Comparative bioinformatics analysis of the mammalian and bacterial glycomes†
Alexander
Adibekian
a,
Pierre
Stallforth
a,
Marie-Lyn
Hecht
a,
Daniel B.
Werz
c,
Pascal
Gagneux
d and
Peter H.
Seeberger
*ab
aMax Planck Institute of Colloids and Interfaces, Department of Biomolecular Systems, Research Campus Golm, D-14424, Potsdam, Germany. E-mail: peter.seeberger@mpikg.mpg.de; Fax: +49 331 567 9302; Tel: +49 331 567 9301
bFreie Universität Berlin, Institute for Chemistry and Biochemistry, Arnimallee 22, D-14195, Berlin
cInstitut für Organische und Biomolekulare Chemie, Georg-August-Universität Göttingen, Tammannstr. 2, D-37077, Göttingen, Germany
dGlycobiology Research Training Center, University of California, San Diego, School of Medicine, 9500 Gilman Drive MC 0687, La Jolla, CA 92093-0687, U.S.A
First published on 5th November 2010
Abstract
A comparative analysis of bacterial and mammalian glycomes based on the statistical analysis of two major carbohydrate databases, Bacterial CarbohydrateStructure Data Base (BCSDB) and GLYCOSCIENCES.de (GS), is presented. An in-depth comparison of these two glycomes reveals both striking differences and unexpected similarities. Within the prokaryotic kingdom, we focus on the glycomes of seven classes of pathogenic bacteria with respect to (i) their most abundant monosaccharide units; (ii) disaccharide pairs; (iii) carbohydrate modifications; (iv) occurrence and use of sialic acids; and (v) class-specific monosaccharides. The aim of this work is to gain insights into unique carbohydrate patterns in bacteria. Data interpretation reveals significant trends in the composition of specific carbohydrate classes as result of evolution-driven structural adaptations of bacterial pathogens and symbionts to their mammalian hosts. The differences are discussed in light of their value for biomedical applications, such as the targeting of unique glycosyl transferases, vaccine development, and devising novel diagnostic tools.
Introduction
Carbohydrates are one of the four major classes of biomolecules, in addition to nucleic acids, proteins and lipids.1 These highly complex macromolecules fulfill a variety of tasks ranging from structural and metabolic functions, to regulating development, cell signaling, cell adhesion, and host–pathogen interactions.2,3 The wide array of diverse functions governed by carbohydrates is reflected in the wealth of structurally distinct carbohydrate molecules. Individual glycan structural and chemical diversity is determined by the specific combination of selected elements from a set of monosaccharide building blocks, different glycosidic linkages used to link these monosaccharides, and by the stereochemical configuration of the glycosidic bonds. Branching and site-specific modifications to particular glycans further increase the complexity of the glycome.The mammalian glycome, namely all glycans found in mammals whether free or bound, is built from a limited number of monosaccharides. In total, just ten monosaccharides are used to cover the entire occupied mammalian glycospace.4 These ten building blocks can give rise to a tremendous number of possible glycans compared to linear macromolecules like nucleic acids or proteins.5 However, only a small subspace of the theoretical glycospace – the theoretically possible combinations of monosaccharides, is occupied in mammals.
This situation changes drastically for prokaryotes.6 The bacterial outer cell surface behaves like the skin of multicellular organisms and mediates all the interactions between bacteria and their changing and sometimes harsh environment. In contrast, most animal cell surfaces have to protect cells only within the relatively constant environment of the body, and mediate cell–cell communication.7 Consequently, higher and more diverse selective pressures are acting on bacterial cell surface molecules, necessitating adaptations in the chemical and structural composition of the bacterial cell surface. The relatively short generation time of bacteria allows cell surface molecules to adapt more quickly to external pressures.8
Different classes of carbohydrates decorate bacterial cell walls that consist of complex and often composite glycoconjugates. Both Gram-positive and Gram-negative bacteria contain a peptidoglycan layer consisting of β(1–4)-linked N-acetylglucosamine and N-acetylmuramic acid residues. Whereas this layer is generally thicker in Gram-positive bacteria, Gram-negative bacteria possess an additional outer layer typically containing lipopolysaccharides (LPS). Finally, some bacteria produce extracellular capsules, consisting mostly of polysaccharides, and often containing highly variable structures that are strongly antigenic to mammals (K-antigens). Besides preventing desiccation9 and their involvement in the adhesion processes during biofilm formation,10extracellular capsule carbohydrates can be important virulence factors in pathogenic bacteria and play key roles in recognition by, or evasion of, host immune systems. In general, many pathogenic bacteria find themselves in a “dual glycan speedway”, having to evolve away from phage recognition from below, and from host recognition from above.11,12 Despite potential biological and clinical implications, little is known at a statistical level about the differences and similarities between bacterial and mammalian repertoires of glycans and monosaccharides.
