Pattern generation with synthetic sensing systems in lipid bilayer membranes†
Toshihide
Takeuchi‡§
,
Javier
Montenegro‡
,
Andreas
Hennig¶
and
Stefan
Matile
*
Department of Organic Chemistry, University of Geneva, Geneva, Switzerland. E-mail: stefan.matile@unige.ch; Web: www.unige.ch/sciences/chiorg/matile/ Fax: +41 22 379 3215; Tel: +41 22 379 6523
First published on 29th September 2010
Abstract
The objective of this study was to introduce differential sensing techniques to synthetic systems that act, like olfactory receptors, as transporters in lipid bilayer membranes. Routine with most alternative chemosensing ensembles, pattern generation has, quite ironically, remained inaccessible in lipid bilayers because the number of available crossresponsive sensor components has been insufficient. To address this challenge, we here report on the use of cationic hydrazides that can react in situ with hydrophobic analytes to produce cationic amphiphiles which in turn can act as countercation activators for polyanionic transporters in fluorogenic vesicles. To expand the dimension of signals generated by this system, a small collection of small peptides containing a positive charge (guanidinium, ammonium) and one to three reactive hydrazides are prepared. Odorants are used as examples for hydrophobic analytes, perfumes to probe compatibility with complex matrices, and counterion-activated calf-thymus DNA as representative polyion–counterion transport system. Principal component and hierarchical cluster analysis of the obtained multidimensional patterns are shown to differentiate at least 21 analytes in a single score plot, discriminating also closely related structures such as enantiomers, cis–trans isomers, single-atom homologs, as well as all tested perfumes. Inverse detection provides access to analytes as small as acetone. The general nature of the introduced methodology promises to find diverse applications in current topics in biomembrane research.
Introduction
In mammalian olfactory systems, about 350 olfactory receptors recognize more than 10,000 different odorants.1 Incompatible with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 recognition by individual receptors, these biological sensing systems use less specific interactions with several receptors to generate patterns that are recognized as unique “fingerprints” by the brain. Attractive because little discriminatory power is needed for signal generation, these lessons from nature have been successfully applied to several chemosensor systems.2–17 The bottleneck of this approach is the establishment of multiple molecular recognition frameworks, which are necessary to generate patterns. So far, this has not been possible with synthetic sensing systems that work, like olfactory receptors, in lipid bilayer membranes.18–31 The objective of this study was to change this situation, using odorants O1–O30 as illustrative collection of challenging analytes with high similarity (Fig. 1).
1 recognition by individual receptors, these biological sensing systems use less specific interactions with several receptors to generate patterns that are recognized as unique “fingerprints” by the brain. Attractive because little discriminatory power is needed for signal generation, these lessons from nature have been successfully applied to several chemosensor systems.2–17 The bottleneck of this approach is the establishment of multiple molecular recognition frameworks, which are necessary to generate patterns. So far, this has not been possible with synthetic sensing systems that work, like olfactory receptors, in lipid bilayer membranes.18–31 The objective of this study was to change this situation, using odorants O1–O30 as illustrative collection of challenging analytes with high similarity (Fig. 1).
