and in fact the cluster demarcation improved for some tissue features. In particular, the dark green cluster is more tightly associated with the caudate-putamen after the RMieS correction, before correction this cluster included some tissue areas belonging to the cortex (yellow cluster).
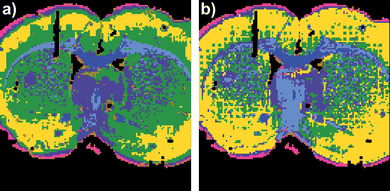 | ||
| Fig. 6 Coronal rat brain tissue section UHC analysis maps generated for seven clusters a) before and b) after RMieS correction. | ||
4. Conclusion
This work has evaluated the effectiveness of the RMieS-EMSC algorithm for correcting FTIR images of tissue sections. The principal findings are given in Table 1.| Test | Result |
|---|---|
| Preliminary single iteration of RMieS algorithm followed by univariate imaging or UHCA multivariate mapping compared with a full 8 iterations run of the RMieS algorithm followed by the same imaging/mapping. | Single iteration RMieS corrected FTIR images had meaningful spatial contrast but the contained spectra were over corrected towards the reference spectrum. Eight iteration RMieS correction showed spectra with much closer correspondence to the feature variations seen in the original uncorrected individual spectra. |
| Use of the data set average spectrumversus use of the default Matrigel spectrum as the reference spectrum for the RMieS algorithm. | The data set average spectrum as reference produced RMieS corrected spectra with much closer correspondence to the original spectra compared to the spectra obtained from using the Matrigel spectrum as the reference. |
| Comparison of UHCA of “raw” (non-derivative) cervical transformation zone tissue section spectra with and without RMieS correction. | No correspondence with known tissue features was found for any UHCA map produced from the uncorrected spectra. RMieS corrected spectra did produce UHCA maps with good correspondence to tissue features but only for maps computed with an unreasonably high number of clusters. |
| Comparison of UHCA of second derivative cervical transformation zone tissue section spectra with and without RMieS correction. | The RMieS corrected UHCA maps gave improved tissue feature demarcation and did so in fewer clusters. |
| Comparison of univariate images of cervical transformation zone tissue section based on the wavenumber value of the amide I band peak with and without RMieS correction. | Amide I wavenumber images from RMieS corrected data showed correspondence with the tissue features whereas there was little correspondence for similar images produced from uncorrected data sets. |
| Comparison of UHCA of second derivative rat brain tissue section spectra with and without RMieS correction. | The RMieS corrected UHCA maps gave better tissue feature demarcation. |
RMieS correction of spectra in FTIR images of tissue sections can produce new FTIR images that are better descriptors of the true chemical content of the tissue samples than are FTIR images comprised of uncorrected spectra. UHCA maps from RMieS corrected FTIR images gave improved tissue feature demarcation compared to UHCA maps from uncorrected image data. Improvements in UHCA clustering efficiency and tissue feature identification accuracy were also observed for both tissue samples in this study but were more apparent for the cervical transformation zone tissue than in the rat brain tissue. We attribute this difference to strongly Mie scattering collagenous fibrils within the connective tissue feature of the cervical tissue section.
Use of the RMieS corrected average spectrum from the FTIR image as the reference spectrum for RMieS baseline correction of all the other spectra contained in the image gave the best preservation of relative peak heights and band profiles. To achieve the best possible correction of spectra towards pure absorbance spectra it was necessary to employ the RMieS correction process iteratively. Uncorrected univariate images of the amide I band peak wavenumber position gave little correspondence with cervical tissue histopathological features whereas after RMieS correction these features became visible. This observation supports the claim11 that RMieS correction will return the true amide I band peak wavenumber value, important for protein secondary structure determination. Future studies will entail a more rigorous test of the effectiveness of RMieS correction applied to FTIR spectra of tissue sections by using a supervised classification model such as linear discriminant analysis clustering24,25 or an artificial neural network.26
Acknowledgements
We are grateful to Professor Michael Quinn of the Department of Obstetrics and Gynecology, Royal Women's Hospital, Parkville, Victoria, Australia for the cervical transformation zone biopsy samples and Dr Elisabeth Schültke of Stereotactic Neurosurgery at University Medical Centre Freiberg, Germany for the rat brain sections. This work is financially supported by an Australian Research Discovery Project Grant DP0878464.References
- K. R. Bambery, B. R. Wood, M. A. Quinn and D. McNaughton, Aust. J. Chem., 2004, 57, 1139–1143 CrossRef CAS.
- K. R. Bambery, E. Schültke, B. R. Wood, S. T. Rigley MacDonald, K. Ataelmannan, R. W. Griebel, B. H. J. Juurlink and D. McNaughton, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 900–907 CrossRef CAS.
- H. Fabian, P. Lasch, M. Boese and W. Haensch, J. Mol. Struct., 2003, 661–662, 411–417 CrossRef CAS.