Here, we present the first in-depth comparative analysis of the glycomes of seven classes of pathogenic bacteria and compare these with the mammalian glycome to reveal both common patterns and striking differences. Bacterial glycome data was extracted from the Bacterial CarbohydrateStructure Data Base (BCSDB)13–15 currently the largest database for bacterial glycans, while information regarding the mammalian glycome was obtained from the database, GLYCOSCIENCES.de (GS).13–15
Our statistical analyses aim at answering five main questions: (i) which monosaccharide units are the most abundant in seven distinct classes of bacteria, as judged by their frequency of occurrence in the BCSDB; and how does this compare to the most abundant mammalian monosaccharides in GLYCOSCIENCES.de; (ii) which disaccharide pairs are found in bacteria and how do they compare to the mammalian glycome; (iii) which carbohydrate modifications are particular to bacteria and how do these differ from mammalian carbohydrate modifications; (iv) given the importance of sialic acids as terminal monosaccharides on most mammalian glycans, what sialic acids or related nine carbon backbone monosaccharides (nonoses) do bacteria utilize; and (v) are there monosaccharides specific for a single bacterial class?
By directly comparing bacterial and mammalian glycomes, we believe we will enhance the understanding of the host–microbial pathogen coevolution at the molecular level. In addition, this statistical analysis can be used when designing tailor-made diagnostic tools for rapid identification of diverse bacterial classes, and when searching for candidates for carbohydrate-based immunoadjuvants16 and vaccines.17,18 Furthermore, illuminating glycosidic linkages and carbohydrate modifications unique to microbes will give hints for the selection of glycosyltransferases that have potential as targets for novel antibacterial drugs.
Results
Monosaccharide analysis
First, we compared the monosaccharide composition of seven classes of bacteria, to that found in mammals (Fig. 1). The graph displays the 25 most common monosaccharides found in the BCSDB (from now on referred to as consensus monosaccharides). For each class, the number in parentheses indicates the proportion of the analyzed glycome that can be constructed from these 25 monosaccharides.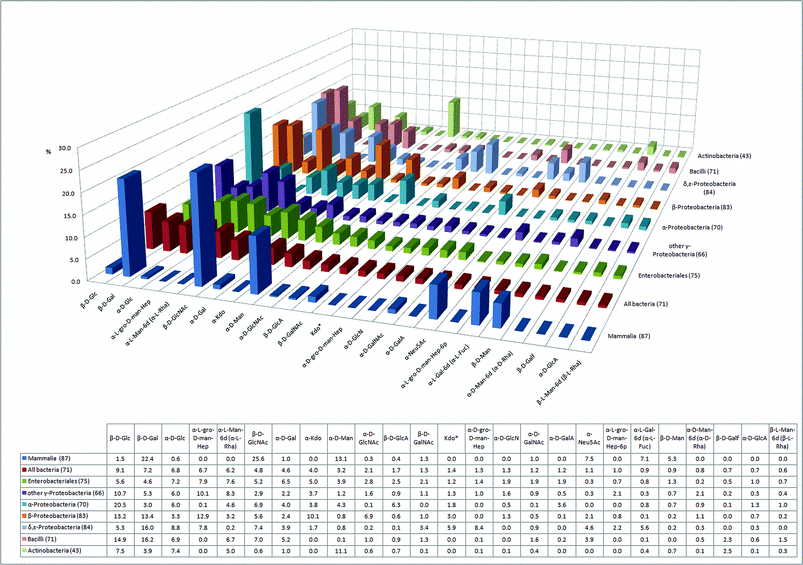 | ||
| Fig. 1 The 25 most abundant monosaccharides in bacteria and their relative abundance in seven human pathogenic bacterial classes and in mammals (percentage of the glycome covered by the 25 monosaccharides is indicated in parentheses). | ||
The 25 consensus monosaccharides constitute a significant portion of the glycome in all classes analyzed. Of the mammalian glycome covered in the database, 87% can be constructed with the 25 consensus monosaccharides. An average coverage of 71% indicates that a large portion of the bacterial glycome is built by the consensus monosaccharides. Actinobacteria constitute an exception where only 43% of the glycome is accounted for by these 25 monosaccharides (Fig. 1). Actinobacteria are, along with Bacilli, the two Gram-positive classes in our study, thus, they are phylogenetically separate from the other five bacterial classes that are all Gram-negative. The glycome coverage of Bacilli by the consensus monosaccharides, however, is significantly higher, with 74%. The underrepresentation of Actinobacteria glycans by the consensus monosaccharides is indicative of the presence of a large proportion of Actinobacteria-specific unusual monosaccharides. Actinobacteria include major players in the carbon cycle of soil decomposition, nitrogen-fixing symbionts of plants,19 and interestingly also prominent human pathogens from the genus Mycobacterium. The observation that the 25 consensus monosaccharides are more widespread in the glycome of Bacilli than Actinobacteria can be explained by the fact that the class of Bacilli, in our statistical analysis, is mostly represented by mammalian pathogens like Streptococcus, Staphylococcus and Bacillus. Mammalian pathogens and mammalian gut symbionts have undergone rapid evolutionary changes in their glycosylation patterns to closely mimic the glycans of their hosts, thus evading the innate and adaptive immune systems of the host.6