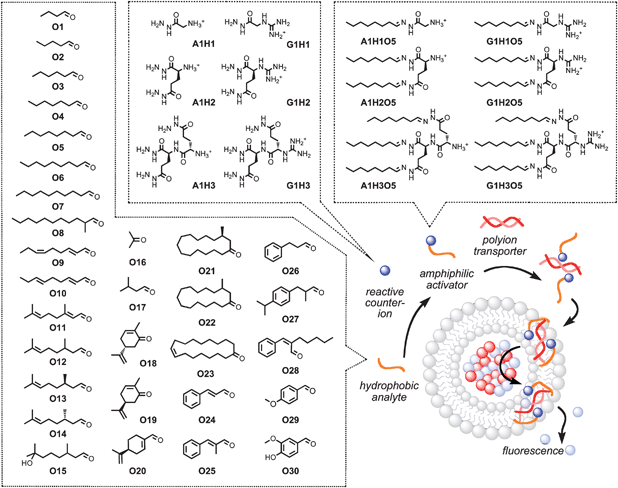 | ||
| Fig. 1 Sensing scheme for fragrance sensing by pattern generation in fluorogenic vesicles. Hydrophobic analytes (e.g., odorants O1–O30) are incubated with hydrophilic reactive counterions (e.g., A1H1–A1H3, G1H1–G1H3) to give amphiphilic counterions (e.g., A1H1O5–A1H3O5, G1H1O5–G1H3O5) that activate polyions (e.g., ctDNA) in lipid bilayer membranes (e.g., fluorogenic vesicles). Citral O11 is a mixture of cis- (neral) and trans-isomers (geranial). | ||
Hydrophobic analytes in general are problematic with established membrane-based synthetic sensing systems because they tend to simply disappear in the hydrophobic membrane instead of interacting with synthetic pores26–28 or activating polyion transporters.29–31 This problem has been solved with the in situ introduction of hydrophilic headgroups that can react with hydrophobic analytes to afford amphiphilic counterions which, in turn, can activate polyion transporters and generate a “turn-on” fluorescent response (Fig. 1).29 The transport processes involved in this sensing scheme have been studied in detail for both polyanion30,31 and polycation transporters.29 Without going into details, polyion–counterion transport systems operate based on the exceptionally strong yet labile binding of counterions by polyions originating from their need to minimize intramolecular charge repulsion.29–31 To cross a lipid bilayer membrane, polyions simply have to exchange their intrinsic, hydrophilic counterions by the amphiphilic counterion activators obtained from the covalent capture of the analyte of choice by the matching reactive counterion. The fluorescent response for active polyion–counterion complexes is generated using the same proximity effect to pick up suitably charged fluorescent probes and move them across the membrane. To introduce differential sensing approaches2–10 to synthetic systems that operate in lipid bilayers,18–31 it occurred to us that a collection of different reactive hydrophilic counterions could already be sufficient to generate analyte-specific fingerprints for hydrophobic analytes. As reported in the following, this turned out to be true.
Results and discussion
Reactive counterions A1H1–A1H3 and G1H1–G1H3 were designed and synthesized to explore differential sensing in lipid bilayer membranes (Fig. 1). They all contain either an ammonium (A1H1–A1H3) or a guanidinium cation (G1H1–G1H3) to interact with polyanionic DNA transporters, and one to three hydrazides to capture odorants by hydrazone formation.27,29,32–34 Details on their synthesis can be found in the Supporting Information (Figs. S1–S3†).35 Incubation of 30 fragrant aldehydes and ketones O1–O30 with the six hydrazides A1H1–A1H3 and G1H1–G1H3 gave rapid access to 180 cationic amphiphiles A1H1O1–G1H3O30 with little synthetic effort. Hydrazone formation was confirmed in all cases by electrospray mass spectrometry (ESI MS, Table S2†). Their ability to activate polyanion transporters was explored under routine conditions, using calf-thymus (ct) DNA as polyanion transporter acting in EYPC-LUVs ⊃ HPTS/DPX (i.e., egg yolk phosphatidylcholine large unilamellar vesicles loaded with the anionic fluorophore 8-hydroxy-1,3,6-pyrenetrisulfonate and the cationic quencher p-xylene-bis-pyridinium bromide).30,31 This assay reports transport activity as fluorescence recovery due to release of HPTS or DPX from the vesicles (Fig. S4A†; previous U-tube analysis has shown