- M. Diem, L. Chiriboga and H. Yee, Biopolymers, 2000, 57, 282–290 CrossRef CAS.
- K. M. Gough, D. Zelinski, R. Wiens, M. Rak and I. M. C. Dixon, Anal. Biochem., 2003, 316, 232–242 CrossRef CAS.
- E. Gazi, J. Dwyer, P. Gardner, A. Ghanbari-Siahkali, A. Wade, J. Miyan, N. Lockyer, J. Vickerman, N. Clarke, J. Shanks, L. Scott, C. Hart and M. Brown, J. Pathol., 2003, 201, 99–108 CrossRef CAS.
- G. Mie, Ann. Phys., 1908, 330, 377–445 CrossRef.
- A. Kohler, J. Sule-Suso, G. D. Sockalingum, M. Tobin, F. Bahrami, Y. Yang, J. Pijanka, P. Dumas, M. Cotte and H. Martens, Appl. Spectrosc., 2008, 62, 259–266 CrossRef CAS.
- H. Martens, J. P. Nielsen and S. B. Engelsen, Anal. Chem., 2003, 75, 394–404 CrossRef CAS.
- P. Bassan, H. J. Byrne, J. Lee, F. Bonnier, C. Clarke, P. Dumas, E. Gazi, M. D. Brown, N. W. Clarke and P. Gardner, Analyst, 2009, 134, 1171–1175 RSC.
- P. Bassan, A. Kohler, H. Martens, J. Lee, H. J. Byrne, P. Dumas, E. Gazi, M. Brown, N. Clarke and P. Gardner, Analyst, 2010, 135, 268–277 RSC.
- P. Bassan, H. J. Byrne, F. Bonnier, J. Lee, P. Dumas and P. Gardner, Analyst, 2009, 134, 1586–1593 RSC.
- P. Bassan, A. Kohler, H. Martens, J. Lee, E. Jackson, N. Lockyer, P. Dumas, M. Brown, N. Clarke and P. Gardner, J. Biophotonics, 2010, 3(8–9), 609–620 CrossRef CAS.
- F. L. Martin, J. G. Kelly, V. Llabjani, P. L. Martin-Hirsch, I. I. Patel, J. Trevisan, N. J. Fullwood and M. J. Walsh, Nat. Protoc., 2010, 5(11), 1748–1760 CrossRef CAS.
- B. R. Wood, L. Chiriboga, H. Yee, M. A. Quinn, D. McNaughton and M. Diem, Gynecol. Oncol., 2004, 93, 59–68 CrossRef CAS.
- J. Kneipp, P. Lasch, E. Baldauf, M. Beekes and D. Naumann, Biochim. Biophys. Acta, Mol. Basis Dis., 2000, 1501, 189–199 CrossRef CAS.
- P. Lasch, W. Haensch, E. N. Lewis, L. H. Kidder and D. Naumann, Appl. Spectrosc., 2002, 56, 1–9 CrossRef CAS.
- M. Jackson and H. H. Mantsch, Crit. Rev. Biochem. Mol. Biol., 1995, 30(2), 95–120 CrossRef CAS.
- K. Belbachir, R. Noreen, G. Gouspillou and C. Petibois, Anal. Bioanal. Chem., 2009, 395, 829–837 CrossRef CAS.
- M. Jackson, L.-P. Choo, P. H. Watson, W. C. Halliday and H. H. Mantsch, Biochim. Biophys. Acta, Mol. Basis Dis., 1995, 1270, 1–6 CrossRef.
- M. A. Bartlett, S. Ramesh, J. M. DeCou, M. Wolff and H. Jiang, in Biomedical Optical Spectroscopy and Diagnostics Miami Beach, Florida, 2000, ed. T. Li, Optical Society of America, 2000, Vol. 38 of OSA Trends in Optics and Photonics, 320–322 Search PubMed.
- I. S. Saidi, S. L. Jacques and F. K. Tittel, Appl. Opt., 1995, 34, 7410–7418 CrossRef CAS.
- M. Banay-Schwartz, A. Kenessey, T. DeGuzman, A. Lajtha and M. Palkovits, Age, 1992, 15, 51–54 CrossRef CAS.
- C. Krafft, K. Thümmler, S. B. Sobottka, G. Schackert and R. Salzer, Biopolymers, 2006, 82, 301–305 CrossRef CAS.
- J. Trevisan, P. P. Angelov, I. I. Patel, G. M. Najand, K. T. Cheung, V. Llabjani, H. M. Pollock, S. W. Bruce, K. Pant, P. L. Carmichael, A. D. Scott and F. L. Martin, Analyst, 2010, 135, 3266–3272 RSC.
- P. Lasch, M. Diem, W. Hänsch and D. Naumann, J. Chemom., 2006, 20, 209–220 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