Striking differences in the glycome composition of the seven bacterial classes directly reflect fundamental differences in the envelope architecture of Gram-positive and Gram-negative bacteria. Kdo‡ and mannoheptoses are integral constituents of outer membrane LPS. The proportion of Kdo in the glycomes of Gram-negative LPS containing bacteria (the sum of α-Kdo and Kdo*) lies between 5% and 13% (Fig. 1). Conversely, the Gram-positive Bacilli and Actinobacteria do not use Kdo. On the other hand, Actinobacteria are rich in α-mannose (11%), the most abundant monosaccharide in this class (Fig. 1). The occurrence of α-mannose in Actinobacteria is comparable to that in the mammalian glycome (13%). This can be attributed to the presence of lipoarabinomannans (LAM), a particularly important class of glycans well-known in Mycobacterium tuberculosis, the causative agent of tuberculosis.20 Such group-specific membrane-associated glycans are often described as “microbial motifs” and include pathogen associated molecular patterns (PAMPs) that are recognized by the mammalian host immune system by means of pattern recognition receptors (PRRs). Toll-like receptors (TLRs) and C-type lectins of dendritic cells (e.g.mannose binding DC-SIGN) are two of the most important classes of PRRs.21
Neu5Ac is a widespread sialic acid that can be considered a characteristic terminal monosaccharide of the vertebrate lineage. In plants and most protostome animals (insects, molluscs and helminths), Neu5Ac is not found. Besides other effects, the presence of sialic acid on cell-surface glycoconjugates induces the binding of Factor H, a complement pathway regulator, which protects the cell from attack by a complement system, a mechanism to recognize and combat pathogens.22 In addition, sialic acids on mammalian cells engage Siglecs, lectins from an immunoglobulin superfamily, which mostly exert inhibitory effects on a variety of immune cells.23 Interestingly, at least five bacterial classes have acquired Neu5Ac in order to prevent activation of the complement system, albeit in lower abundance than in mammals (Fig. 1). Although the presence of sialic acids in bacteria was traditionally interpreted as a result of the horizontal transfer of sialic acid biosynthesis genes from metazoa to bacteria, recent results suggest that microbial sialic acids are more likely a result of adaptations in an ancestral biosynthetic pathway for nonulosonic acids.24
Another very common mammalian terminal monosaccharide is L-fucose (6-deoxy-L-galactose). In contrast to Neu5Ac, the content of L-fucose is very low in the analyzed bacterial classes, with the exception of δ,ε-proteobacteria, where L-Fuc constitutes 5.6% of the glycome. Indeed, previous work has demonstrated that certain strains of Helicobacter pylori, an ε-proteobacterium, express a relatively large proportion of Lewis A glycan. Lewis A glycan is a fucosylated O-glycan otherwise commonly found in mammalian glycomes as yet another example of bacterial mimicry of host glycans.25
The glycomes of the two classes of Gram-positive bacteria, Bacilli (2.3%) and Actinobacteria (2.5%), contain a larger proportion of the monosaccharide galactofuranose (Galf) than the Gram-negative classes (<0.7%) (Fig. 1). For all Gram-negative bacteria, galactofuranose is primarily found in Enterobacteriales (0.7%). Galactofuranose is a particularly interesting glycan since it is absent from the human glycome. The galactofuranose metabolic pathways have been suggested as novel targets for antimicrobial therapy.26,27 Our analysis indicates that such a therapeutic tool could be effective against Gram-positive bacteria and Enterobacteriales, but most probably not against other classes of Gram-negative bacteria.
We have determined the 20 most abundant bacterial monosaccharides with corresponding glycosidic linkages (Fig. S1†). Compared to the mammalian glycome, where a relatively small set of building blocks is needed for the construction of most oligosaccharides, more than 700 different bacterial monosaccharides are listed in the BCSDB. This large number can be explained by a combination of the following factors: (i) the rapid rate of evolution in bacteria due to short generation times, less efficient DNA proofreading, and ubiquitous horizontal gene transfer, (ii) fewer constraints on integrated development than encountered by multicellular metazoans including mammals, (iii) the long evolutionary history and diverse environments of bacteria, (iv) bacteria are both host and pathogens at the same time. However, 20 building blocks are sufficient for the construction of 30% of the known bacterial glycome (Diagram S1†). A comparison of the bacterial glycome with the entire eukaryotic glycome would be more revealing, but this information does not yet exist in the databases. It should be mentioned that the glycan structures currently found in the database are primarily derived from bacteria that can be cultured and, as a consequence, they have been studied intensively. BCSDB contains entries for only about half of the bacterial phyla and nine bacterial classes have less than 10 records. However, the majority of bacteria cannot be cultured in vitro, and thus access to their undoubtedly rich glycan diversity is extremely limited and solely accessible via metagenomic approaches such as characterizing their glycan metabolic genes and synthesizing their products.28,29 Similar limitations also apply to mammalian carbohydrate entries in GLYCOSCIENCES.de, which are heavily biased towards the well studied mammalian N-glycans.