that countercation-activated ctDNA transports the cationic DPX but not the anionic HPTS across bulk liquid membranes).31 Before the addition of DNA polyions, cationic amphiphiles A1H1O1–G1H3O30 did not cause a fluorescence response and were thus membrane-inactive under the selected, optimized conditions (Figs. S4A, S6A, S7A, S8A†). The fluorescent response to the addition of DNA varied from amphiphile to amphiphile (Figs. S4A, S6A, S7A, S8A†). Characteristic parameters are EC50, the effective concentration needed to reach 50% of YMAX, the maximum normalized response as determined after 190 s, and n, the Hill coefficient, which can all be obtained from Hill analysis of single dose response curves (Figs. S4B, 2A, etc†).Pattern generation was exemplified first for octanal O5, a citrus odorant used for flavor production in the food industry. The dose response curves for A1H1O5–A1H3O5 and G1H1O5–G1H3O5 revealed an increasing ability to activate DNA transporters with increasing number of tails and decreasing acidity of the cation (Fig. 2A, for other odorants, see Fig. S5†). For pattern generation, dose response curves were recorded for increasing concentrations of odorant O5 after incubation with A1H2, A1H3, G1H2 and G1H3 at constant concentrations (Fig. S6†). The three independent readouts EC50, YMAX and n obtained for the four counterion activators yielded a 12-dimensional pattern for O5 (Fig. 2B, row 1). Application of the same routine to O11, O12, O20, O24 and O26 gave a collection of 12D fingerprints for 6 analytes (Fig. 2B). Among odorants O3–O30, this method of pattern generation worked under identical conditions. However, odorants O3, O18, O19 and O21–O23 are shown as examples where slightly higher hydrazide concentrations produce nicer patterns. Odorants O15–O17, O29 and O30 are examples for analytes that gave hydrazones which are too hydrophilic to activate DNA transporters with reasonable sensitivity under meaningful conditions. These more hydrophilic analytes were excluded from pattern generation by direct detection and analyzed separately with a back-up inverse detection method (Fig. S7, see below). Three independent experiments were performed per analyte to determine reproducibility and experimental error; the result was excellent (Fig. 2D, 3A, 3C; Tables S2, S3†). Data available from monovalent A1H1 and G1H1 counterion activators (Table S1†) were not used for pattern generation (Table S2†) because the weak sensitivity obtained for most analytes (e.g., Fig. 2A, + and △) imposed less favorable experimental conditions (higher concentrations, increasing experimental errors, occasional membrane disruption without DNA).
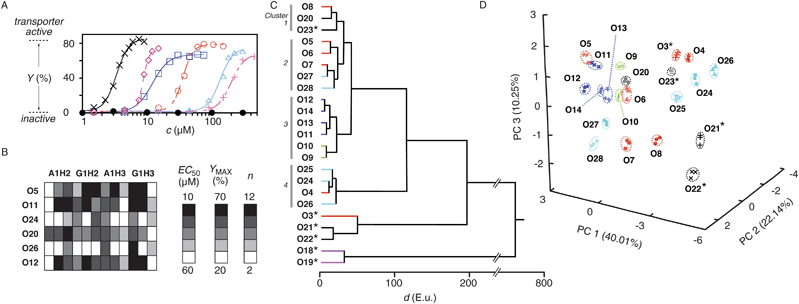 | ||
| Fig. 2 Pattern generation and pattern recognition with cationic hydrazide activators and DNA transporters. (A) Dose response curves for the activation of ctDNA (1.25 μg ml−1) in EYPC-LUVs ⊃ HPTS/DPX with amphiphiles A1H1O5 (+), G1H1O5 (△), A1H2O5 (○), G1H2O5 (◇), A1H3O5 (□), G1H3O5 (×) and O5 (●), (B) 12D pattern generated for 6 odorants with 4 reactive counterions (A1H2, G1H2, A1H3 and G1H3) and three readouts each (from left to right: Effective odorant concentration EC50, the concentration needed to reach YMAX/2, maximal activity YMAX, Hill coefficient n). All data were obtained from Hill analysis of dose response curves for odorants O (variable concentrations) coupled with reactive counterions (constant concentrations, 50 μM (A1H2), 15 μM (G1H2), 30 μM (A1H3) and 5 μM (G1H3) final concentrations) and detected with ctDNA in HPTS/DPX-LUVs (constant concentrations, analog to (A)). (C) HCA dendrogram for 23 odorants, showing the Euclidian distances d between average values from three trials. (D) PCA score plot for 21 odorants (data points are from three independent experiments per analyte, made to determine reproducibility and experimental error, see Table S2†). *Measured with 50 μM (A1H2), 30 μM (G1H2), 50 μM (A1H3) and 30 μM (G1H3, final concentrations), off-scale carvones O18/O19 are omitted in (D), see ref. 35 for details. | ||
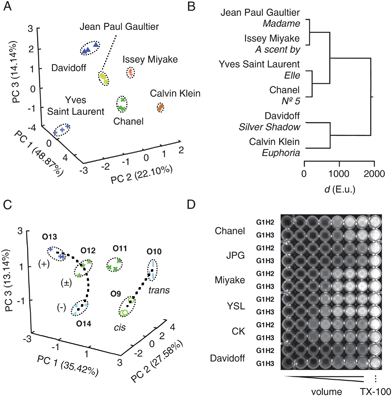 | ||
| Fig. 3 Compatibility with complex matrices, enantiodiscrimination, discrimination of cis–trans isomers and high-throughput formats. (A) PCA score plot and (B) HCA dendrogram for arbitrarily selected perfumes. See Fig. 2 and ref. 35 for details. (C) Focused PCA score plot to corroborate detectability of enantiomers (O12–O14) and cis–trans isomers (O9–O10, dotted lines; from global PCA in Fig. 2D). Data points in (A) and (C) are from three independent experiments per analyte, determining experimental error, see Tables S2, S3†). (D) Fluorescent image of multiwell plate. See ref. 35 for experimental details. | ||
Triplicates of the obtained patterns (Fig. 2B, S11, Table S2†) were subjected to hierarchical clustering (HCA) and principal component analysis (PCA).2 HCA is an unsupervised method of multivariant analysis that converts interpoint Euclidean distances between all samples in the n-dimensional space into 2D dendrograms (Fig. 2C). PCA concentrates the most significant characteristics (variance) of the multidimensional pattern into lower dimensional space by calculating eigenvectors (principal components, PC) in the direction of maximal variances. This reduces the n-dimensional pattern to a single score that is then plotted in the new PC space (Fig. 2D). Both 2D HCA dendrogram and 3D PCA score plot obtained for 23 odorants demonstrated overlap-free discrimination. The HCA dendrogram showed clustering of some of the aromatic odorants (O24–O26, cluster 4) around 20 Euclidian distance units (E.u.). Other clusters found around 20 E.u. contained odorants with mostly unsaturated (O9–O14, cluster 3), saturated (O5–O7, etc, cluster 2) and cyclic alkyl tails (O23 and perillaldehyde O20, main odorant of perilla leaves (or Shiso in Sashimi dishes)). Consistent with the structural similarities, the latter two clusters 1 and 2 merged around 30 E.u., whereas discrimination from unsaturated (cluster 3) and aromatic odorants (cluster 4) occurred around 45 E.u. and 120 E.u., respectively. Several exceptions from these trends concerned either mixed motifs, present in the cyclamen aldehyde cyclosal O27 and jasminaldehyde O28, or odorants with borderline activity such as the shortish heptanal O4 (lost among aromatics) or the longish 2-methylundecanal O8 (within cyclic odorants). Other odorants with weak activity such as hexanal O3, muscone O21/O22 and carvone O18/O19 (but not civetone O23) clustered far apart from the rest. The most distinct carvones O18/O19 were discriminated at nearly 800 E.u. These two outliers were omitted in global PCA score plots because their unique characteristics caused an apparent clustering of all other analytes, making the inherent patterns less presentable and recognizable (Fig. 2D). This adverse effect was naturally not observed in more focused PCA score plots (Fig. S9†).