Disaccharide pair analysis
The distribution of disaccharide pairs in the bacterial and mammalian glycomes is depicted in Fig. 2. These data provide particularly useful information for the analysis of motifs in polysaccharides such as glycosaminoglycans or cell surface arabinomannans. Considering the substrate-specificity of glycosyltransferases, a structurally distinct set of disaccharides in a certain bacterial class indicates the presence of unique glycosyltransferases involved in the synthesis of the respective disaccharide motif. A tailor-made chemical inhibitor of these enzymes could play an important role for future antibacterial therapy.30 From this point of view, the disaccharide α-L-Man-6d/α-L-Man-6d (α-L-Rha/α-L-Rha) is of particular interest as it is, at 4.5% occurrence, the most abundant disaccharide sequence found in bacteria (Fig. 2). This particular disaccharide motif is present in six of seven analyzed bacterial classes, and it is not found in mammals. This disaccharide pair is mostly present as polyrhamnose, a polysaccharide often found as side chain in bacterial peptidoglycans. Interestingly, the presence of polyrhamnose in cell walls of different Gram-positive bacteria was reported to be crucial for the induction of chronic arthritis in rats.31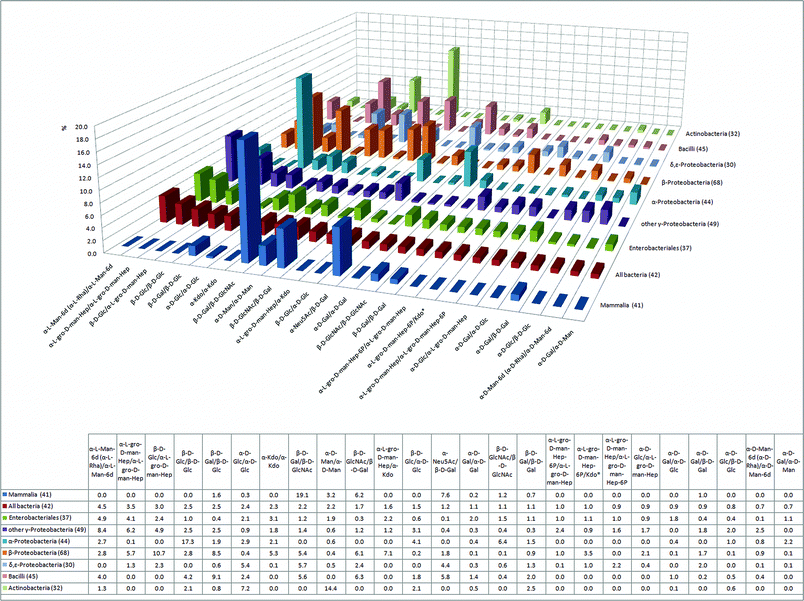 | ||
| Fig. 2 The 25 most abundant disaccharide pairs in bacteria and their relative abundance in six human pathogenic bacterial classes and in mammals (percentage of the glycome covered by the 25 disaccharides is indicated in parentheses). | ||
The sugarL-glycero-D-manno-heptose is an important constituent of the LPS inner core in Gram-negative bacteria where it is directly attached to 3-deoxy-D-manno-2-octulosonate (Kdo). Both L-glycero-D-manno-heptose and Kdo are absent in mammals and Gram-positive bacteria (Fig. 1, Fig. 2). The importance of these two monosaccharides for the vitality of Gram-negative bacteria is illustrated by the fact that eight out of the 25 most abundant bacterial disaccharide pairs, including the second and third most abundant pairs, contain Kdo or L-gro-D-man-Hep (Fig. 2).
Bacterial polysaccharides are traditionally discussed in the context of toxicity and pathogenicity, yet some of them display other features such as immunomodulatory and anticancer activity. Mammals lack β-glucans, polysaccharides composed of β-linked D-glucose monosaccharides (β-D-Glc/β-D-Glc, Fig. 4), and β-glucanases, enzymes with hydrolytic activities against β-glucans. β-Glucose disaccharide pairs are ubiquitous in bacteria with the exception of δ,ε-Proteobacteria, where they are not present (Fig. 2). β-Glucose disaccharides constitute 17% of all disaccharides in α-Proteobacteria (Fig. 2), which can be mostly accounted for by curdlan, a linear β(1-3)-glucan commonly found in the capsules of non-pathogenic Agrobacterium and Rhizobium species.32 Bacterial curdlan exhibits promising immune modulating activity as it activates macrophages and neutrophils in a similar way to the β(1-3)-glucans of fungal and algal origin.33
Carbohydrate modifications
After their assembly by glycosyl transferases, the glycans in eukaryotes and prokaryotes can be modified in a site-specific manner, i.e. they can be acylated, sulfated or epimerized. We have divided the mammalian and bacterial monosaccharides into 15 distinct monosaccharide/modification classes based on: (i) hydroxyl or amine modifications; (ii) sugar ring size; or (iii) number of carbon atoms in the monosaccharide backbone, and compared them (Fig. 3). Our analysis shows that sulfation, a common mammalian glycan modification, is rarely found in bacteria and is mostly restricted to the class of α-Proteobacteria. In mammals, sulfation is found on various O- and N-linked glycans, as well as on glycosaminoglycans (GAGs), a major class of glycopolymers consisting of uronic acids and 2-aminosugars.34 