Overlap-free recognition of at least 21 odorants in HCA and PCA was highly remarkable for a solution-based supramolecular sensor array considering the similarity of the involved structures. The result demonstrated that nearly all challenges in supramolecular recognition (enantiodifferentiation, cis–transisomerization, single-atom homologues, etc.) have been successfully addressed with a single, simple and straightforward to optimize system. For example, discrimination of close derivatives of cinnamaldehyde O24 including 2-methylcinnamaldehyde O25 or hydrocinnamaldehyde O26 was no problem (Fig. 2D, top right; focused PCA, Fig. S9C†). Discrimination of homologous linear alkyl aldehydes, from hexanal O3 over heptanal O4, octanal O5, nonanal O6 and decanal O7 up to 2-methylundecanal O8 occurred with unproblematic single-carbon resolution (global, Fig. 2D; focused, Fig. S9B†).
Detectability of cis–trans stereoisomers was explored with 2E,6Z-nonadienal O9, the cucumber aldehyde with the diffusely “green” odor, and its 2E,6E-isomer O10. Their hydrazones obtained with A1H2, A1H3, G1H2 and G1H3 resembled cis- and trans-isomers of unsaturated phospholipids. Considering how the small structural difference between the latter has a big impact in lipid bilayer structure and function, we presumed that these isomers could be easily discriminated. In fact, HCA dendrogram and PCA score plots revealed overlap-free differentiation between the cis-isomer O9, the trans-isomer O10 as well as the parent nonanal O6 (Fig. 2C, 2D and 3C).
Enantiodiscrimination, a hallmark of the mammalian olfactory system, was explored first with (R)-(+)-citronellal O13, an antifungal insect repellent that accounts for the lemon scent of citronella oil. HCA dendrograms revealed full discrimination between (R)-(+)-citronellal O13, the (S)-(-)-enantiomer O14, their racemic mixture (±)-citronellal O12 and nearly identical citral O11 (Fig. 2C). Note that apparent close proximities in the global score plot (Fig. 2D, center left) are optical illusions due to graphical limitations as confirmed by focused PCA score plots (Fig. 3C).
Attached to A1H2, A1H3, G1H2 and G1H3, the 15-membered ring of (R)-(-)-muscone O21, the legendary primary contributor to the odor of musk,36 looked like two alkyl tails of a phospholipid that are linked together at the end. In the muscone hydrazone G1H3O21, this adds up to an amphiphile with six tails and a single charged head, creating in situ a complexity comparable to lipids such as cardiolipin. However, macrocyclic alkyl tails gave weaker responses than linear alkyl tails. Slightly varied initial hydrazide concentrations were thus used for pattern generation. In the resulting PCA score plot, enantioenriched (R)-(-)-muscone O21 (61% ee) was clearly separated from racemic (±)-muscone O22 (global, Fig. 2; focused, Fig. S9E†). Differentiation from the structurally related civetone O23, a pheromone of the African civet composed of 17-membered cyclic ketone with a cis-alkene in position 8, was no problem (Fig. 2D, S9E†). General validity of enantiodiscrimination with our supramolecular sensing system was corroborated with carvone. R-(-)-carvone O18 smells like caraway, S-(+)-carvone O19 smells like spearmint. Measured at slightly higher activator concentrations like muscone because of weak responsiveness, both enantiomers appeared cleanly separated in HCA dendrograms (Fig. 2C) and PCA score plots (Fig. S9E†).
Compatibility of our new sensing system with samples from daily life was explored with randomly selected perfumes. A 12D pattern was generated as above, with A1H2, A1H3, G1H2 and G1H3 for analyte capture and DNA activation in DPX-vesicles (Fig. S8†). PCA afforded a score plot where Chanel N° 5, Jean Paul Gaultier's Madame, a scent by Issey Miyake, Elle from Yves Saint Laurent, Calvin Klein's Euphoria, and Silver Shadow from Davidoff are separated without overlap (Fig. 3A). In the HCA dendrogram, close similarity was found for Chanel N° 5 and Elle around 100 E.u., whereas Euphoria and Silver Shadow (700 E.u.) were most distinct from the rest (1900 E.u., Fig. 3B).
To sense an analyte as small as acetone O16, direct detection failed because amphiphiles A1H2O16, A1H3O16, G1H2O16 and G1H3O16 were not hydrophobic enough to activate DNA transporters. Therefore, an inverse detection scheme was devised, wherein hydrazides A1H2, A1H3, G1H2 and G1H3 were incubated with constant concentrations of active octanal O5 and increasing concentrations of inactive acetone O16 serving as a competitor. Decreasing concentrations of A1H2O5, A1H3O5, G1H2O5 and G1H3O5 in the presence of increasing concentrations of acetone O16 were then monitored as a decreasing fluorescence emission (Fig. S7†). The same was also true for isovaleraldehyde O17, and the more hydrophilic hydroxycitronellal O15, anisaldehyde O29 and vanillin O30. HCA dendrograms and PCA score plots obtained by inverse detection cleanly separated all tested analytes (Fig. S10†).
Compatibility with multiwell assays was explored as a hint toward potential for practice (Fig. 3D). The obtained dose response pattern could be subjected directly to HCA and PCA or treated by Hill analysis to give EC50, YMAX and n for the generation of the more reliable patterns from continuous response as described above. Preliminary HCA and PCA revealed that perfume recognition is possible also from the simplified patterns generated by less refined “high-throughput” methods under adjusted, clearly different conditions (not shown).
Conclusions
Whereas many wonderful, much more practical differential chemosensors have been reported over the past two decades,2–10 this is the first synthetic differential sensing system that operates, like olfactory receptors, in lipid bilayer membranes. Experimental evidence is delivered for generation and recognition of composite responses or “fingerprints” at highest possible resolution (Fig. S11†). This includes the simultaneous differentiation of at least 21 closely related odorants, covering enantiomers, cis–trans isomers, single-atom homologues, and so on. As an example for compatibility with analyte mixtures in complex matrices, we show that, like a human nose, perfumes can be smelled with neither knowing nor identifying their molecular composition. A more systematic analysis of the impact of analyte mixtures on PCA score plots beyond mixtures of enantiomers (O12–O14, Fig. 3C) is ongoing and will be reported in due course.The key conceptual advance of the introduced approach is that subtle structural differences are magnified covalently in dynamic oligomers as well as non-covalently on polyions and in lipid bilayers. Quite similar amplification of subtle structural differences is known from the distinct impact of enantiomers, cis–trans isomers and single-atom homologues of biological phospholipids and steroids on structure and function of biomembranes. For enantiodiscrimination, this effect is exploited in a highly “chiral” environment, including the use of chiral reactive counterions A1H2, A1H3, G1H2 and G1H3 to produce diastereomeric counterion activators for chiral polyion transporters in chiral lipid bilayer membranes.
HCA dendrograms and PCA score plots turn out to be ideal to demonstrate reproducibility as well as the superb discrimination power of the sensing system. However, specific information on the characteristics of individual polyion-counterion complexes is naturally lost. This information is preserved in the untreated patterns generated with EC50, YMAX and n which in turn fail to inform quantitatively on discrimination power (Fig. S11†). These raw patterns confirm that, inter alia, guanidinium counterions are more active than ammonium counterions. This effect originates from the well-established, preorganized and hydrogen-bond assisted recognition of phosphates by guanidinium cations.31 Intermediate hydrophobicity of the counterion amphiphiles gives the best results. Too hydrophilic counterions fail to partition into the membrane, too hydrophobic counterions precipitate before reaching the bilayer or end up buried in the middle of the membrane. Surprisingly, the best overall activity with all different headgroups was observed for jasmine aldehyde O28 and cyclamen aldehyde O27 with branched aromatic tails (Fig. S11†). This finding was unpredictable and is expected to be of highest importance in applications toward cellular uptake of oligonucleotides (DNA, RNA).