In GAGs, sulfation occurs at hydroxyls of both uronic acids and aminosugars, or at the amine of the aminosugar. Sulfation is catalyzed by different mammalian GAG-modifying enzymes in the endoplasmic reticulum – Golgi secretory system. GAGs are also present in bacterial capsules, however, sulfation has not yet been described in these structures.35Sulfation on the mammalian GAG heparan sulfate creates an important recognition element for P- and L-selectins, mammalian C-type lectins involved in leukocyte trafficking to the site of inflammation (P-selectin), or leukocyte homing to lymph nodes (L-selectin).2,36 Thus, sulfation of GAGs, along with the epimerization of glucuronic to iduronic acid, is considered a combinatorial trick of vertebrates that not only allows GAG-binding proteins to distinguish between multiple endogenous GAGs, but also excludes binding to bacterial GAG-mimics.37,38 On the other hand, sulfated glycosaminoglycans on mammalian cell surfaces are used by some viruses as recognition and attachment sites.39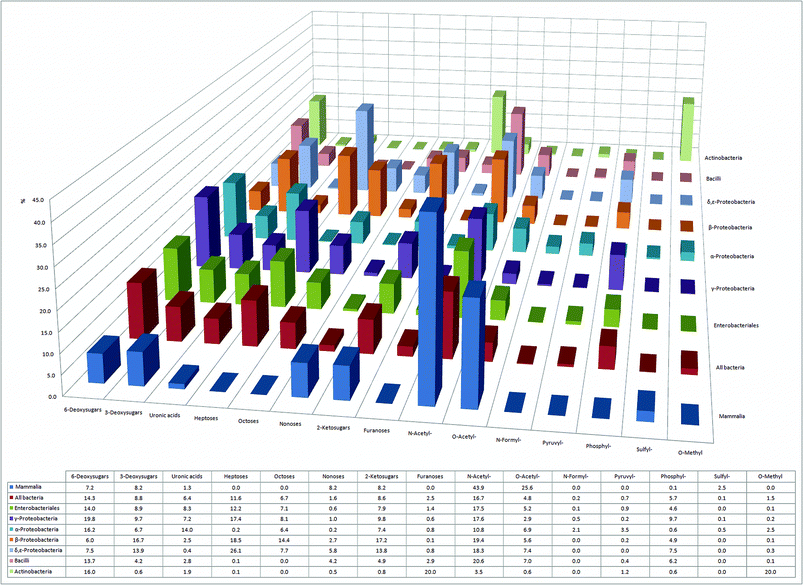 | ||
| Fig. 3 Overview of 15 different classes and modifications of monosaccharides with their relative abundance in six bacterial classes and in mammals. | ||
Monosaccharide modifications like N- and O-acylation, 6-deoxygenation, and 6-oxidation to uronic acids appear to be ubiquitous to mammals and all classes of bacteria (Fig. 3). Intriguingly, our analysis also reveals that certain bacterial classes have unique structural signatures in their glycomes. Decoration of sugars with formyl- and pyruvyl-residues appears to be typical for α-Proteobacteria, whereas 20% of all sugars in Actinobacteria are methylated (Fig. 3). Moreover, although phosphorylation of mammalian glycans is very unusual, it appears to be a common modification in all seven bacterial classes, being most abundant in γ-Proteobacteria, where 10% of all monosaccharides are phosphorylated (Fig. 3).
Sialic acids in the prokaryotic kingdom
In the constant evolutionary race to evade attack by mammalian defense mechanisms, the ability to mimic the mammalian sialic acid glycan cap represents a selective advantage and major challenge for pathogens.12,24 Many bacterial pathogens were able to face this challenge by adopting the mammalian sialic acid Neu5Ac (D-gro-D-gal-Non5NAc) as the predominant nonose in their glycomes (Fig. 4 and Fig. S2†), whereas many other bacteria attempt to mimic Neu5Ac with two structurally related nonulosonic acids: legionaminic (Leg)40,41 (e.g. Legionella species, γ-Proteobacteria) and pseudaminic acid (Pse) (e.g. Pseudomonas species, γ-Proteobacteria; Campylobacter species,42–45 ε-Proteobacteria). Furthermore, our analysis suggests that all seven classes of bacteria have at least one nonose present in their glycomes (Fig. 4).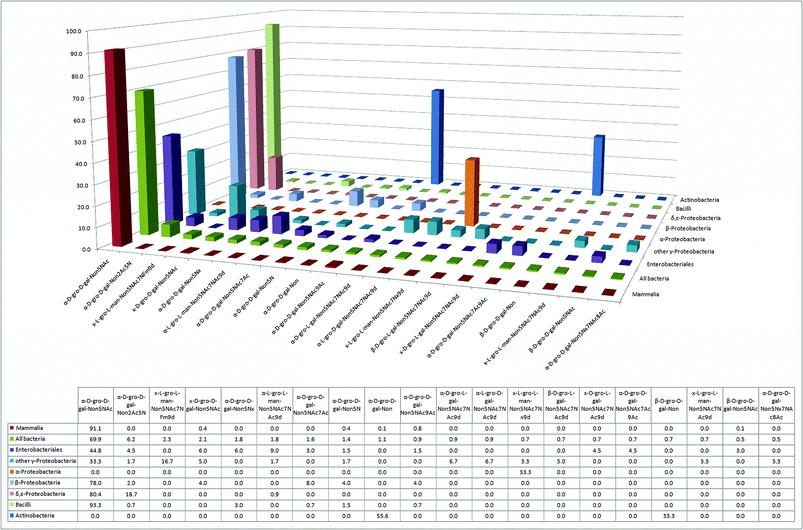 | ||
| Fig. 4 The 20 most abundant sialic acid derivatives and their relative abundance in six bacterial classes and in mammals. | ||
Strikingly, despite the large number of nonoses synthesized by bacteria, there are no entries for N-glycolylneuraminic acid (Neu5Gc)46 in the bacterial glycan database. Thus, Neu5Gc appears to be a monosaccharide nonose exclusively used by metazoans, although there is literature evidence for at least one bacterium having the capacity to incorporate mammalian Neu5Gc into its glycolipids.47 The two bacterial classes with the smallest relative amount of nonoses (Fig. 3), α-Proteobacteria and Actinobacteria, include several prominent obligate or facultative intracellular parasites like Rickettsia and Brucella (both α-Proteobacteria) or Mycobacterium (Actinobacteria), which, by virtue of their intracellular localization, would not benefit from the presence of sialic acid analogs. Indeed, both of these classes lack Neu5Ac, in contrast to all other bacterial classes that were analyzed (Fig. 4).