The introduced approach excels with facile accessibility, exceptional modularity and broad applicability. This will allow us to explore variations with regard to counterion activators (numbers of heads and tails, nature and position of charges, including charge inversions to add cell-penetrating peptides as transporters,29,37 covalent capture chemistry (boronates,11,28enzyme-/aptamer-coupled capture,26–30etc32–34), polyion and membrane composition (including polymersomes38), readouts (including color,39 circular dichroism, current, multiwell assays (Fig. 3D), chips, etc) and other applications (fragrant cellular uptake,29,37,40 controlled release,34etc). For example, boronates are attractive to covalently capture analytes with diol or catechol groups such as sugars or polyphenols and have already been used successfully to sense polyphenols in green tea with synthetic multifunctional pores.28
Acknowledgements
We thank J. Praz for contributions to synthesis, D. Jeannerat, A. Pinto and S. Grass for NMR measurements, the Sciences Mass Spectrometry (SMS) platform at the Faculty of Sciences, University of Geneva, for mass spectrometry services, H. Riezman and M. Umebayashi for access to the fluorescence imaging system, L. Wunsche and A. Herrmann (Firmenich, Geneva) for odorant samples, and the University of Geneva and the Swiss NSF for financial support.References
- L. B. Buck, Angew. Chem., Int. Ed., 2005, 44, 6128–6140 CrossRef CAS.
- J. C. Jurs, G. A. Bakken and H. E. McClelland, Chem. Rev., 2000, 100, 2649–2678 CrossRef CAS.
- J. J. Lavigne and E. V. Anslyn, Angew. Chem., Int. Ed., 2001, 40, 3118–3130 CrossRef.
- S. H. Shabbir, L. A. Joyce, G. M. da Cruz, V. M. Lynch, E. Sorey and E. V. Anslyn, J. Am. Chem. Soc., 2009, 131, 13125–13131 CrossRef CAS.
- M. A. Palacios, R. Nishiyabu, M. Marquez and P. Anzenbacher Jr, J. Am. Chem. Soc., 2007, 129, 7538–7544 CAS.
- B. A. Suslick, L. Feng and K. S. Suslick, Anal. Chem., 2010, 82, 2067–2073 CrossRef CAS.
- A. Buryak and K. Severin, Angew. Chem., Int. Ed., 2005, 44, 7935–7938 CrossRef CAS.
- H. Zhou, L. Baldini, J. Hong, A. J. Wilson and A. D. Hamilton, J. Am. Chem. Soc., 2006, 128, 2421–2425 CrossRef CAS.
- D. C. Gonazlez, E. N. Savariar and S. Thayumanavan, J. Am. Chem. Soc., 2009, 131, 7708–7716 CrossRef.
- N. T. Greene and K. D. Shimizu, J. Am. Chem. Soc., 2005, 127, 5695–5700 CrossRef CAS.
- G. Mirri, S. D. Bull, P. N. Horton, T. D. James, L. Male and J. H. Tucker, J. Am. Chem. Soc., 2010, 132, 8903–8905 CrossRef CAS.
- A. Satrijo and T. M. Swager, J. Am. Chem. Soc., 2007, 129, 16020–16028 CrossRef CAS.
- C. Guarise, L. Pasquato, V. De Filippis and P. Scrimin, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 3978–3982 CrossRef CAS.
- A. Wada, S. Tamaru, M. Ikeda and I. Hamachi, J. Am. Chem. Soc., 2009, 131, 5321–5330 CrossRef CAS.
- M. Florea and W. M. Nau, Org. Biomol. Chem., 2010, 8, 1033–1039 RSC.
- J.-L. Reymond, V. S. Fluxa and N. Maillard, Chem. Commun., 2009, 45, 34–46 Search PubMed.
- L. Vial and P. Dumy, New J. Chem., 2009, 33, 939–946 RSC.
- A. P. Davis, D. N. Sheppard and B. D. Smith, Chem. Soc. Rev., 2007, 36, 348–357 RSC.
- C. P. Wilson and S. J. Webb, Chem. Commun., 2008, 44, 4007–4009 Search PubMed.
- A. Satake, M. Yamamura, M. Oda and Y. Kobuke, J. Am. Chem. Soc., 2008, 130, 6314–6315 CrossRef CAS.
- X. Li, B. Shen, X. Q. Yao and D. Yang, J. Am. Chem. Soc., 2009, 131, 13676–13680 CrossRef CAS.
- G. W. Gokel and N. Barkey, New J. Chem., 2009, 33, 947–963 RSC.
- J. T. Davis, P. A. Gale, O. A. Okunola, P. Prados, J. C. Iglesias-Sanchez, T. Torroba and R. Quesada, Nat. Chem., 2009, 1, 138–144 CrossRef CAS.
- R. E. Dawson, A. Hennig, D. P. Weimann, D. Emery, V. Ravikumar, J. Montenegro, T. Takeuchi, S. Gabutti, M. Mayor, J. Mareda, C. A. Schalley and S. Matile, Nat. Chem., 2010, 2, 533–538 CrossRef CAS.
- J. K. W. Chui and T. M. Fyles, Chem. Commun., 2010, 46, 4169–4171 RSC.
- G. Das, P. Talukdar and S. Matile, Science, 2002, 298, 1600–1602 CrossRef CAS.
- S. Litvinchuk, H. Tanaka, T. Miyatake, D. Pasini, T. Tanaka, G. Bollot, J. Mareda and S. Matile, Nat. Mater., 2007, 6, 576–580 CrossRef CAS.
- S. Hagihara, H. Tanaka and S. Matile, J. Am. Chem. Soc., 2008, 130, 5656–5657 CrossRef CAS.
- S. M. Butterfield, T. Miyatake and S. Matile, Angew. Chem., Int. Ed., 2009, 48, 325–328 CrossRef CAS.
- T. Takeuchi and S. Matile, J. Am. Chem. Soc., 2009, 131, 18048–18049 CrossRef CAS.
- T. Takeuchi, V. Bagnacani, F. Sansone and S. Matile, ChemBioChem, 2009, 10, 2793–2799 CrossRef CAS.
- O. Ramström and J.-M. Lehn, Nat. Rev. Drug Discovery, 2002, 1, 26–36 CrossRef CAS.
- P. T. Corbett, J. Leclaire, L. Vial, K. R. West, J. L. Wietor, J. K. M. Sanders and S. Otto, Chem. Rev., 2006, 106, 3652–3711 CrossRef CAS.
- A. Herrmann, Org. Biomol. Chem., 2009, 7, 3195–3204 RSC.
- See Supporting Information.
- L. Ruzicka, Helv. Chim. Acta, 1926, 9, 715–729 CAS.
- J. B. Rothbard, T. C. Jessop, R. S. Lewis, B. A. Murray and P. A. Wender, J. Am. Chem. Soc., 2004, 124, 9506–9507 CrossRef.
- M. Kumar, M. Grzelakowski, J. Zilles, M. Clark and W. Meier, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20719–20724 CrossRef CAS.
- S. M. Butterfield, A. Hennig and S. Matile, Org. Biomol. Chem., 2009, 7, 1784–1792 RSC.
- S. Bhattacharya and A. Bajaj, Chem. Commun., 2009, 45, 4632–4656 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details. See DOI: 10.1039/c0sc00386g |
| ‡ These two authors contributed equally to this work. |
| § Current address: Department of Degenerative Neurological Diseases, National Institute of Neuroscience, National Center of Neurology and Psychiatry, Tokyo, Japan. |
| ¶ Current address: BAM Federal Institute for Materials Research and Testing, Bioanalytical Chemistry and Fluorescence Spectroscopy, Berlin, Germany. |
| This journal is © The Royal Society of Chemistry 2011 |