Bacteria-specific monosaccharides
Finally, we identified the ten most abundant monosaccharides that are present in bacteria but that are not found in mammals. The structures of these sugars are presented in Fig. 5. These monosaccharides either have an unusual chain length and configuration, as in the case of octose Kdo or L-glycero-D-manno-heptose, an inverted configuration as in the case of D-rhamnose, or unusual modifications such as phosphorylations.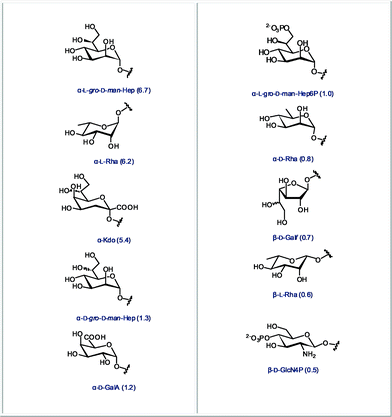 | ||
| Fig. 5 The 10 most abundant monosaccharides found in bacteria, but not in mammals (relative abundance in all bacteria indicated in parentheses). | ||
Rapid detection of bacterial contamination is undoubtedly of great importance. In order to choose an appropriate antimicrobial treatment, it is useful to assign the detected contamination to a specific bacterial class. Thus, we also identified monosaccharides that are abundant in a certain class of bacteria, but completely absent in other classes. These unique diagnostic sugars are shown in Fig. 6. Some of these sugars are highly abundant in their corresponding glycome, such as the methylated derivatives of glucose and mannose in Actinobacteria, or the 6-deoxy-4-formamido-mannose in α-Proteobacteria. Many of these structures also represent a fascinating future challenge for synthetic chemists. Efficient stereoselective synthesis of monosaccharides that bear amines, phosphates or deoxygenated carbons within the same scaffold, is not straightforward and will require novel synthetic strategies and methodologies.
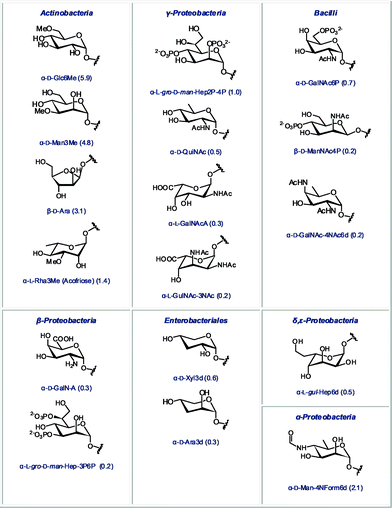 | ||
| Fig. 6 Monosaccharides found in only one bacterial class (relative abundance in the glycome of this class indicated in parentheses). | ||
Conclusions
We have analyzed the most comprehensive carbohydrate databases available to date with respect to questions of relevance both to chemists and biologists. Although such a database analysis suffers from the fact that not all existing carbohydrate structures are listed, we believe that with more than 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures available, important trends can be extracted. Why should a careful analysis of a carbohydrate database not have the potential to lead us to new insights into the glycomics area? An ultimate goal of bacterial glycomics is to determine the exact role of specific bacterial glycans during the recognition, infection, and/or manipulation of a mammalian host. Such understanding could greatly assist the design of carbohydrate-based vaccines and novel chemotherapies targeting bacterial glycan metabolism.
000 structures available, important trends can be extracted. Why should a careful analysis of a carbohydrate database not have the potential to lead us to new insights into the glycomics area? An ultimate goal of bacterial glycomics is to determine the exact role of specific bacterial glycans during the recognition, infection, and/or manipulation of a mammalian host. Such understanding could greatly assist the design of carbohydrate-based vaccines and novel chemotherapies targeting bacterial glycan metabolism.
The monosaccharide analysis shown in Fig. 1 has revealed significant differences in the use of galactofuranose between mammals and bacteria. Galactofuranose-containing microbial glycans, such as arabinogalactan in M. tuberculosis, have been shown to be highly antigenic and render galactofuranose an excellent vaccine candidate. M. tuberculosis, despite several decades of successful chemotherapeutic treatment, has re-emerged through the evolution of multidrug resistance. Consequently, tuberculosis has once again become one of the leading causes of death, with approximately 3 million fatalities annually worldwide.48 The disaccharide pair analysis gives hints that the glycosyl transferases involved in the incorporation of α-L-Rha/α-L-Rha, Kdo or L-gro-D-man-Hep are promising targets for new antibiotics. Finally, the ten most abundant monosaccharides that are present in bacteria but not found in mammals (Fig. 5) are promising molecular markers – if unambiguously detected with immuno- or lectin assays – for the presence of bacteria in a mammalian system.
In conclusion, our report represents an important step towards a quantitative analysis of the bacterial glycome. Not only does this direct comparison between the mammalian and bacterial glycomes illuminate significant differences between these two kingdoms, it also unveils the unique nature of glycomes from different bacterial classes. Striking differences in the repertoire and usage of monosaccharides by several distinct prokaryote classes is evident from this comparison. The results presented in this report provide the first panoramic overview of lineage-specific glycome evolution of bacteria and mammals, and contributes to a better understanding of glycan structural diversity in bacteria, particularly from the point of view of molecular evolution.
Methods
All sequences from the Bacterial CarbohydrateStructure Data Base (BCSDB)13–15 and the database GLYCOSCIENCES.de13–15 (GS) were translated into GlycoCT, a uniform XML-based format, and added in a nonredundand fashion to GlycomeDB, an open source meta-database of carbohydrate structures. Data extraction and analysis were performed as previously described.49 The detailed database composition is represented in Fig. S3.†BCSDB contains a total of 8504 bacterial glycan entries, 8479 of those correspond to bacteria with an assigned taxonomy. GS contains 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 records for pro- and eukaryotes, with 13
120 records for pro- and eukaryotes, with 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 entries related to organisms with assigned taxonomical information. The statistical analyses use the combined data from BCSDB and GS. Eukaryotic glycans are further divided in accordance with the standard eukaryotic class system. Mammalian glycan structures comprise 78% of all the eukaryotic glycans, and constitute by far the largest group of eukaryotes analyzed. Primates and rodents represent the two largest mammalian subgroups. Our analysis of the data available for the bacterial glycome focuses on the six best studied bacterial classes: γ-Proteobacteria, Bacilli, Actinobacteria, β-Proteobacteria, α-Proteobacteria and δ,ε-Proteobacteria. Each class contains prominent human pathogens, which are listed in parentheses. Since the order Enterobacteriales is particularly well-studied, and since they exhibit significant differences to other γ-Proteobacteria, they have been analyzed separately. Class-specific mono- and disaccharide compositions represent percent proportions of a mono- or disaccharide in all entries for this bacterial class available in BCSDB or GS.
704 entries related to organisms with assigned taxonomical information. The statistical analyses use the combined data from BCSDB and GS. Eukaryotic glycans are further divided in accordance with the standard eukaryotic class system. Mammalian glycan structures comprise 78% of all the eukaryotic glycans, and constitute by far the largest group of eukaryotes analyzed. Primates and rodents represent the two largest mammalian subgroups. Our analysis of the data available for the bacterial glycome focuses on the six best studied bacterial classes: γ-Proteobacteria, Bacilli, Actinobacteria, β-Proteobacteria, α-Proteobacteria and δ,ε-Proteobacteria. Each class contains prominent human pathogens, which are listed in parentheses. Since the order Enterobacteriales is particularly well-studied, and since they exhibit significant differences to other γ-Proteobacteria, they have been analyzed separately. Class-specific mono- and disaccharide compositions represent percent proportions of a mono- or disaccharide in all entries for this bacterial class available in BCSDB or GS.
Acknowledgements
This work was supported by the Max Planck Society, ETH Zürich, a PhD fellowship from the Studienstiftung des Deutschen Volkes (to P.S.), and the graduate school of the Competence Center for Systems Physiology and Metabolic Diseases, Zurich (to M-L.H.). D.B.W. thanks the Fonds der Chemischen Industrie for a Liebig Fellowship and the Deutsche Forschungsgemeinschaft (DFG) for an Emmy Noether Fellowship.Notes and references
- J. D. Marth, Nat. Cell Biol., 2008, 10, 1015–1016 CrossRef CAS
.
- A. Varki, Glycobiology, 1993, 3, 97–130 CrossRef CAS
.
- A. Helenius and M. Aebi, Science, 2001, 291, 2364–2369 CrossRef CAS
.
- T. Feizi, Glycoconjugate J., 2000, 17, 553–565 CrossRef CAS
.
- D. B. Werz, R. Ranzinger, S. Herget, A. Adibekian, C. W. von der Lieth and P. H. Seeberger, ACS Chem. Biol., 2007, 2, 685–691 CrossRef CAS
.
- J. R. Bishop and P. Gagneux, Glycobiology, 2006, 17, 23R–34R CrossRef
.
- A. S. Cross, Curr. Top. Microbiol. Immunol., 1990, 150, 87–95 Search PubMed
.
- S. A. Newman, J. Biosci., 2005, 30, 75–85 Search PubMed
.
- E. B. Roberson and M. K. Firestone, Appl. Environ. Microbiol., 1992, 58, 1284–1291 CAS
.
- J. W. Costerton, K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta and T. J. Marrie, Annu. Rev. Microbiol., 1987, 41, 435–464 CrossRef CAS
.
- I. W. Suthereland, K. A. Hughes, L. C. Skillman and K. Tait, FEMS Microbiol. Lett., 2004, 232, 1–6 CrossRef
.
- P. Gagneux and A. Varki, Glycobiology, 1999, 9, 747–755 CrossRef CAS
.
- C.-W. von der Lieth, J. Carbohydr. Chem., 2004, 23, 277–297 CrossRef CAS
.
- T. Lütteke, A. Bohne-Lang, A. Loss, T. Goetz, M. Frank and C.-W. von der Lieth, Glycobiology, 2006, 16, 71R–81R CrossRef
.
- S. Herget, P. V. Toukach, R. Ranzinger, W. E. Hull, Y. A. Knirel and C. W. von der Lieth, BMC Struct. Biol., 2008, 8, 35 CrossRef
.
- S. Boonyarattanakalin, X. Liu, M. Michieletti, B. Lepenies and P. H. Seeberger, J. Am. Chem. Soc., 2008, 130, 16791–16799 CrossRef CAS
.
- P. Stallforth, B. Lepenies, A. Adibekian and P. H. Seeberger, J. Med. Chem., 2009, 52, 5561–5577 CrossRef CAS
.
- P. H. Seeberger and D. B. Werz, Nature, 2007, 446, 1046–1051 CrossRef CAS
.
- M. Goodfellow and S. T. Williams, Annu. Rev. Microbiol., 1983, 37, 189–216 CrossRef CAS
.
- J. S. Schorey and L. Sweet, Glycobiology, 2008, 18, 832–841 CrossRef CAS
.
- N. W. Palm and R. Medzhitov, Immunol. Rev., 2009, 227, 221–233 CrossRef CAS
.
- S. Rodríguez de Córdoba, J. Esparza-Gordillo, E. Goicoechea de Jorge, M. Lopez-Trascasa and P. Sánchez-Corral, Mol. Immunol., 2004, 41, 355–367 CrossRef CAS
.
- P. R. Crocker, J. C. Paulson and A. Varki, Nat. Rev. Immunol., 2007, 7, 255–266 CrossRef CAS
.
- A. L. Lewis, N. Desa, E. E. Hansen, Y. A. Knirel, J. I. Gordon, P. Gagneux, V. Nizet and A. Varki, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13552–13557 CrossRef CAS
.
- C. Nilsson, A. Skoglund, A. P. Moran, H. Annuk, L. Engstrand and S. Normark, PLoS One, 2008, 3, e3811 CrossRef
.
- P. S. Schmalhorst, S. Krappmann, W. Vervecken, M. Rohde, M. Muller, G. H. Braus, R. Contreras, A. Braun, H. Bakker and F. H. Routier, Eukaryotic Cell, 2008, 7, 1268–1277 CrossRef CAS
.
- L. L. Pedersen and S. J. Turco, Cell. Mol. Life Sci., 2003, 60, 259–266 CrossRef CAS
.
- M. R. Rondon, P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman and R. M. Goodman, Appl. Environ. Microbiol., 2000, 66, 2541–2547 CrossRef CAS
.
- C. S. Riesenfeld, P. D. Schloss and J. Handelsman, Annu. Rev. Genet., 2004, 38, 525–552 CrossRef CAS
.
- C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS
.
- T. J. Lehman, J. B. Allen, P. H. Plotz and R. L. Wilder, Rheumatol. Int., 1985, 5, 163–167 CrossRef CAS
.
- M. McIntosh, B. A. Stone and V. A. Stanisich, Appl. Microbiol. Biotechnol., 2005, 68, 163–173 CrossRef CAS
.
- T. H. Hida, K. Ishibashi, N. N. Miura, Y. Adachi, Y. Shirasu and N. Ohno, Inflammation Res., 2009, 58, 9–14 CrossRef CAS
.
- H. Yu and X. Chen, Org. Biomol. Chem., 2007, 5, 865–872 RSC
.
- P. L. DeAngelis, Anat. Rec., 2002, 268, 317–326 CrossRef CAS
.
- A. Varki, J. Clin. Invest., 1997, 99, 158–162 CrossRef CAS
.
- T. F. Meyer, Folia Microbiol., 1998, 43, 311–319 Search PubMed
.
- Y. Chen, M. Gotte, J. Liu and P. W. Park, Mol. Cells, 2008, 26, 415–426
.
- E. Crublet, J. P. Andrieu, R. R. Vivès and H. Lortat-Jacob, J. Biol. Chem., 2008, 283, 15193–15200 CrossRef CAS
.
- Y. A. Knirel, E. T. Rietschel, R. Marre and U. Zähringer, Eur. J. Biochem., 1994, 221, 239–245 CrossRef CAS
.
- Y. A. Knirel, A. S. Shashkov, Y. E. Tsvetkov, P. E. Jansson and U. Zähringer, Adv. Carbohydr. Chem. Biochem., 2003, 58, 371–417 CAS
.
- P. Guerry and C. M. Szymanski, Trends Microbiol., 2008, 16, 428–435 CrossRef CAS
.
- P. Guerry, Trends Microbiol., 2007, 15, 456–461 CrossRef CAS
.
- P. Guerry, C. P. Ewing, I. C. Schoenhofen and S. M. Logan, J. Bacteriol., 2007, 189, 6731–6733 CrossRef CAS
.
- M. I. Kanipes, X. Tan, A. Akelaitis, J. Li, D. Rockabrand, P. Guerry and M. A. Monteiro, J. Bacteriol., 2008, 190, 1568–1574 CrossRef CAS
.
- O. T. Keppler, R. Horstkorte, M. Pawlita, C. Schmidt and W. Reutter, Glycobiology, 2001, 11, 11R–18R CrossRef CAS
.
- K. L. Fox, A. D. Cox, M. Gilbert, W. W. Wakarchuk, J. Li, K. Makepeace, J. C. Richards, E. R. Moxon and D. W. Hood, J. Biol. Chem., 2006, 281, 40024–40032 CrossRef CAS
.
- S. Borrell and S. Gagneux, Int. J. Tuberc. Lung
Dis., 2009, 13, 1456–1466 Search PubMed
.
- R. Ranzinger, S. Herget, T. Wetter and C. W. von der Lieth, BMC Bioinformatics, 2008, 9, 384 CrossRef
.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S3, Diagram S1 and list of carbohydrate-related abbreviations. See DOI: 10.1039/c0sc00322k |
| ‡ For a list of carbohydrate-related abbreviations see Supplementary Information List 1. |
| This journal is © The Royal Society of Chemistry 2011 |
